
CONSOLIDATED TREE CONSTRUCTION ALGORITHM:
STRUCTURALLY STEADY TREES
J.M. Pérez, J. Muguerza, O. Arbelaitz, I. Gurrutxaga
Informatika Fakultatea, Basque Country University, po. Box 649, 20080 Donostia, Spain
Ke
ywords: decision trees, steadiness, explaining capacity, structural diversity measure
Abstract: This paper presents a new methodology for building decision trees or classification trees (Consolidated
Trees Construction algorithm) that faces up the problem of unsteadiness appearing in the paradigm when
small variations in the training set happen. As a consequence, the understanding of the made classification is
not lost, making this technique different from techniques such as bagging and boosting where the
explanatory feature of the classification disappears. The presented methodology consists on a new meta-
algorithm for building structurally more steady and less complex trees (consolidated trees), so that they
maintain the explaining capacity and they are faster, but, without losing the discriminating capacity. The
meta-algorithm uses C4.5 as base classifier. Besides the meta-algorithm, we propose a measure of the
structural diversity used to analyse the stability of the structural component. This measure gives an
estimation of the heterogeneity in a set of trees from the structural point of view. The obtained results have
been compared with the ones get with C4.5 in some UCI Repository databases and a real application of
customer fidelisation from a company of electrical appliances.
1 INTRODUCTION
Several pattern recognition problems need an
explanation of the made classification together with
a good performance of the classifier related to its
discriminating capacity. Diagnosis in medicine,
fraud detection in different fields, customer
fidelization, resource assignation, etc, are examples
of applications where an explanation of the
classification made becomes as important as the
accuracy of the system. Among the set of
classification techniques that are able to give an
explanation for the classification made, we can find
the decision trees [Quinlan 93]. However,
classification trees have a problem; they are too
sensitive to the sample used in the induction process.
This weakness of decision trees is called
unsteadiness or instability [Dietterich 00a, Chawla el
al 02]. Due to this behaviour, several algorithms for
building classification trees with greater
discriminating capacity and steadiness have been
developed.
Bagging [Breiman 96], boosting [Freund et al 96],
and different variants of them [Chawla el al 02]; are
examples of techniques used to face the problem. All
of them use resampling techniques. Bagging uses a
set of trees induced from different bootstrap
subsamples, extracted from the initial sample
(training set). In the multiple classifier built,
classification is made by simple voting or weighted
voting [Duin et al 00, Bauer et al 99]. When the used
technique is boosting, the different trees are built
sequentially and the patterns are reweigthed
depending on the error made by the preceding
classifier. This makes the next tree in the sequence
concentrate in the hardest zones of the classification
field, so, in the decision boundaries. The final
decision is based on a weighted vote that takes into
account the error estimated during the learning
process of each one of the trees composing the
system.
These techniques obtain improvements in the
behaviour of the global system but they lose the
explaining capacity of the classifier, because in
every case the final classifier is a combination of
different trees with different structures. Our
approximation introduces a new methodology that
brings together both characteristics: stability and
14
M. Pérez J., Muguerza J., Arbelaitz O. and Gurrutxaga I. (2004).
CONSOLIDATED TREE CONSTRUCTION ALGORITHM: STRUCTURALLY STEADY TREES.
In Proceedings of the Sixth International Conference on Enterprise Information Systems, pages 14-21
DOI: 10.5220/0002602200140021
Copyright
c
SciTePress
explaining capacity. We have developed a new
algorithm for building trees. We denominate the
built trees consolidated trees. This methodology
uses different training subsamples and builds a
single accorded tree among all of them. Every
technique mentioned before to make more stable
classification trees, loses its explanatory component,
because a set of trees with very different structures is
used to make the final decision. Our aim has been to
design a new methodology for building trees that
maintaining the performance of standard
classification trees, reduces the complexity and adds
structural stability to the induced tree. This makes
the explanation related to classification much more
robust and steady, so, less sensible to small changes
in the sample used to build the classifier. In order to
analyse the structural stability of classification trees,
we have defined a diversity measure or distance that
measures how similar or different a set of trees is.
The paper proceeds with the description of our new
methodology for building consolidate trees, Section
2. In Section 3, the structural diversity measure is
introduced, followed in Section 4 by the description
of the data sets and the experimental set-up. The
results of our experimental study are discussed in
section 5. Finally, Section 6 is devoted to summarise
the conclusions and future research.
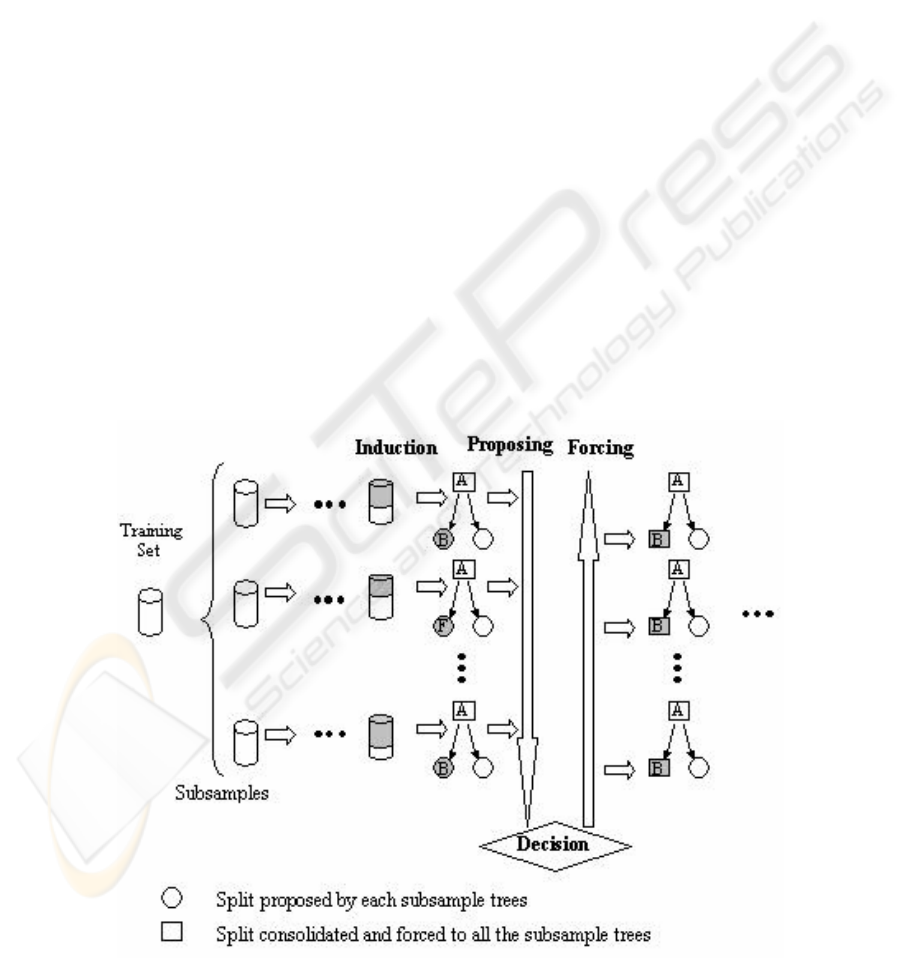
2 CTC LEARNING ALGORITHM
The tree building methodology we propose,
Consolidated Trees Construction (CTC) algorithm,
is based on resampling techniques. Several training
subsamples are extracted from the original training
set and a single tree is built based on consensus
among the partial trees that are being built from each
subsample. The main difference with bagging,
which makes it radically different, is that the
consensus is achieved in each step of the trees’
building process. The decision about which variable
will be used to make the split in a node is accorded
among the different proposals coming from the trees
(using the base classifier: C4.5 in our case) being
built from the different subsamples. The decision is
made by a voting process. Once the decision about
the variable selected to split the trees is made, all the
trees (each one associated to a different subsample)
are forced to use that variable to make the split. The
process is repeated iteratively until no more
divisions are possible, due to some stop criterion.
An schema of the algorithm can be found in (Figure
1) and it proceeds the following way:
Figure 1: Consolidation of a node.
CONSOLIDATED TREE CONSTRUCTION ALGORITHM: STRUCTURALLY STEADY TREES
15

1. Extract a set of subsamples (Number_Samples)
from the original training set using the desired
resampling technique (Resampling_Mode): size
of the subsamples (100%, 75%, 50%, etc; with
regard to the original set’s size), with replacement
or without replacement, stratified or not, etc.
2. The final tree is built node by node in preorder.
Each consolidated node is built the following
way:
a. For each subsample, induce the variable that
would be used to make the split at that level of
the partial tree; in the example (B, F,…,B).
b. Analyse the number of partial trees that
propose to make an split and decide depending
on the established criteria Crit_Split (ex:
simple majority, absolute majority, etc.)
whether to split or not. If the decision is not to
split (a leaf node is created), jump to the next
node to consolidate and go to step 2a.
c. Analyse the number of votes that has the most
voted variable (votes of variable B in Figure 1).
If based on Crit_Variable the variable has not
enough votes, consolidate the node as a leaf
node and go to step 2a. When the number of
votes is enough this variable will be the one
used to split the consolidated node.
d. Decide the branches the node to split will have,
depending on Crit_Branches criteria. If the
variable to split is continuous, determine the
cutting point (ex: using the mean or the median
of the values proposed for that variable). If the
variable to split is discrete, decide the set of
categories for each branch (ex: a branch for
each category, using heuristics such as C4.5’s
subset option, etc.).
e. Force the accorded split (variable and
stratification) in every tree asociated to each
subsample (every partial tree in the example is
forced to make the split with the consolidated
variable B). Jump to next node and go to step
2a.
The different decisions can be made by voting,
weighted voting, etc. Once the consolidated tree has
been built, its behaviour is similar to the one of the
used base classifier. Section 5 will show that the
trees built using this methodology, have similar
discriminating capability (the differences are not
statistically significant) but they are structurally
more steady and less complex. In order to analyse
this second aspect, we have defined the structural
diversity measure that we present in next section.
3 STRUCTURAL DIVERSITY
MEASURE
We will use this section to define the diversity
measure or structural distance, that will allow us to
analyse the stability of the consolidated trees and
compare them to the standard ones. The aim is to be
able to measure the heterogeneity existing in sets of
trees built using each of the methodologies to be
compared. The estimation of the degree of structural
diversity in a group, is made analysing the structural
differences among each possible pair of trees in the
group, and, calculating average values of the
differences obtained.
The defined metric or distance (Structural_Distance,
SD) is based on a vector (M
0
, M
1
, M
2
) with three
values used to compare two trees (T
i
, T
j
). Both trees
are looked through in preorder, node by node, and
the corresponding split variables are compared to
know whether they match or not. The three
components are calculated the following way:
• M
0
: Number of common nodes in T
i
and T
j
.
We understand as common nodes the ones that
being in the same position in both trees, make
the split based on the same variable.
• M
1
: measures the number of times that
looking through a common branch, a tree
makes a split in a node and the other does not.
Each increment is weighted depending on the
complexity of the subtree beginning in this
node.
• M
2
: measures the number of times that
looking through a common branch and arriving
to a common node, the variables chosen to
make the split in both trees are different. Each
increment is weighted depending on the
complexity of both subtrees.
The pseudo-code of the algorithm used for
calculating each one of the components of the
proposed measure is in Appendix.
The definitions show that to increase the value of
M
0
, the same variable has to appear in the same
node, so that the variable used to make the split has
been chosen at the same level in both trees. Once a
different split appears, the remaining subtrees of the
compared trees have an effect on the value of M
1
or
M
2
. This is important because in the tree
construction process (top-down), the variables used
to split a node are selected depending on their
statistical importance (entropy, p-value, ...). As a
consequence, this measure of similarity/diversity
takes also into account when making the
ICEIS 2004 - ARTIFICIAL INTELLIGENCE AND DECISION SUPPORT SYSTEMS
16
comparison, the statistical importance each tree has
given to each of the independent variables.
We have defined two measures, M
1
and M
2
, because
M
2
indicates higher diversity level among the trees
than M
1
does, and we think that it is important to
differentiate both cases.
Evidently, when greater is the first component and
smaller the two others, more homogeneous is the set
of trees used to calculate the values.
In order to calculate the diversity in the set of trees
we estimate the mean of the vector:
2,1,0
),(
)1(
2
)(
1
0,
=
−
=
∑
−
<
=
xwith
TTSD
mm
TSD
m
lk
lk
lk
Mx
set
mean
Mx
(1)
Once the vector has been obtained, the comparison
between groups can be done at vector level or at
scalar level making any combination of the different
components depending on the importance we want
to give to each of the measures get.
In this case we define a scalar measure
(%Common
mean
) based in three percentages
calculated from the three components normalised in
respect to the sum of the number of nodes of the two
trees compared. The scalar is computed with a lineal
combination of the three percentages with the vector
of weights (1,-1,-1). In this experimentation we
don’t take into account the different level of
importance of the two last components. If the scalar
measure is positive, the compared trees are more
similar than different among them, and, when the
value is negative the common part of the trees is
smaller than the different one. The range of values
the scalar can take is between –100 and +100. This
measure allows the comparison of groups of trees
with different complexities (at the structural level),
because the components are normalised in respect to
the number of nodes of the comparison.
4 EXPERIMENTATION
Six databases of real applications have been used for
the experimentation. Most of them belong to the
well known UCI Repository benchmark [Blake et al
98], widely used in the scientific community. Table
1 shows the characteristics of the databases used in
the comparison. The last database is a real data
applications from our environment, and does not
belong to UCI. The data set called Faithful is centred
in the electrical appliance’s sector. In this case, we
try to analyse the profile of the customers during the
time, so that a classification related to their fidelity
to the brand can be done. This will allow the
company to follow different strategies to try to
increment the number of customers that are faithful
to the brand, so that the sales increase. In this kind of
applications is very important to use a system that
provides information about the factors taking part in
the classification (the explanation), because it is
nearly more important to analyse and explain why a
customer is or is not faithful, than the own
categorisation of the customer. This is the
information that will help the corresponding
department to make good decisions.
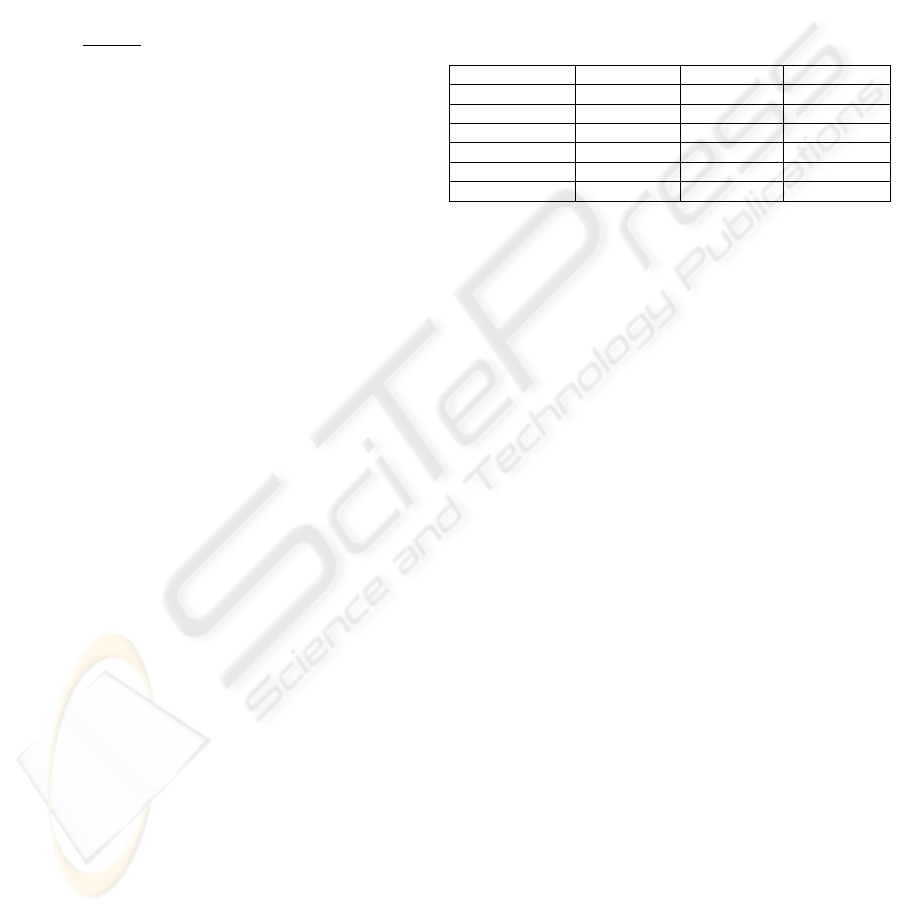
Table 1: Description of experimental domains
Domain
N. of patterns N. of features N. of classes
Breast cancer W 699 10 2
Heart disease C 303 13 2
Hypothyroid 3163 25 2
Lymphography 148 18 4
Segment 210 19 7
Faithful 24507 49 2
The CTC methodology has been compared to the
C4.5 tree building algorithm Release 8 of Quinlan
[Quinlan 93], using the default parameters’ settings.
The methodology used for the experimentation
[Hastie et al 01] is a 10-fold stratified cross
validation. Cross validation is repeated five times
using a different random reordering of the examples
in the data set. This methodology has given us the
option to compare 5 times groups of 10 trees CTC
and C4.5 in both senses the structural point of view
and the discriminating capacity (50 executions for
each instance of the analysed parameters). In each
run we have calculated the average error and its
standard deviation, together with the SD
mean
(T
set
) of
each of the groups of trees and the values
Comp
mean
(estimated as the number of internal nodes
of the tree) and %Common
mean
(explained in Section
3).
In order to evaluate the structural improvement
achieved in a given domain by using the algorithm
CTC compared to the algorithm C4.5, we have
calculated, (%Comm
CTC
-%Comm
C4.5
)/%Comm
C4.5
,
the relative improvement.
For every result we have tested the statistical
significance [Dietterich 98, Dietterich 00] of the
differences of the results obtained with the two
algorithms using the paired t-test (with significance
level of 95%).
CONSOLIDATED TREE CONSTRUCTION ALGORITHM: STRUCTURALLY STEADY TREES
17
5 RESULTS
We have analysed several parameters of the CTC
meta-algorithm. The ranges of the different
parameters analysed are:
1. Number_Samples: 3, 5, 10, 20, 30, 40, 50, 75,
100, 125, 150, 200. The obtained results show
that 10 is the minimum number of samples to
achieve satisfactory results with CTC. So, the
average results presented in this section do not
take into account the values 3 and 5, except for
Faithful where due to the size of the database the
study has been done from 3 to 40 subsamples.
2. Resampling_Mode: due to the importance of the
diversity of the subsamples used to build a
classifier [Skurichina et al 00], different options
have been proved (samples of size 50% and 75%
of the original training set drawn without
replacement, and bootstrap samples). The best
results have been obtained with the 75% (results
shown in the paper), probably because this is the
case where each subsample has larger amount of
information of the original sample.
3. Crit_Split and Crit_Variable: simple majority
among the Number_Samples for both. The
possibility of introducing thresholds to these
criteria has not been considered in this paper
because the comparison of our consolidated trees
(CTC) with the simple classification trees C4.5
has been done pruning the trees with the pruning
algorithm of the C4.5 Release 8 software. The
pruning has been done in order to obtain two
systems with similar complexity level (the same
zone in the learning curve) and also to be able to
make the structural comparison with similar
development conditions for all trees. We can not
forget that developing too much a classification
tree leads to a greater probability of overtraining
and on the other hand, the fact that a variable
appears in the tree is less significant because the
system takes into account very specific details of
the training set.
4. Crit_Branches: the selection of the new branches
to be created when the variables are continuous
has been made based on the median.
Experimentation with the mean has also been
done, but the results were worse, probably
because this measure has smaller stability. When
the variables were discrete we have not used the
subset option of C4.5 and as many branches as
categories has the variable selected to make the
split have been proposed.
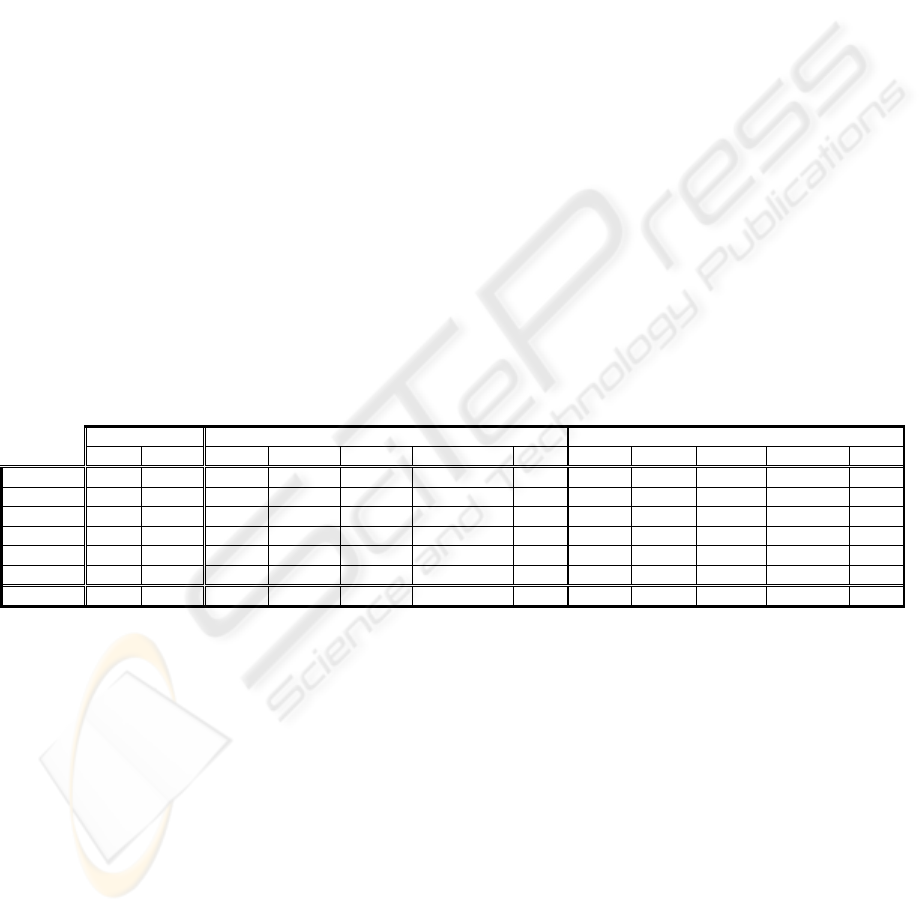
Table 2: Error and complexity comparison among C4.5 and CTC. The best values related to Number_Samples in each data
set are shown
C4.5 CTC (ERR
MIN
) CTC (COMP
MIN
)
Err Comp Err R.Dif Comp R.Dif N_S Err R.Dif Comp R.Dif N_S
Breast-W 5.63 3.16 5.40 -4.12 3.11 -1.41 125 5.49 -2.49 3.04 -3.52 30
Heart-C 23.96 15.11 22.85 -4.61 13.51 -10.59 20 22.92 -4.35 13.20 -12.65 30
Hypo 0.71 5.38 0.72 0.56 4.51 -16.12 20 0.72 0.84 4.40 -18.18 30
Lymph 20.44 8.84 19.65 -3.88 9.24 4.52 30 19.93 -2.51 9.09 2.76 40
Segment 13.61 11.71 11.52 -15.31 13.82 18.03 50 13.51 -0.75 12.64 7.97 10
Faithful 1.48 39.47 1.48 -0.13 32.44 -17.79 20 1.50 0.94 28.56 -27.65 05
Average
10.97 13.94 10.27 -4.58 12.77 -3.89 10.68 -1.38 11.82 -8.54
Table 2 shows the results related to error (Err) and
complexity (Comp) for the different data sets. The
comparison among C4.5 and CTC we present in the
table has been calculated using the best value of the
parameter Number_Samples (N_S) for CTC in order
to minimise the error or the complexity of the
classifier. The relative differences among the two
algorithms (R.Dif) are also presented, for both
parameters, the error and the complexity. The last
row shows the mean of the results obtained with the
two algorithms, for the different domains. It can be
observed that in average, the CTC algorithm obtains
a relative improvement in the error of 4.58%. In this
case, when talking about the complexity of the
generated trees, the relative improvement is 3.89%.
Even when the best Number_Samples for
minimising the complexity has been selected, the
CTC has smaller errors than the C4.5, being the
improvement 1.38%. The relative complexity
reduction obtained in this case is 8.54%. In the
larger database we have used (Faithful), where the
reduction of the complexity becomes more
important, the obtained improvement is 27.65%.
This reduction is statistically significant for every
value of the parameter Number_Samples.
To confirm the robustness of the algorithm when the
parameter Number_Samples is changed, Table 3 (left
side) shows the average values of the error and the
complexity obtained with all the analysed values for
that parameter.
ICEIS 2004 - ARTIFICIAL INTELLIGENCE AND DECISION SUPPORT SYSTEMS
18
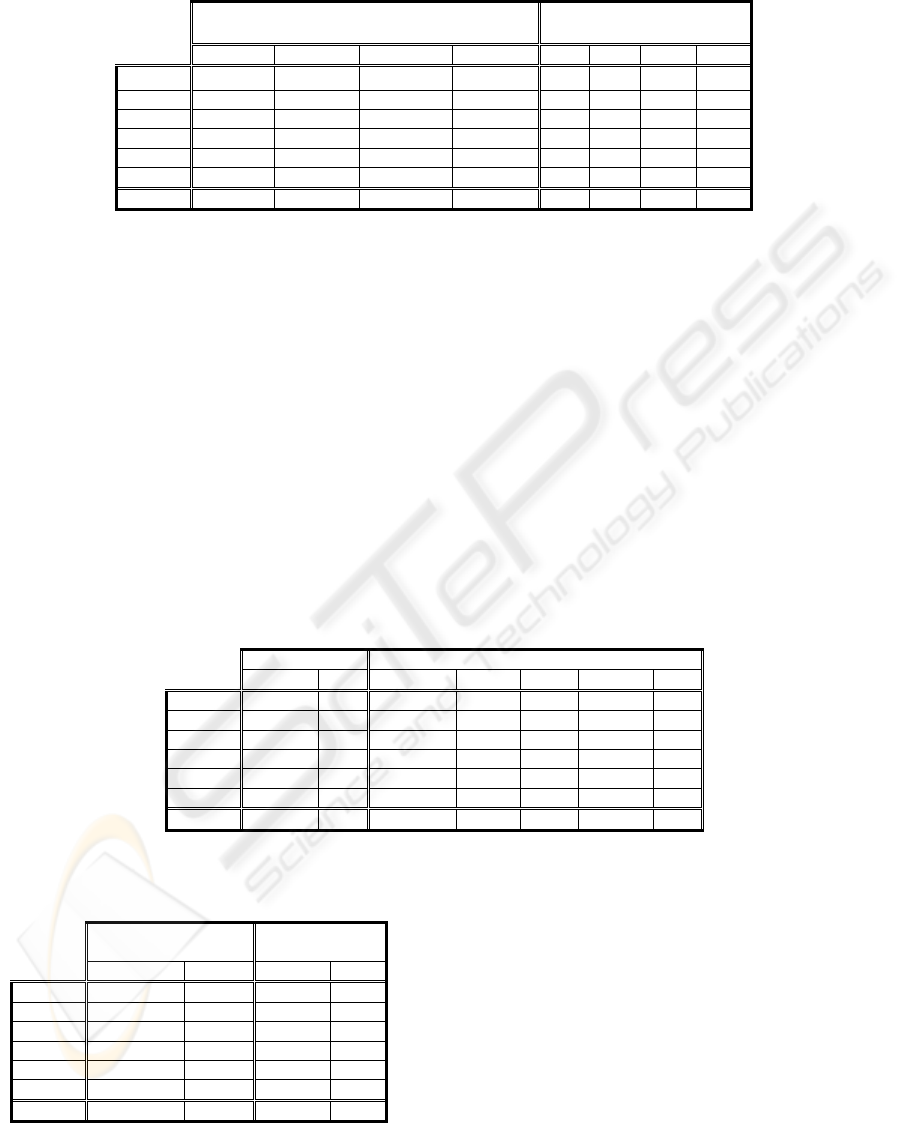
Table 3: Average results of the error and the complexity in CTC taking into account the whole range of N_S (left side) and
for N_S=30 (right side)
CTC
(Average N_S=10,20,30,40,50,75,100,125,150,200)
CTC
N_S=30
Err R.Dif Comp R.Dif Err R.Dif
Comp
Dif
Breast-W
5.56 -1.21 3.09 -2.11 5.49 -2.49 3.04 -3.52
Heart-C 23.43 -2.21 13.84 -8.41 22.92
-4.35 13.20 -12.65
Hypo 0.73 2.30 4.65 -13.60 0.72 0.84 4.40 -18.18
Lymph 20.03 -2.01 9.18 3.74 19.65
-3.88 9.24 4.52
Segment 12.42 -8.70 14.08 20.23 13.13
-3.48 13.47 14.99
Faithful 1.49 0.84 30.03 -23.92 1.49 0.67 29.53 -25.17
Average
10.61 -1.83 12.48 -4.01 10.57
-2.11 12.15 -6.67
It can be observed that a relative improvement of
1.83% in the error is maintained and the complexity
improves in a 4.01%. If we would like to tune the
parameter in order to optimise the error/complexity
trade off in the analysed domains, the results address
us to N_S=30. The results for N_S=30 appear in
Table 3 (right side). They do not differ substantially
from the results obtained finding in each domain the
best value for Number_Samples. The CTC maintains
its improvement if compared to C4.5 in both cases.
If we analyse the statistical significance of the
differences among the two algorithms, we won’t
find significant differences in the error parameter for
none of the values of Number_Samples. However,
when analysing the complexity significant
differences in favour of CTC are found in three of
the six databases (Heart-C, Hypo, Faithful).
Regarding to the structural stability of the trees
obtained with each algorithm, and taking into
account the metric presented in Section 3, Table 4
shows that except in one of the databases (Lymph),
the trees built using CTC algorithm, are more similar
among them than the ones generated with C4.5. This
means that the induction mechanism is able to
extract more information about the explanation of
the classification, and besides, in a more steady way.
The improvement is in average of 16.82%, and, the
differences are statistically significant in every case
where better results are obtained.
Table 4: Comparison of C4.5 and CTC related to the structural diversity measure, using the best value of Number_Samples
in each domain
C4.5 CTC (%COMM
MIN
)
%Comm
Err %Comm R.Dif Err R.Dif N_S
Breast-W
56.99 5.63 65.89 15.62 5.49 -2.49 30
Heart-C -59.01 23.96
-57.06 3.30 23.16 -3.32 150
Hypo 27.99 0.71 41.11 46.90 0.73 2.53 75
Lymph -27.72 20.44
-35.50 -28.08 20.18 -1.25 75
Segment -67.09 13.61
-36.19 46.06 12.76 -6.26 20
Faithful -57.95 1.48 -48.01 17.15 1.50 0.94 05
Average
-21.13 10.97
-11.63 16.82 10.64 -1.64
Table 5: Average results of the structural metric in the
CTC algorithm. The whole range of N_S is taken into
account in left side and N_S=30 in right side
CTC
(Average N_S=10..200)
CTC
N_S=30
%Comm R.Dif %Comm
R.Dif
Breast-W
60.51 6.19 65.89 15.62
Heart-C -61.10 -3.53 -58.57 0.75
Hypo 35.15 25.60 37.14 32.69
Lymph -36.71 -32.43 -36.76 -32.62
Segment -47.16 29.70 -43.33 35.41
Faithful -50.71 12.49 -50.94 12.09
Average
-16.67 6.33 -14.43 10.66
Table 5 shows in the left side the mean of the
differences in the structural metric of both
algorithms, for all the experimented values with
parameter Number_Samples. The average value
favours CTC, and, we should not forget that the
error also favours it. This proves again the stability
of the meta-algorithm in respect to the tuning of the
parameter Number_Samples.
The same table, right side, presents results of the
structural diversity among both algorithms, when
N_S=30. The results in this case are near the optimal
results we found and better than the average results
obtained (left side). As a consequence we can ensure
that the influence of the parameter Number_Samples
in the final result is not critical in any of the
analysed criteria.
CONSOLIDATED TREE CONSTRUCTION ALGORITHM: STRUCTURALLY STEADY TREES
19

6 CONCLUSIONS AND FURTHER
WORK
A new algorithm to build classification trees that are
structurally more steady and with smaller
complexity level has been presented (Consolidated
Trees Construction, CTC). This algorithm achieves
and even improves the discriminating capacity of
C4.5. The proposed algorithm maintains the
explanatory feature of the classification and this is
very important in many real life’s domains. The
algorithm builds trees that reduce the error rate and
the complexity of the classifier if compared to the
C4.5.
Besides, and to prove the goodness and the stability
of the explanation related to the classification, a
measure of the structural diversity of two trees is
proposed. This measure analyses the stability of the
variables and their statistical importance (level
where they appear in the tree). The measure allows
the analysis of the heterogeneity of a set of trees
from the structural or explanatory point of view.
In this paper we have proven that for the analysed
domains, the CTC is able to extract more
information about the explanation of the
classification, and, in a more steady way. The
differences are statistically significant in most of the
analysed domains.
On the other hand, the stability of the meta-
algorithm when varying the parameter
Number_Samples has been proved; N_S=30 is an
adequate value for all the databases used in the
experimentation.
The first work to do in the future is to enlarge the set
of domains analysed. We are also thinking on
experimenting with other possibilities for the
parameter Resampling_Mode (different amount of
information or variability of the subsamples) as
further work.
Other interesting possibility is to generate new
subsamples dynamically, during the building process
of the CTC, where the probability of selecting each
of the cases is modified based on the error (similar
to boosting).
We are also analysing a possibility where the own
meta-algorithm builds trees that do not need to be
pruned. With this aim, we would make a tuning of
the parameters Crit_Split and Crit_Variable, so that
the generated trees are situated in a better point of
the learning curve and the computation load of the
training is minimised. Heuristic techniques for the
stratification of the discrete variables can also be
studied in order to build trees with greater
explaining capacity.
This methodology can be very useful when
resampling is compulsory (large databases, class
imbalance, ...).
APENDIX
int CalculateSD (Tree Ti, Tree Tj,
int VMetric[])
{
if ((Ti->NodeType != LEAF) &&
(Tj->NodeType != LEAF))
if (Ti->Variable == Tj->Variable)
{VMetric[0]++;
if (Ti->Forks != Tj->Forks)
return (–1);
ForEach(k,1, Ti->Forks)
CalculateSD (Ti->Branch[k],
Tj->Branch[k],VMetric);
}
else //different division variables
{Vmetric[2]+=
CalcNumberDescendents(Ti);
Vmetric[2]+=
CalcNumberDescendents(Tj);
return (0);
}
else
if ((Ti->NodeType == LEAF) &&
(Tj->NodeType == LEAF))
return (0);
else
if (Ti->NodeType = LEAF)
{Vmetric[1]+=
CalcNumberDescendents(Tj);
return (0);
}
else
{Vmetric[1]+=
CalcNumberDescendents(Ti);
return (0);
}
return (–1);
}
ICEIS 2004 - ARTIFICIAL INTELLIGENCE AND DECISION SUPPORT SYSTEMS
20

ACKNOWLEDGEMENTS
The work described in this paper was partly done
under the University of Basque County / Euskal
Herriko Unibertsitatea (UPV/EHU) project: 1/UPV
00139.226-T-14882/2002.
We would like to thank the company Fagor
Electrodomesticos, S. COOP. for permitting us the
use, in this work, of their data (Faithful) obtained
through the project BETIKO.
The lymphography domain was obtained from the
University Medical Centre, Institute of Oncology,
Ljubljana, Yugoslavia. Thanks go to M. Zwitter and
M. Soklic for providing the data.
REFERENCES
Quinlan J. R., 1993. C4.5: Programs for Machine
Learning. Morgan Kaufmann Publishers Inc.(eds), San
Mateo, California.
Dietterich T.G., 2000. Ensemble Methods in Machine
Learning. Lecture Notes in Computer Science, Vol.
1857. Multiple Classifier Systems: Proc. 1st. Inter.
Workshop, MCS, Cagliari, Italy, 1-15
Chawla N.V., Hall L.O., Bowyer K.W., Moore Jr.,
Kegelmeyer W.P., 2002. Distributed Pasting of Small
Votes. Lecture Notes in Computer Science Vol. 2364.
Multiple Classifier Systems: Proc. 3th. Inter.
Workshop, MCS, Cagliari, Italy, 52-61
Breiman L., 1996. Bagging Predictors. Machine Learning,
24, 123-140
Freund, Y., Schapire, R. E., 1996. Experiments with a
New Boosting Algorithm. Proceedings of the 13th
International Conference on Machine Learning, 148-
156
Duin R.P.W, Tax D.M.J., 2000. Experiments with
Classifier Combining Rules. Lecture Notes in
Computer Science 1857. Multiple Classifier Systems:
Proc. 1st. Inter. Workshop, MCS, Cagliari, Italy, 16-
29
Bauer E., Kohavi R., 1999. An Empirical Comparison of
Voting Classification Algorithms: Bagging, Boosting,
and Variants. Machine Learning, 36, 105-139
Blake, C.L., Merz, C.J., 1998. UCI Repository of Machine
Learning Databases. University of California, Irvine,
Dept. of Information and Computer Sciences
http://www.ics.uci.edu/~mlearn/MLRepository.html
Hastie T., Tibshirani R. Friedman J., 2001. The Elements
of Statistical Learning. Springer-Verlang (es). ISBN:
0-387-95284-5
Dietterich T.G., 1998. Approximate Statistical Tests for
Comparing Supervised Classification Learning
Algorithms. Neural Computation, 10, 7, 1895-1924
Dietterich T.G., 2000. An Experimental Comparison of
Three Methods for Constructing Ensembles of
Decision Trees: Bagging, Boosting, and
Randomization. Machine Learning, 40, 139-157
Skurichina M., Kuncheva L.I., Duin R.P.W., 2002.
Bagging and Boosting for the Nearest Mean Classifier:
Effects of Sample Size on Diversity and Accuracy.
Lecture Notes in Computer Science 2364. Multiple
Classifier Systems: Proc. 3th. Inter. Workshop, MCS,
Cagliari, Italy, 62-71
CONSOLIDATED TREE CONSTRUCTION ALGORITHM: STRUCTURALLY STEADY TREES
21
