
A VIRTUAL LABORATORY FOR
WEB AND GRID ENABLED SCIENTIFIC EXPERIMENTS
Francesco Amigoni, Mariagrazia Fugini
Dipartimento di Elettronica e Informazione, Politecnico di Milano, Milano, Italy
Diego Liberati
Istituto di Elettronica e Ingegneria dell’Informazione e delle Telecomunicazioni, Consiglio Nazionale delle Ricerche
Dipartimento di Elettronica e Informazione, Politecnico di Milano, Milano, Italy
Keywords: Virtual Organizations, Cooperative Information Systems, e-Science, Bioinformatics, e-Collaboration, Web
Services, Multiagent Systems.
Abstract: Interactions and organization models tend to be more and more oriented to flexibility, heterogeneity and
collaboration in the style of Virtual Organizations. This paper is framed in the context of e-science and
presents an approach to the definition and execution of distributed scientific experiments as a pool of
services executed on collaborating sites at different heterogeneous organizations. The focus is on flexibility,
reuse, orchestration and interoperability of services within a cooperation process. It is discussed how the
workflow of the experiment can be specified by actors with low information technology, but high domain,
knowledge and how an agent-based framework can enhance the flexibility of experiment execution.
Examples are given in the bioinformatics context. A prototype environment is described.
1 INTRODUCTION
The concept of Virtual Organization, nowadays
boosted also thanks to the Grid computing (Foster
and Kesselman, 2004), is a general model, free from
specific technical solutions: a set of individuals
and/or institutions having direct access to computers,
software, data, tools, knowledge, services and other
resources in a dynamic heterogeneous way, share the
aim of achieving a common goal through
collaboration. The basic idea is the virtualization of
resources, consisting in creating and associating to
resources a generic interface to allow services to be
used through remote control. In this paper, we face
such issue linked to orchestration of services in the
context of e-Science (Hey and Trefethen, 2004), (De
Roure et al., 2004), where the concept is known as
Collaborative Environment, to denote a
Collaborative Laboratory (Bosin et al., 2005). The
aim is to enable a set of scientific (but possibly also
commercial) partners to design the workflow of
distributed experiments, and to support the execution
of the experiment on distributed cooperative nodes,
each providing and using a set of services.
Virtualization of resources is achieved using Web
Service technologies (Alonso et al., 2004
).
Orchestration is achieved by taking each virtualized
resource as a component of a distributed cooperative
process, modelled in terms of workflows (van der
Alst and van Hee, 2002). The general process of
problem solving strategy in a Virtual Organization
can be seen as a whole of single collaborative
procedures or services (Travica, 2005), each
composed of input data, a computing core, and
output data. The suitable orchestration (Peltz, 2003)
of services is the aggregate workflow related to a
business activity, which is globally a service whose
resources (services) are dynamically found through a
discovery mechanism, and executed, through
dynamic binding, on the network (Figure 1). In the
resulting scientific experiment, modelled as a Web
Service, the resources are: functions, seen in terms
of services according to the Service Oriented
Architecture paradigm (Comm. of the ACM, 2003);
data; computational power; ICT components; or
humans. Global-scale experimental networking
initiatives, developed in the last years, aim to
227
Amigoni F., Fugini M. and Liberati D. (2007).
A VIRTUAL LABORATORY FOR WEB AND GRID ENABLED SCIENTIFIC EXPERIMENTS.
In Proceedings of the Ninth International Conference on Enterprise Information Systems - SAIC, pages 227-230
DOI: 10.5220/0002369902270230
Copyright
c
SciTePress
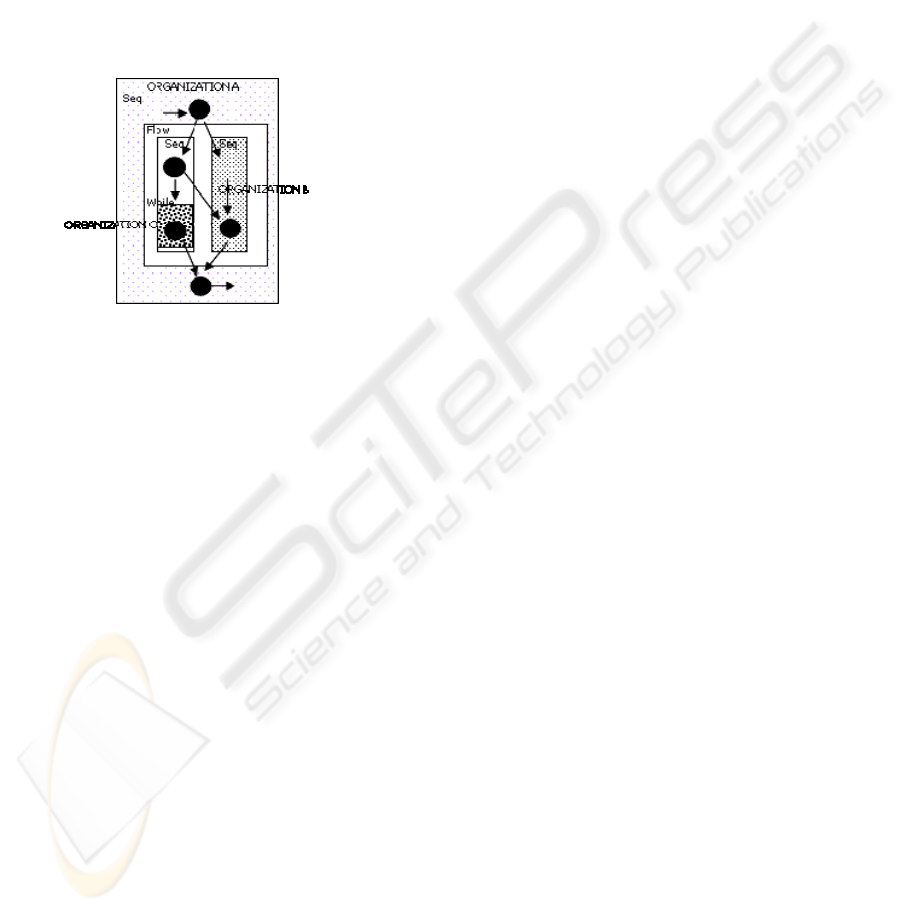
provide advanced infrastructures for e-scientists
through the collaborative development of
networking tools, advanced grid services, and data-
intensive applications (Newman et al., 2003). Web
Services and the Grid are then merged in the often
called Semantic Grid, in order to design Virtual
Laboratories, that is, Virtual Organizations where
cooperation to execute a scientific experiment is
supported. The paradigmatic distributed experiment
used in the paper refers to a process of DNA-
microarrays clustering, based on a technique
illustrated in (Liberati et al., 2005).
Figure 1: Plan of problem solving as a whole of connected
collaborative procedures.
2 SETTING THE VIRTUAL
LABORATORY
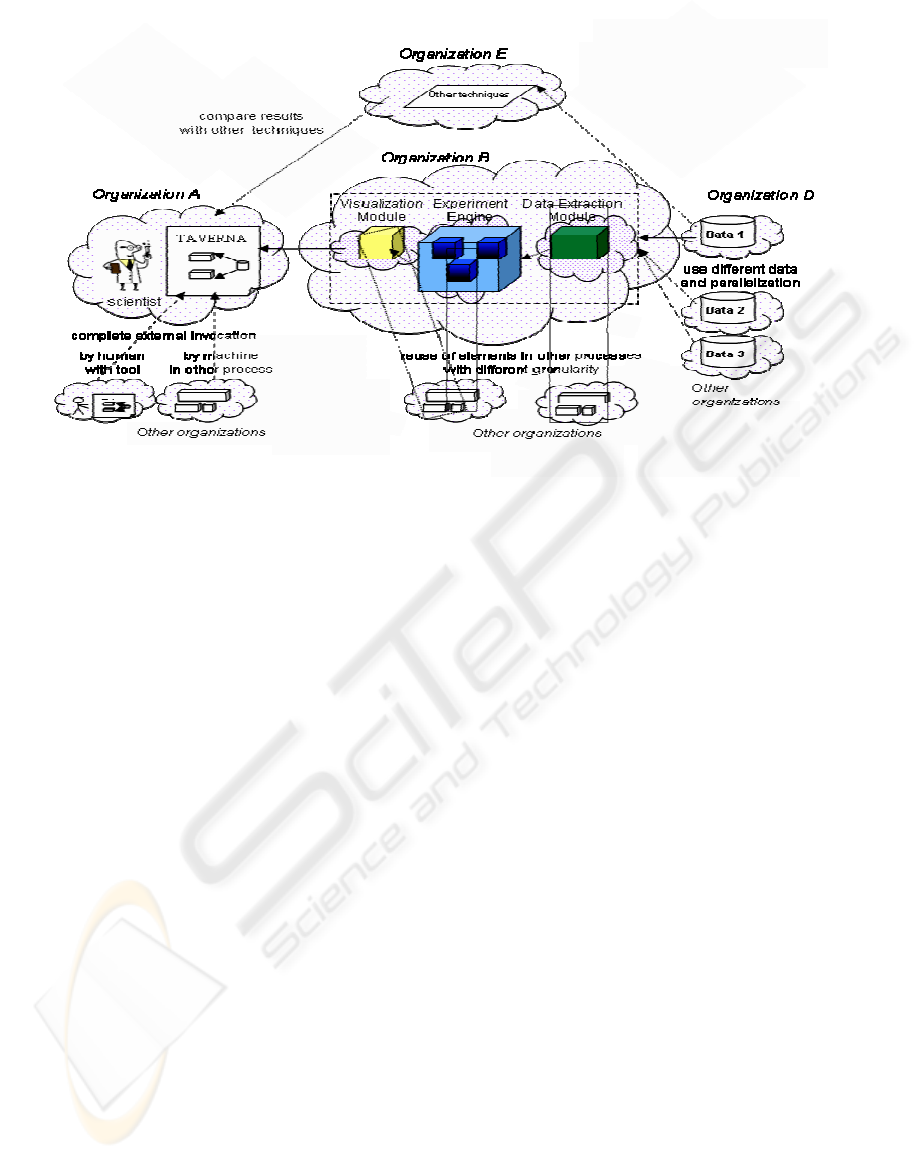
The functional architecture of our proposed Virtual
Laboratory is represented in Figure 2.
“Organization” identifies each member of a Virtual
Laboratory who cooperates to a specific experiment
run. The “clouds” denote distinct physical sites. We
mainly distinguish, besides sites where experiments
are invoked and visualised, the following sites:
Site where the experiment is defined. The
Workflow Designer associated to Organization A
creates the workflow model that characterizes the
distributed experiment by selecting the needed
resources and specifying their choreography in the
experiment workflow. The tool used to support this
actor in specifying the experiment workflow is a
workflow editor, in particular the Taverna tool
(http://taverna.sourceforge.net/), allowing scientists
to define and execute their workflows, and to
analyse the deriving outputs, through operations of
Web Service discovery, selection, and link, which
can be executed through a graphical support. Once
the definition of the experiment has been completed,
an instance of a workflow model is created and
produces results (e.g., usually in the Scufl format
XML file). Scufl (Simple Conceptual Unified Flow
Language) (Oinn, 2004) is a workflow description
language similar to BPEL (Business Process
Execution Language), used in the commercial and
software engineering environments (Andrews et al.,
2003).
Site/sites where data are placed. We assume
that the site of Organization D stores (and has
published as available, according to the Web Service
style (Booth et el., 2004)) all the data necessary for
the experiment. The data resources can also be
drawn from various sites (possibly belonging to
different organizations); this means that several
instances of the experiment have to be launched at
the same time, in order to achieve the best
parallelism and resource allocation policy. Data can
be found in any location on the Web, identified
through an URL, and published in a registry in the
UDDI style.
Site/sites where the computation takes place.
In our example, processing uses the Matlab tool
(http://www.mathworks.com), itself modelled as a
Web Service performing the clustering process
described in (Liberati et al., 2005). The computation
can be partitioned in functional sub-modules. The
granularity and the terms of the contract set up in the
Virtual Laboratory before starting an experiment and
regarding collaboration are negotiable upon specific
demands and according to the specification of
complexity, performance, and costs of the
experiment. Additional modular potentialities are
available through foreseen interfaces, that can be
linked and referred to existing services, using simply
the Web Service Definition Language (WSDL)
interfaces, with possibly additional information
regarding the modalities of execution of the
experiment (e.g., security, performance, and in
general Quality of Service related parameters).
3 ARCHITECTURAL ISSUES
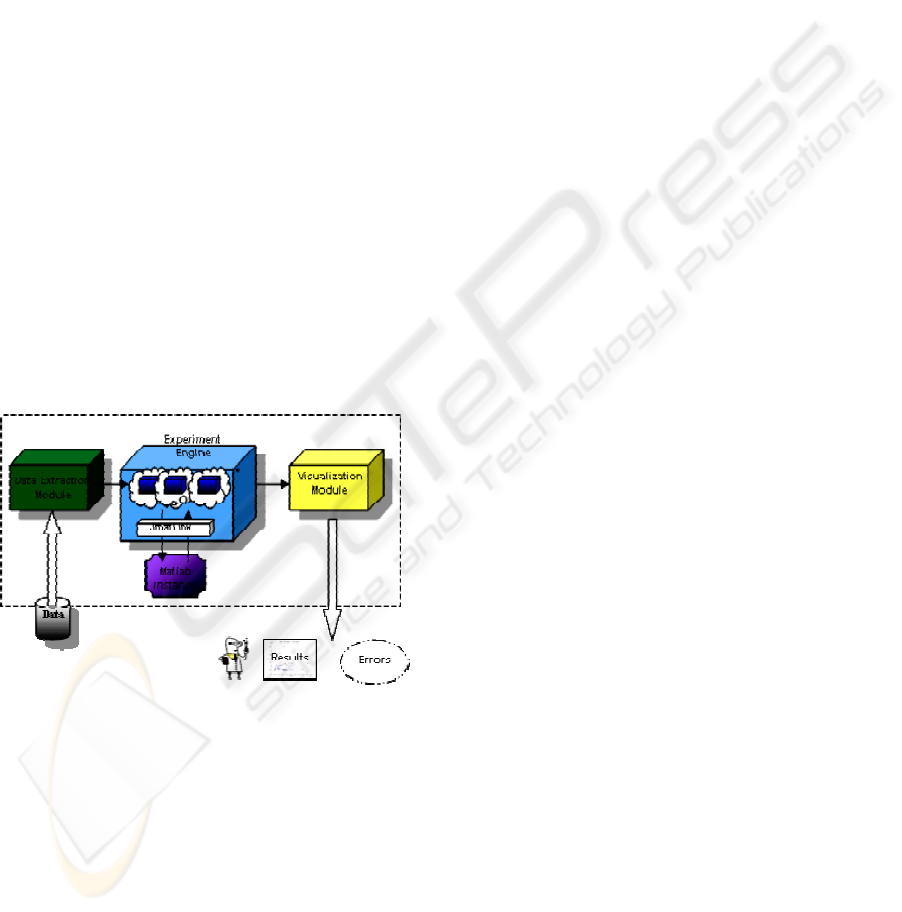
Figure 3 shows the implementation of some
components of the Virtual Laboratory support
system in our specific experiment, that is, a process
of DNA-microarrays clustering. The Experiment
Engine interfaces the Matlab environment and
executes the code for the clustering procedure. The
core of the elaboration is a Matlab function. This is
made active by Java code through a suitable
JMatLink interface (http://jmatlink.sourceforge.net/).
ICEIS 2007 - International Conference on Enterprise Information Systems
228
The whole Java code is exposed as a Web Service.
Single functions can be isolated as independent Web
Services, and allocated on different sites, allowing
reuse. Web Services are created and deployed via
Apache Axis, installed on the Apache Tomcat
container (http://tomcat.apache.org/). The Data
Extraction Module is used as follows. Nearly the
whole totality of DNA-microrrays data is available
in a few standard formats. Each can be further
distinguished in different variants. A wide choice of
data is available for example on the Cancer Program
Data Set of Broad Institute
(http://www.broad.mit.edu/cgi-
bin/cancer/datasets.cgi). The Data Extraction
Module is a Java module that can express these
different kinds of formats into a unified
representation.
4 FUTURE WORKS
An interesting evolution can promote the flexibility
of the Virtual Laboratory concepts presented in the
paper. In particular, the use of agents (Wooldridge,
2002) allows for dynamic selection, at run time, of
the services that execute the activities of an
experiment. An improved architecture can make use
of two kinds of agents. A Supervisor Agent (SA) is
in charge of supervising the overall execution of the
experiment. For each activity of the experiment, the
SA discovers from a Yellow Pages directory service
the set of agents able to provide the needed
service(s). These agents are called Resource Agents
(RAs) and represent and manage the actual services.
The services are still modelled as Web Services,
described with various properties, among which the
non-functional QoS parameters (availability,
robustness, time, cost, security) that are needed to
select the Web Services. The basic role of the SA is
to assign each activity to the most appropriate RA
and to supervise its execution. For example, the
activity assignment can be done according to the
well-known contract net protocol (Smith, 1980), in
which an SA announces an activity to some RAs and
awards it to the best offering RA. A given
organization of the collaborative network can have
more SAs (i.e., more experiments currently running)
and more RAs (i.e., more resources made available).
In this scenario, the Workflow Designer does not
even need to be aware of the physical structure of
the network that will be used to execute the
experiment in a distributed collaborative way. For
example, the Workflow Designer does not need to
specify, as in Figure 1, that the initial activity of the
workflow will be executed by a service from
Organization A. The Workflow Designer has only to
specify the kind of the initial activity and, then, the
SA in charge of supervising the execution of the
experiment will find the most appropriate service to
execute the activity, according to Quality of Service
parameters. Note that the service from Organization
A will execute the activity if it is the only available
Figure 2: General architecture of the Virtual Laboratory.
A VIRTUAL LABORATORY FOR WEB AND GRID ENABLED SCIENTIFIC EXPERIMENTS
229
service for the activity or if it is considered by the
SA to be the best service (e.g., requiring minimum
time and with high degree of security) to execute the
activity.
Setting up this improved architecture requires
addressing a number of issues, including the
dynamic negotiation of contracts between agents,
and the interface between agents and the workflow
engine and between agents and Web Services.
When the Grid Services (Foster and Kesselman,
2004) will have reached a better stability on the
market, it will be possible to build a prototype of the
proposed system by using also this technology.
The approach can be generalized, with the
opportune tuning, to wider areas of business
interaction. The evolutions in the commercial
domain will be offered by the virtual marketplaces
of services, where the modalities of distribution are
characterised by ways of payment (for subscription
or for amount consumed) for single transactions.
Also privacy and data security are major
concerns in our current research, considering both
methods to select trusted nodes within the
cooperation network and to obscure or encrypt both
the transmitted and stored data and portions of the
experiment workflow, to preserve sensitivity,
according to user security requirements.
Figure 3: Prototype system modules.
ACKNOWLEDGEMENTS
We thank prof. Nicoletta Dessì, dr. Andrea Bosin,
and dr. Barbara Pes, for common ideas and work.
We acknowledge the contribution of Alberto
Trovato in the prototype implementation.
REFERENCES
Alonso, G., Casati, F., Kuno, H., Machiraju, V. (2004).
Web Services – Concepts, Architectures, and
Applications. Springer Verlag.
Andrews, T., et al. (2003). BPEL4WS, Business Process
Execution Language for Web Services version 1.1.
Booth, D., Haas, H., McCabe, F., Newcomer, E. I.,
Champion, C., Ferris, C., Orchard, D. (2004). Web
Service Architecture W3C Specification. W3C
Working Group Note, 11 February 2004,
http://www.w3.org/TR/ws-arch/
Bosin, A., Dessì, N., Fugini, M., Liberati, D., Pes, B.
(2005). Supporting Distributed Experiments in
Cooperative Environments. Proc. Int’l Workshop on
Enterprise and Networked Enterprises
Interoperability, ENEI'2005, Nancy, France, Lecture
Notes in Computer Science 3812.
Comm. of the ACM (2003). Special Issue on Service
Oriented Architectures, 46(10).
De Roure, D., Gil, Y., Hendler, J. A. (eds.) (2004). IEEE
Intelligent Systems, Special Issue on E-Science, 19(1).
Foster, I., Kesselman, C. (2004). The GRID2: blueprint for
a new computing infrastructure. Morgan Kaufmann.
Hey, T., Trefethen, A. (2004). e-Science and its
implications. UK e-Science Core Programme,
Engineering and Physical Sciences Research Council,
Polaris House, Swindon SN 1ET, UK.
Liberati, D., Garatti, S., Bittanti, S. (2005). Unsupervised
mining of genes classifying leukemia, Encyclopedia of
data warehousing and mining, 1155-1159, J. Wang
(ed.), Idea Book.
Newman, H., et al. (2003). Data-intensive for e-Science.
Communications of the ACM, 46(11).
Oinn, T. (2004). Xscufl Language Reference, European
Bioinformatics Institute, http://www.ebi.ac.uk/~tmo/
mygrid/XScuflSpecification.html.
Peltz, C. (2003). Web services orchestration and
choreography. IEEE Computer, 36(10).
Smith, R. G. (1980). The Contract Net Protocol: High-
Level Communication and Control in a Distributed
Problem Solver. IEEE Transactions on Computers,
29(12).
Travica, B. (2005). Virtual organization and electronic
commerce, ACM SIGMIS Database, 36(3).
van der Aalst, W., van Hee, K. M. (2002). Workflow
Management: Models, Methods, & Systems. The MIT
Press.
Wooldridge, M. (2002). An Introduction to Multiagent
Systems. John Wiley and Sons.
ICEIS 2007 - International Conference on Enterprise Information Systems
230
