
AUTOMATIC HEART LOCALIZATION
IN ULTRASOUND FETAL IMAGES
Mozart Lemos de Siqueira and Philippe Olivier Alexandre Navaux
Institute of Informatics, Federal University of Rio Grande do Sul
PO Box: 15064, ZIP: 91501-970, Porto Alegre-RS, Brazil
Keywords:
Medical Imaging, Fetal Cardiology, Ultrasound, Bhattacharyya Coefficient, Texture Feature.
Abstract:
This paper presents the research developed in order to detect the cardiac structure in echocardiography gray
images from the fetal heart. It is based on patten recognition and use a density probability function with the
scales of gray. The function is also used for search of the similar cardiac structure, where it is applied on the
whole image, and then compared with the pattern of structure in which one interested. In order to obtain the
similarity that defines the choice of the structure of interest we use the Bhattacharyya coefficient.
The method uses texture features to isolate the region of interest inside the ultrasound image to improve the
results and performance. A prototype was developed to evaluate the proposed method. The results of the
experiments are also presented in this paper.
1 INTRODUCTION
Scientific Research in medical imaging area grows
constantly and its results generate many benefits to
people’s health. Usually, these researches cover many
aspects of image processing and medicine such as dis-
ease predicting and more accurate diagnostics. In this
context, this article presents a computational tech-
nique for automatic localization of cardiac structure
in images. The images used is fetal echocardiography
and, more specific, fetal’s ultrasound images (Duncan
and Ayeche, 2000; Sheehan, 2000). Such images are
important to the prenatal phase, because an early di-
agnosis of congenital cardiopathy can help the med-
ical treatment. Therefore, this work may be used to
help automatic analysis, mainly when the physician
involved is not a heart specialist.
Although ultrasound images provide a lot of in-
formation about cardiac structures, the resulting im-
ages are contaminated by speckle noise, which cor-
rodes the borders of the cardiac structures (Kang and
Hong, 2002; Zong et al., 1998; Crimmins, 1985; Bur-
ckhardt, 1978). This characteristic turns difficult the
automatic image processing, and specially the pattern
recognition. Besides this kind of noise, other factors
influence the outcome of fetal ultrasound image. For
instance, the transducer
1
and the fetus positioning, the
rotation and the scale variations in images of different
patients and the composition of the tissue separating
the fetus heart are issues that must be taken into ac-
count when dealing with heart images (Mattos, 1999).
The method presented in this paper makes use of a
density probability function to obtain the mapping of
the region of interest and generate a searching mold to
be used in other target images. This mold is based on
scales of gray image in the region where the cardiac
structure is positioned. Besides, the mold calculation
considers the distance of the pixels of the center.
The search for the region of interest begins with
the mapping of the candidate regions in the target
image. Each candidate region is compared to the
searching mold using the Bhattacharyya coefficient
(Djouadi et al., 1990). This calculated coefficient pro-
vides a index that defines the degree of similarity be-
tween the candidate region and the searching mold.
A prototype for the cardiac structure localization
was developed in order to evaluate the automatic
searching pattern. The search of the candidate re-
gions was implemented using the mold and local-
1
The electronic device used to capture ultrasound im-
ages
107
Lemos de Siqueira M. and Olivier Alexandre Navaux P. (2007).
AUTOMATIC HEART LOCALIZATION IN ULTRASOUND FETAL IMAGES.
In Proceedings of the Second International Conference on Computer Vision Theory and Applications, pages 107-112
DOI: 10.5220/0002059101070112
Copyright
c
SciTePress
ization in different images that was analyzed with a
moving window shifted in 10 pixels along the image
columns and lines. The moving window size was de-
fined based on the mold size. Although this search
covers the whole image and can provide an accurate
result, the time to process on each image is usually
greater than one minute considering a resolution of
640x480. Because this, it was necessary to find new
manners to increase the search performance.
Allowing the search performance in considera-
tion, we developed a module to images preprocessing
and to isolate the region of interest (ROI). The entropy
texture feature was used for selection of region of in-
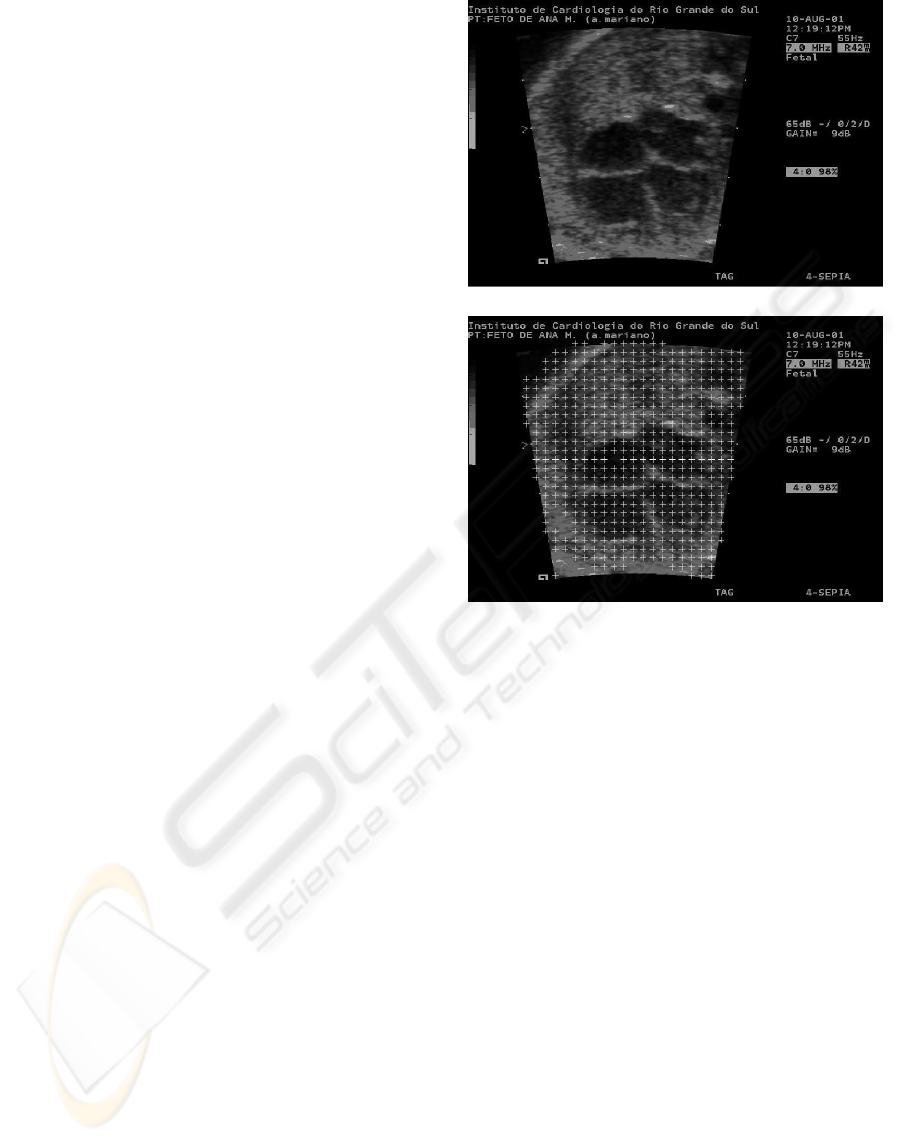
terest. The Figure 1 (a) presents an original image and
a preprocessed image (b) with entropy texture feature
to select the ROI. This approach improves the search
performance because it does not need search regions
out of ultrasound, i.e. the black frame on images. The
use of texture features on this ultrasound images is
justified by the specific characteristics of them, there-
fore many works about ultrasound processing using
this features (Valdes-Cristerna et al., 2004; Brusseau
et al., 2004; Hope et al., 2005).
The paper is organized in five sections. After this
introduction, we present a brief section with related
research. The Section 3 present the description of the
proposed model. The Section 4 show the obtained re-
sults. Finally, the paper presents the conclusion sec-
tion.
2 RELATED RESEARCH
The pattern recognition in images is an area that has
offered many results. The goal of pattern recogni-
tion is to automatically spot specific objects inside
images, without the intervention from the user. There
are many application possibilities for pattern recogni-
tion in medicine, since several medical routines gen-
erate images. In some routines, physicians look for
image patterns in nodules, intern structures, the be-
havior of the heart (i.e. systole and diastole), for ex-
ample. There are works that already address this rou-
tines (Lee et al., 2001; Brown et al., 2001; Bruijne
et al., 2003; Salvadorls et al., 2003).
Jacob et al. (Jacob et al., 2002) and Sugioka et
al (Sugioka et al., 2003) developed research using
patterns to detect cardiac structures using active con-
tours (snakes) in echocardiographic images. Comani-
ciu (Comaniciu et al., 2004) proposed a methodology
to tracking cardiac edges in echocardiographic im-
ages using several information extract of the images.
One of the principal ways of searching objects in
images is through patterns. In most of the cases, the
(a)
(b)
Figure 1: Fetal echocardiography obtained with ultrasound
device. (a) Original image; (b) The regions of interest iso-
lated with entropy.
objects of interest must be known, and their charac-
teristics are searching in the image. When the object
of interest is not previously known, the complexity
of the search increases. The search algorithm perfor-
mance is important to the processing time do not to
be a bottleneck to the system.
3 PROPOSED MODEL
To developing the automatic localization of the car-
diac structure was studied the approach proposed
by Comaniciu (Comaniciu et al., 2003) for tracking
down dynamic objects. We proposed a model inspired
on Comaniciu and implement a prototype to evaluate
our proposal. The prototype was applied in 640x480
fetal cardiac ultrasound images with four chamber cut
plans (Nelson, 1998).
The searching pattern is calculated based on the
gray scale histogram and the space location of pixels
of a given object of interest. This gray scale was used
VISAPP 2007 - International Conference on Computer Vision Theory and Applications
108

as parameter in the calculation of the searching mold.
The method process is composing by three stages:
the first, the second and the third. The first stage of
the process consists in the selection of the region of
interest to be found. This structure region is isolated,
and then used in the calculation of the searching mold.
This task is done by the user, who interacts with the
prototype in order to select the limits of the structure
inside of an image.
The second stage of the localization process is the
search for regions that looks like the searching mold.
In this search it is used the same calculation that gen-
erated the mold, applied to several regions of the im-
age, which are called candidates. From the distri-
butions generated in the candidate regions, it is es-
timated the position of the pattern that is more similar
among the candidate regions and the searching pattern
used.
The last stage of the process is the search for the
region of interest inside the image. At this stage, the
similarity is calculated through the Bhattacharyya co-
efficient. The whole process is shown in Figure 2.
2. Candidate molds
p
u2
p
1u
un
p
1. Searching mold
Result
3. Similarity
qu
Figure 2: Prototype model (q
u
is the searching mold and
p
un
, the candidate regions).
3.1 Calculation of the Searching Mold
To determine the mold of the cardiac structure, the
method follows this procedure: a circular region, ra-
dius h, with center x
c
, placed in the center of position
of the desired structure of the region. For each point
(or pixel), x = (x
1
,x
2
), in the region, a vector of char-
acteristics is extracted and categorized according to
a discrete number of characteristics. This point re-
ceives an index of that characteristic, u = b(x). The
distribution of characteristics, q = {q
u
}
u=1...m
, which
computes the occurrence of a given characteristic u in
the region of the desired structure, is calculated by:
q
u
=
∑
n
i=1
k(|x
i
−x
c
|/h)δ(b(x
i
),u)
∑
n
i=1
k(|x
i
−x
c
|/h)
(1)
Where x
c
is the center of the region, and δ is the
delta Kronecker function
2
. Notice that the distribu-
tion satisfies
∑
n
i=1
q
u
= 1.
The function k(x) is an isotropic kernel that re-
duces the importance of characteristics removed from
the center, in the distribution calculation q. Specif-
ically, the important characteristic is the gray scale
of the pixel. The distribution (q) represents an his-
togram of gray q = {q
u
}
u=1...,m
which incorporates
spatial and color information of the image pixels.
Figure 3 shows a scheme with the stages of the
generation process of the mold. In this figure, it
is possible to see the window of the system, devel-
oped at Matlab (Gonzalez, 2004), with an echocar-
diographic image where the region of the image used
for the calculation of the mold is selected. To this
mold, it is applied the equation 1, and the vector q
u
is calculated and stored for later use in searching for
images of different patients. Figure 4 shows the distri-
bution characteristic of the molds of cardiac structure
of different patients.
uq
Figure 3: Calculation of searching mold.
3.2 Search in the Image of Interest
At this stage of the process, it is necessary to scan
given image searching for a similar mold to the
searching mold generated in the previous stage. The
solution developed for searching was preprocessing
the image with entropy texture feature to select only
the ROI.
To search for the candidate region, it must be as-
sumed that in this region of the image the distribution
2
The Kronecker delta function returns 1 if its arguments
are equal and 0, otherwise.
AUTOMATIC HEART LOCALIZATION IN ULTRASOUND FETAL IMAGES
109
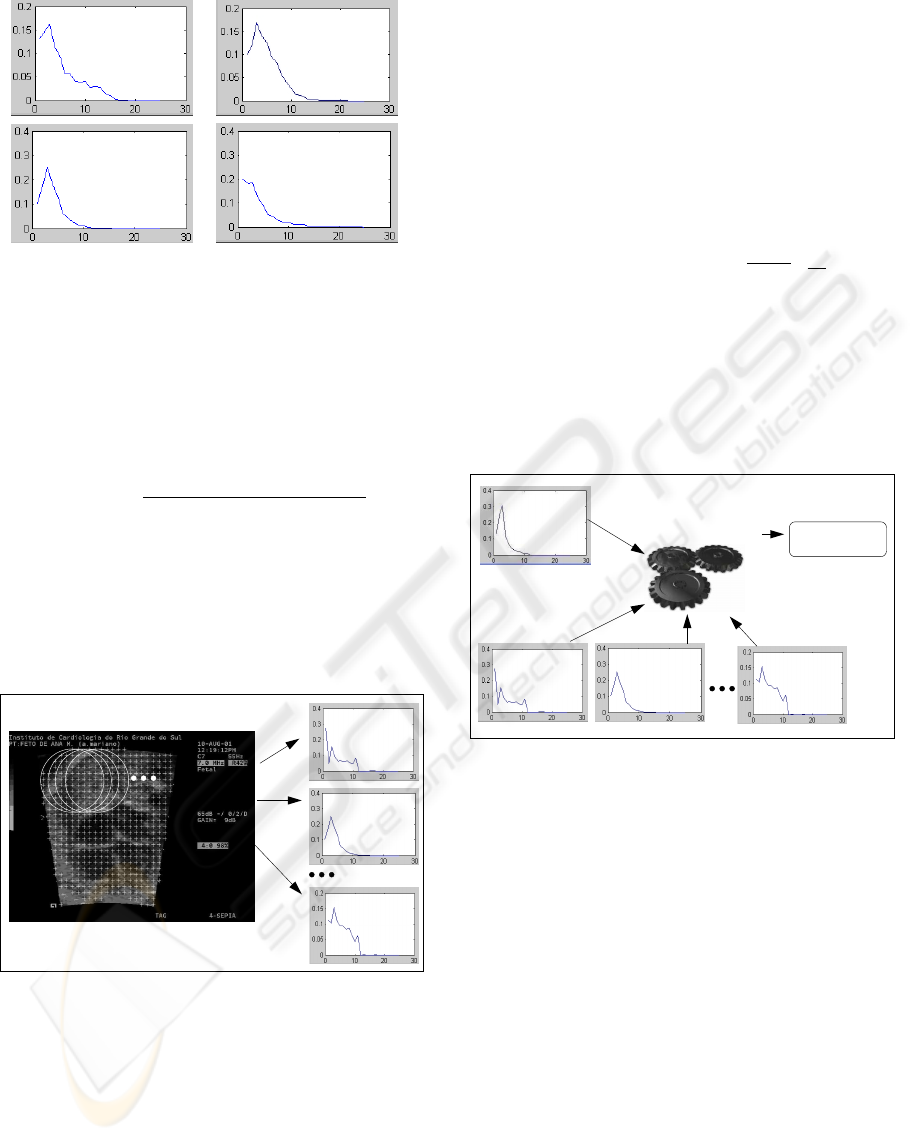
Figure 4: Distribution characteristic of the molds in differ-
ent patients.
is similar to the searching mold of the structure. By
doing this, the searching mold equation was used for
the calculation of the candidate regions as in Equa-
tion 2.
p
u
(y, h) =
∑
n
i=1
k(|x
i
−y|/h)δ(b(x
i
),u)
∑
n
i=1
k(|x
i
−y|/h)
(2)
In the search, the image is examined with a mov-
ing window and a set of candidates generated for later
comparison with the mold. Figure 5 shows the pro-
cess of searching in an image. The number of candi-
date patterns depends on the diameter of the searching
pattern and the image processed.
pu n
pu 2
pu 1
Figure 5: Process of searching for structures.
3.3 Finding of the Structure in the
Image of Interest
Spotting the cardiac structure takes place by compar-
ing the candidate mold found at the previous stage
with the structure mold generated during the first
stage of the process. The comparison is done using
the Bhattacharyya coefficient (Djouadi et al., 1990),
which provides an index of similarity between the
mold distribution and candidate mold.
The Bhattacharyya coefficient is a measure of the
statistical separability of classes, and gives an esti-
mate of the probability of correct classification. It is
a divergence-type measure that has a straightforward
geometric interpretation. It is the cosine of the angle
between n-dimensional vectors. The closer to 1 the
provided value is, the more similar the vectors are.
The calculation is presented in the Equation 3.
ρ(y) = ρ[p(y),q] =
m
∑
u=1
p
p
u
(y)
√
q
u
(3)
The method spots the searching structure through
the patterns bearing more similarity. Only the can-
didate region bearing more similarity is selected; all
the others are ignored. Even so, the calculation of the
candidate region is done considering the whole image.
The Figure 5 shows this process.
qu − mold
Bhattacharyya coefficient
pu 2 pu npu 1 (candidate mold)
candidate mold = 2
More Similar
Figure 6: Finding of the cardiac structure.
4 RESULTS
The tests was applied in an image sample where the
searching mold has been calculated from an image
and the search occurred over 33 different images. In
the sample showed in Table 1 we classified the results
in three types: success (second column of table 1),
find two chamber (third column of table 1) and fail
(fourth column of table 1). The first class is the objec-
tive of work, i.e. success on the search for the heart
structure on the image. When the method finds the
heart, but not totally we classified with two chamber
and when the method fail, the third class is select. The
last line in the table shows the means of all tests re-
sults.
The Figure 7 shows images with the results, the
first image showed in the Figure 7(a) is a instance of
VISAPP 2007 - International Conference on Computer Vision Theory and Applications
110
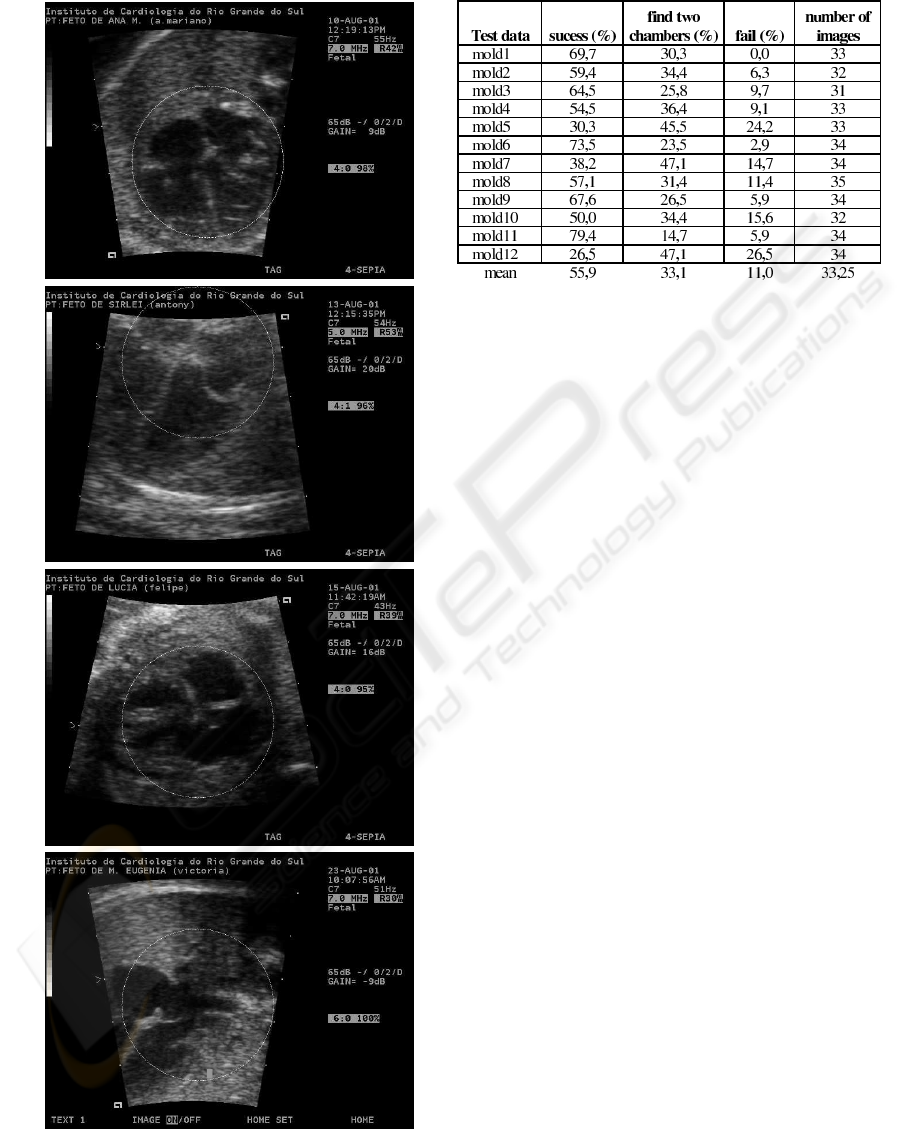
(a)
(b)
(c)
(d)
Figure 7: Results obtained on tests.
Table 1: Results obtained with the prototype tests.
search mold and it localization on the image, the oth-
ers two, Figure 7 (b) and (c) are instances of success,
and the Figure 7 (d) is a search result instance of class
”find two chamber”.
5 CONCLUSION
This paper presents a model developed for pattern
recognition of cardiac structure in echocardiographic
images, as well as the necessary modifications for
its improvement. One of reason that motives the re-
search about echocardiographic images has been the
dynamics feature. In reality, these images are extract
of videos of the cardiac dynamics that allow work-
ing with these sequences on future. This approach
can take advantage of the dynamics information on
the heart test that nowadays does not be used.
The images used were kindly provided by the fe-
tal cardiology team at Institute of Cardiology of Porto
Alegre. Those images were captured with an echocar-
diographic machine produced by Siemens (Aspen).
That machine allows the recording of images in DI-
COM format. The resolution of the images was
640x480 pixels.
It has been observed that the size of the search-
ing mold influenced the results; larger molds showed
better performance. A small region may not represent
adequately the structure. This happens because of the
noise, the size of the heart is variable, and the search
is based exclusively on the intensity of gray.
The use of texture feature, to select the region
of interest had been important to increase the perfor-
mance of the method, because texture can separate the
ultrasound image of the background.
The prototype developed can automatically ex-
tract the pattern of cardiac structure of echocardio-
AUTOMATIC HEART LOCALIZATION IN ULTRASOUND FETAL IMAGES
111
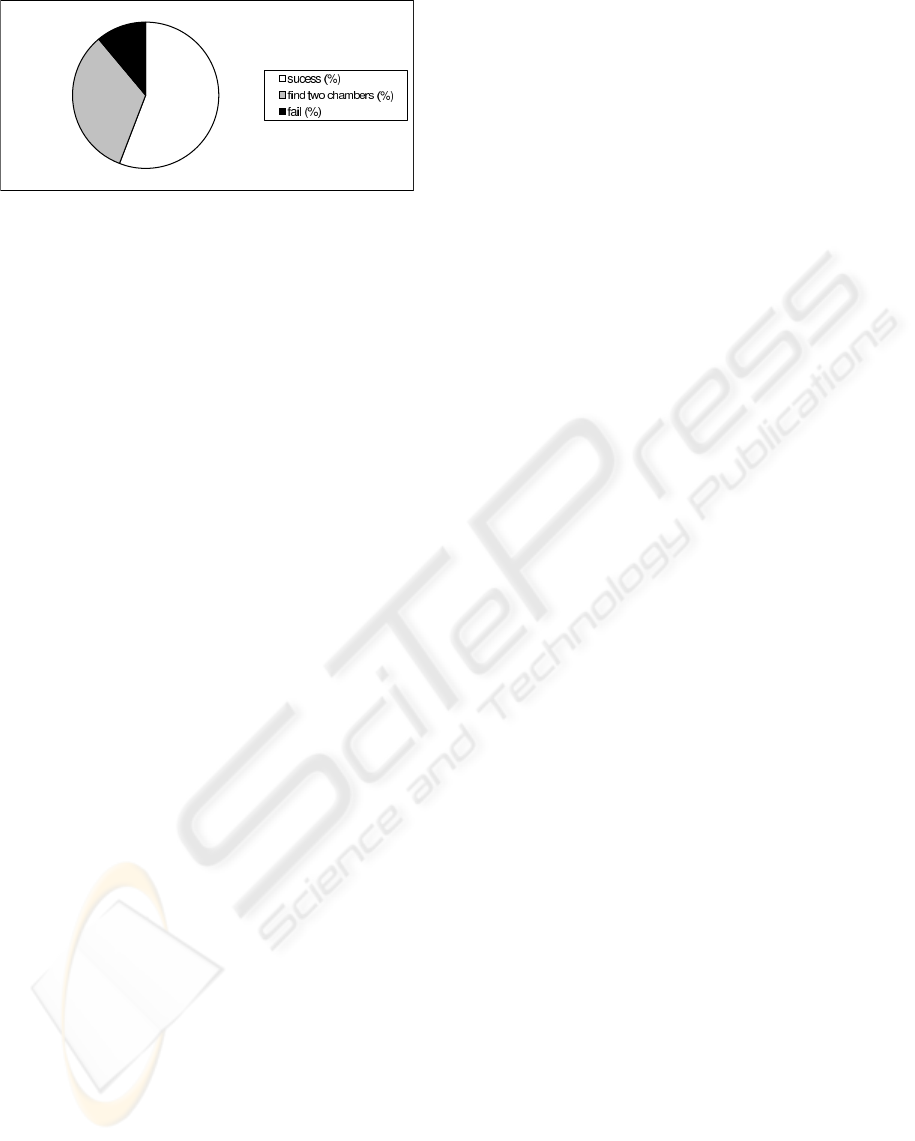
Figure 8: Trial obtained, only mean.
graphic images. Considering the graphic depicted in
Figure 8 and the class ”find two chambers” as ob-
jective, we can confirm the success of the proposed
method.
REFERENCES
Brown, M. S. et al. (2001). Patient-specific models for lung
nodule detection and survellience in ct images. IEEE
Transactions on Medical Imaging, 20(12):1242–1250.
Bruijne, M., Niessen, W. J., Maintz, J. B. A., and Viergever,
M. A. (2003). Localization and segmentation of aortic
endografts using marker detection. IEEE Transaction
on Medical Imaging, 22(4):473–482.
Brusseau, E., de Korte, C. L., Mastik, F., Schaar, J., and
van der Steen, A. F. W. (2004). Fully automatic lumi-
nal contour segmentation in intracoronary ultrasound
imaging a statistical approach. IEEE Transactions on
Medical Imaging, 23(5):554–566.
Burckhardt, C. B. (1978). Speckle in ultrasound b-mode
scans. IEEE Transactions on Sonics and Ultrasonics,
SU-25(1):1–6.
Comaniciu, D., Ramesh, V., and Meer, P. (2003). Kernel-
based object tracking. IEEE Transactions on Pattern
Analysis and Machine Intelligence, 25(5):564–577.
Comaniciu, D., Zhou, X. S., and Krishnan, S. (2004). Ro-
bust real-time myocardial border tracking for echocar-
diography: An information fusion approach. IEEE
Transaction on Medical Imaging, 23(7):849–860.
Crimmins, T. R. (1985). Geometric filter for speckle reduc-
tion. Applied Optics, 24(10):1438–1443.
Djouadi, A., Snorrason, O., and Garber, F. D. (1990).
The quality of training-sample estimates of the bhat-
tacharyya coefficient. IEEE Transactions on Pattern
Analysis and Machine Intelligence, 12(1):92–97.
Duncan, J. S. and Ayeche, N. (2000). Medical image anal-
ysis: Progress over two decades and the challenges
ahead. IEEE Transactions in Pattern Analysis and
Machine Intelligence, 22(1):85–105.
Gonzalez, R. C. (2004). Digital image processing : using
matlab. Pearson Prentice Hall, Upper Saddle River, 1
edition.
Hope, T., Linney, N., and Gregson, P. (2005). Using the
local mode for edge detection in ultrasound images.
In Proc of. Canadian Conference on Electrical and
Computer Engineering, pages 374–377, Canada. Los
Alamitos: IEEE.
Jacob, G., Noble, J. A., Behrenbruch, C., Kelion, A. D.,
and Banning, A. P. (2002). A shape-space-based ap-
proach to tracking myocardial borders and quantifying
regional left-ventricular function applied in echocar-
diography. IEEE Transaction on Medical Imaging,
21(3):226–238.
Kang, S. C. and Hong, S. H. (2002). A speckle reduction fil-
ter using wavelet-based methods for medical imaging
application. In Proc of. 14th International Conference
on Digital Signal Processing, DSP2002, volume 2,
pages 1169–1172, Santorini, Greece. Los Alamitos:
IEEE.
Lee, Y., Ishigaki, T., et al. (2001). Automated detection of
pulmonary nodules in helical ct images based on an
improved template-matching technique. IEEE Trans-
action on Medical Imaging, 20(7):595–604.
Mattos, S. S. (1999). O Corac¸
˜
ao Fetal. Revinter, Rio de
Janeiro/RJ.
Nelson, T. R. (1998). Ultrasound visualization. In Advances
in Computers, volume 47, pages 185–253. Academic
Press, New York.
Salvadorls, A., .Maingourd, Y., Ful, S., and LeralluT, J.-
F. (2003). Optimizaton of an edge detection algo-
rithm for echocardiographic images. In Proc. of the 25
Annual International Conference of the IEEE EMBS,
Cancun, Mexico. Los Alamitos: IEEE.
Sheehan, F. (2000). Echocardiography. In Handbook of
Medical Imaging, volume 2, pages 609–674. Spie,
Bellingham.
Sugioka, K. et al. (2003). Automated quantification of
left ventricular function by the automated contour
tracking method. ECHOCARDIOGRAPHY: A Jor-
nal of Cardiovascular Ultrasound and Allied Tech.,
20(4):313–318.
Valdes-Cristerna, R., Jimenez, J., Yanez-Suarez, O., Leral-
lut, J., and Medina, V. (2004). Texture-based echocar-
diographic segmentation using a non-parametric esti-
mator and an active contour model. In Proc. of the
26th Annual International Conference of the IEEE
EMBS, San Francisco, US. Los Alamitos: IEEE.
Zong, X., Laine, A., and Geiser, E. (1998). Speckle reduc-
tion and contrast enhancement of echocardiograms via
multiscale nonlinear processing. IEEE Transactions
on Medical Imaging, 17(4):532–540.
VISAPP 2007 - International Conference on Computer Vision Theory and Applications
112
