
Cell Image Segmentation by Feature Random Enhancement Module
Takamasa Ando
∗
and Kazuhiro Hotta
†
Meijo University, 1-501 Shiogamaguchi, Tempaku-ku, Nagoya 468-8502, Japan
Keywords:
Cell Image, Semantic Segmentation,U-Net, Feature Random Enhancement Module.
Abstract:
It is important to extract good features using an encoder to realize semantic segmentation with high accuracy.
Although loss function is optimized in training deep neural network, far layers from the layers for computing
loss function are difficult to train. Skip connection is effective for this problem but there are still far layers
from the loss function. In this paper, we propose the Feature Random Enhancement Module which enhances
the features randomly in only training. By emphasizing the features at far layers from loss function, we can
train those layers well and the accuracy was improved. In experiments, we evaluated the proposed module
on two kinds of cell image datasets, and our module improved the segmentation accuracy without increasing
computational cost in test phase.
1 INTRODUCTION
In recent years, the development of deep learning
technology has been remarkable, and there is a de-
mand to use it in various situations. Since the seg-
mentation of cell images obtained by microscopes is
performed manually by human experts, it tends to
be subjective results. Objective results are required
by the same criteria using deep learning technology
(Ciresan et al.,2012). However, the optimal network
for segmentation using deep learning has not been es-
tablished yet. Even if the accuracy is not so high, it
is actually used in the field of cell biology to obtain
objective results. Therefore, automatic segmentation
method with high accuracy is desired. U-Net is still
widely used for segmentation of microscope images
because it works well for small number of training
images and high accuracy is obtained without adjust-
ing hyperparameters. For this reason, many improve-
ments of U-Net have been proposed for microscope
images (Jiawei Zhang et al.,2018; Qiangguo Jin et
al.,2019; Qiangguo Jin et al.,2018).
This study belongs to one of those variations and
improves the accuracy of U-Net. Although conven-
tional improvement is done by deepening, the pro-
posed method does not require any additional com-
putational resources at all during inference. There-
fore, it retains the advantage of U-Net that it requires
fewer computing resources. Therefore, it is a verysig-
nificant proposal in the segmentation of cell images
where there is a demand for lightweight and accurate
model.
A neural network such as CNN basically uses
backpropagation for training. For this reason, there
is a phenomenon that near layers to the layer for com-
puting loss are more updated in comparison with far
layers from the loss (Y. Bengio et al.,1994). In or-
der to solve the problem, ResNet (Kaiming He et
al.,2016) used skip connection and improved the ac-
curacy. U-Net (Philipp Fischer et al.,2015) is the fa-
mous deep neural network for cell segmentation task.
U-Net also has skip connection between encoder and
decoder, and it contributes to improve the accuracy.
In general, it is well known that skip connection gives
the information of location and fine objects which
were lost in encoder to decoder. However, we con-
sider that the same theory as ResNet is used in skip
connection to improve the accuracy. By using skip
connection, the loss is propagated to encoder well,
and weights are successfully updated. This is also the
reason why U-net improved the accuracy in compari-
son with standard Encoder-Decoder CNN.
Figure 1 shows the structure of U-net. We see that
skip connection is effective to propagate the loss to
encoder. However, the layers shown as yellow in the
Figure 1 are the farthest from the loss at the final layer.
Therefore, in the case of U-net, the yellow layer in the
Figure is the most difficult to train though the layer
has semantic information. In this paper, we propose
new module to train the layer effectively. We consider
to enhance the feature map at yellow layer which is
the farthest layer from the loss function. In conven-
tional methods, the yellow layer is difficult to learn,
and network learns to decrease the feature values at
520
Ando, T. and Hotta, K.
Cell Image Segmentation by Feature Random Enhancement Module.
DOI: 10.5220/0010326205200527
In Proceedings of the 16th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2021) - Volume 4: VISAPP, pages
520-527
ISBN: 978-989-758-488-6
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

Figure 1: U-Net and the problem.
the yellow layer not to affect the output. This phe-
nomenon is shown in section 3.1. Feature values at
the yellow layer are smaller than those at skip connec-
tion from encoder, and features at the yellow layer are
not effective for segmentation result. Therefore, the
model without yellow layer sometimes has higher ac-
curacy than the model with yellow layer as shown in
section 4.3. To train yellow layers effectively, we se-
lect some feature maps randomly at yellow layer and
increase the absolute value of the feature maps multi-
plied by a large constant value. Although the features
at yellowlayer and features from encoder are concate-
nated, the features at yellow layer are used mainly be-
cause the yellow layer has larger values by constant
multiplication. If the filters selected by our module
are not effective for classification, network has a large
loss. Therefore, the selected feature maps are trained
well by gradient descent.
In experiments, we evaluated our method on two
kinds of cell image datasets. Intersection over Union
(IoU) is used as an evaluation measure. The effective-
ness of the proposed module was shown in compari-
son with the conventional U-Net without our module
and U-net with SuperVision that loss function is com-
puted at yellow layer.
The structure of this paper is as follows. Section
2 describes related works. Section 3 describes the de-
tails of the proposed method. Experimental result on
two kinds of cell image datasets are shown in section
4. Finally, we summarize our work and discusses fu-
ture works in section 5.
2 RELATED WORKS
U-Net is a kind of Encoder-Decoder CNN (Heng-
shuang Zhaon et al.,2017). Unlike the PSPNet (Heng-
shuang Zhao et al.,2017), the Encoder-Decoder CNN
does not use features in parallel but features are ex-
tracted in series. Thus, in Encoder-Decoder CNN, far
layers from the layer for computing loss are not up-
dated well. ResNet and U-net solved this problem by
skip connection.
There is also a technique called Deep supervision
proposed in Deeply-Supervised Nets (Chen-Yu Lee et
al.,2015) to address the problem. In deep supervision,
loss is also computed at middle layer. Far layers from
final layer are updated well by supervision. U-Net++
(Zongwei Zhou et al.,2018) also used this technique.
However, forcing loss from the ground truth in the
middle of U-Net may not obtain an intermediate rep-
resentation for better inference. In addition, U-Net++
has a structure in which the output image is restored
by the decoder from various parts of the encoder, and
multiple decoders are connected to each other. How-
ever, the advantage of U-Net which is a small com-
putational resource is vanished. This is accompanied
by a large number of parameters due to multiple de-
coders. In this paper, we propose new methods based
on the merits and demerits of these previous studies.
There are some methods that we referred to con-
sider a new method. In the proposed method, feature
enhancement is performed on some feature maps dur-
ing training. There are many techniques for weight-
ing feature maps. SENet (Jie Hu et al.,2018) pro-
posed to weight important channels. Attention which
has been proposed in the field of natural languages
(Ashish Vaswani et al.,2017) is also used in the field
of images. In recent years, many methods have
been proposed that focus on channels (Yulun Zhang
et al.,2018; Sanghyun Woo et al.,2018; Yanting Hu
et al.,2018). Attention-U-net used attention for skip
connection (Ozan Oktay et al.,2018).
Dropout (Nitish Srivastava et al.,2014) is also re-
lated to our approach. Dropout sets a part of the fea-
ture map to 0 in only training. This prevents overfit-
ting by randomly removing elements in only training.
Our method randomly enhances some feature maps at
the farthest layer from loss function. We do not set
some elements to 0 and enlarge some feature maps.
When some elements are set to 0, backpropagation
from the element is stopped. In our method, features
Cell Image Segmentation by Feature Random Enhancement Module
521
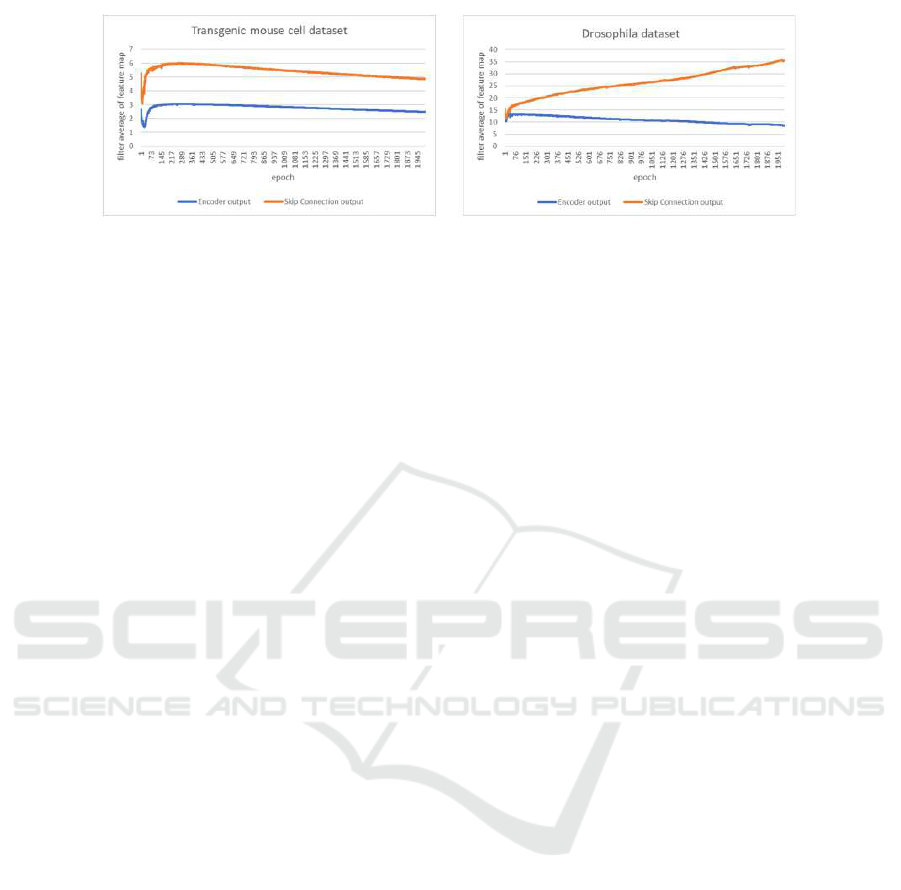
Figure 2: Comparison of feature values at the yellow layer and skip connection.
are enlarged randomly to use backpropagation effec-
tively for the farthest layer.
3 PROPOSED METHOD
This section describes the proposed method. Section
3.1 gives the overview of the proposed method. Sec-
tion 3.2 mentions the implementation details of our
method.
3.1 Overview of the Proposed Method
When we obtain segmentation result by the U-Net,
the magnitude of features at yellow layer as shown in
Fig. 1 is often smaller than that of features at skip
connection from encoder to decoder. Fig.2 shows the
fact when U-net is trained on two different datasets.
Two lines in each graph show the average feature val-
ues at yellow layer in Fig.1 and those at skip connec-
tion from encoder to decoder. Note that both features
are extracted after ReLU function and those two fea-
ture maps have positive values. The magnitude of
these values in Fig.2 means the magnitude of influ-
ence for the network’s outputs because convolution is
adopted after both features in Fig.2 are concatenated.
The Figure shows that the encoder’s output features
are obviously smaller than the features at skip con-
nection, and the features at yellow layer are not used
effectively. Batch renormalization is used before we
compute ReLU function in Fig 2. If we do not use
backpropagation, both feature maps have similar av-
erage values because the features are normalized by
batch renormalization. Therefore, the training of U-
net makes the features at yellow layer small.
Does this fact show that yellow layer is not re-
quired? Our answer is “NO”. This phenomenon is
caused from that near layers to the layers for com-
puting loss are updated well and far layers are not
updated well. Yellow layer in Fig.1 is the farthest
layer from loss function because encoder is updated
through skip connection. Therefore, network learns
the layers connected by a skip connection in compar-
ison with the yellow layer because it is difficult to up-
date the yellow layer.
Although the easy solution for this problem is to
use new normalization for yellow layers using aver-
age and variance of skip connection, it prevents the
yellow layer’s values from being small. However, it
generates new problem that can not learn appropri-
ate ratio of yellow layer and skip connection values.
Therefore, we propose feature enhancement module
to solve the problem. Our method is soft constraint
as emphasis some feature maps in comparison with
normalization. The soft constraint means emphasiz-
ing ”a part of feature maps” at yellow layer. The new
normalization described above increases the values in
”all feature maps” at yellow layer to match the skip
connection. Specifically, we select some feature maps
at yellow layer randomly at each epoch and increase
the absolute values of the features by multiplying a
large constant value. This allows to use the features
at yellow layer effectively. If the features enlarged by
our module are not effective, network has a large loss.
Therefore, the selected filters are trained well by gra-
dient descent. The reason why we select some feature
maps randomly is to prevent vanishing gradient.
Our method is not used in test phase. This is be-
cause it able to learn the appropriate magnitude of
values in non-selected feature maps. The evidence
for this is from experimental results, and it is shown
in section 4.4. Since our method is soft constraint, it
solve the problem that the normalization at the con-
catenation of two feature maps can not solve. Thus,
the accuracy is improved without changing the in-
ference time or computational resources because our
method is not used in test phase. This is an advan-
tage of our method though many methods deepened
the network to improve the accuracy.
3.2 Implementation Details
To describe the proposed method for U-Net, the en-
coder’s output is enhanced by multiplying feature
maps selected randomly at each epoch by X. The
VISAPP 2021 - 16th International Conference on Computer Vision Theory and Applications
522
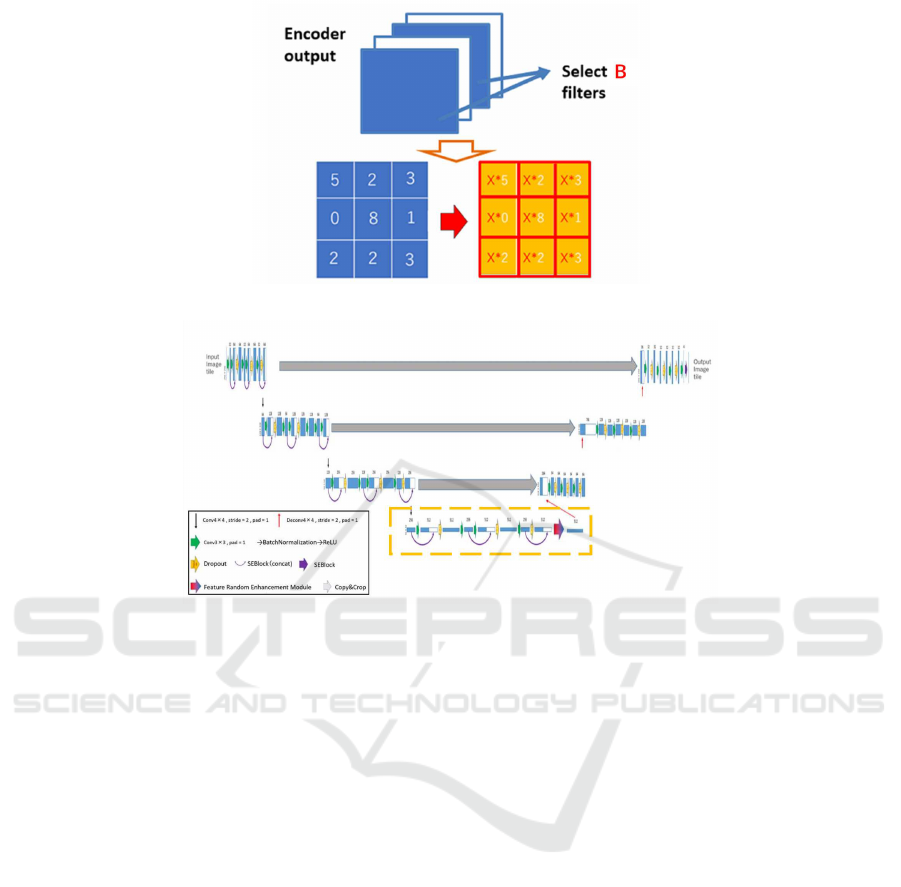
Figure 3: Feature Random Enhancement Module.
Figure 4: U-Net with SE block.
number of feature maps selected randomly is denoted
as B. The feature maps are re-selected each epoch and
the network weight is updated during training.
The closest approaches is Dropout. Similarly,
dropout is used only during training, and some neu-
rons are randomly set to 0. If there is an element set
to 0, the backpropagation stops at that element. It is a
method to allow the ensembles. The proposed method
differs from Dropout. We use an adjustable feature
emphasis not setting to 0. This is to improve the case
where there is a difference in the ease of updating be-
tween the near and far layers.
The proposed method can be implemented in ad-
dition to Dropout. However, this does not mean that
Dropout will be replaced by the proposed method.
Our method is difficult to implement in many layers
because we determine adequate parameters X and B
at each layer. Implementing it at the farthest layer
from the loss function solves the problem presented
in this paper and is the most effective.
Figure 3 shows the detailed description of the pro-
posed method. In the proposed method, we multiply
X by all values in the selected feature maps which
are the end of encoder shown as yellow layer in Fig-
ure 1. This operation is performed only in training
phase. The enhanced feature maps are selected ran-
domly. Thus, all channels in encoder’s output are
not enhanced. We need to select hyperparameters X
and B appropriately. Hyperparameters were searched
by using the optimization with Tree-structured Parzen
Estimator (TPE) (J. Bergstra et al.,2011) which uses
Bayesian optimization.
4 EXPERIMENTS
This section shows the experimental results of the
proposed method. Section 4.1 and 4.2 describe the
dataset and the network used in experiments. Exper-
imental results are shown in section 4.3. In section
4.4, additional experiments are conducted for consid-
erations.
4.1 Dataset
In this paper, we conduct experiments on two kinds of
cell image datasets. The first dataset includes only 50
fluorescent images of the liver of a transgenic mouse
expressing a fluorescent marker on the cell membrane
and nucleus (A. Imanishi et al.,2018). The size of the
image is 256 times 256 pixels and consists of three
classes; cell membrane, cell nucleus, and background.
Cell Image Segmentation by Feature Random Enhancement Module
523
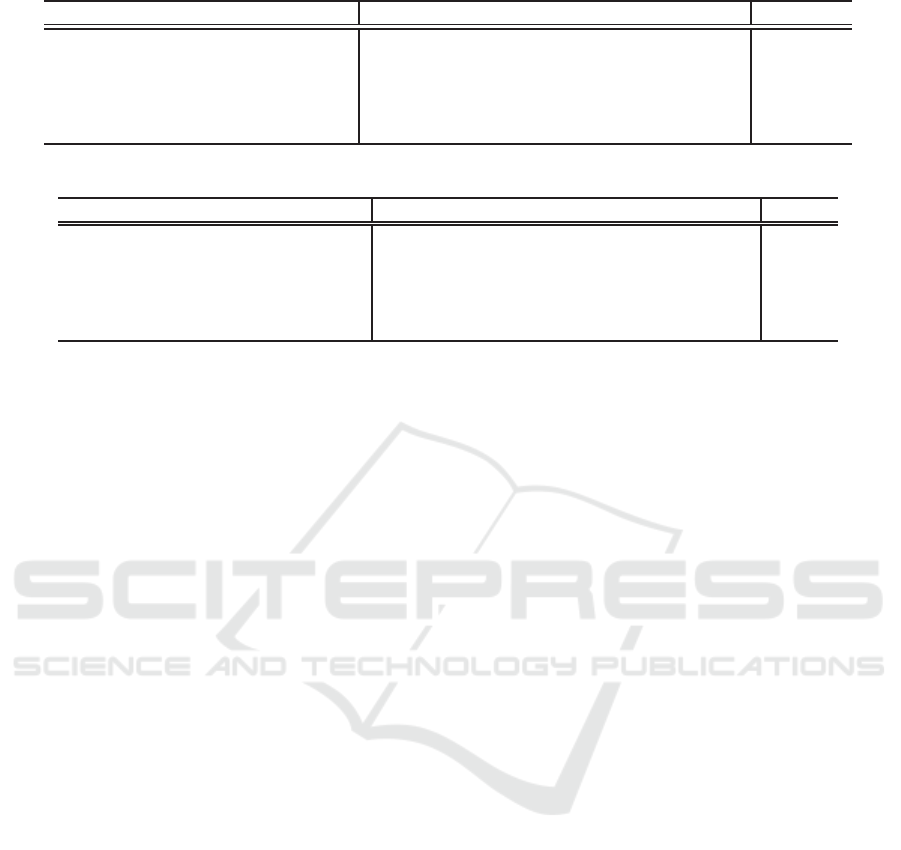
Table 1: IoU of Transgenic mouse cell dataset.
menbrane[%] nuclear[%] background[%] mIoU[%]
U-Net + SEblock 37.78 65.75 74.96 59.50
U-Net + SEblock without deep layers 39.15 66.84 75.11 60.36
U-Net + SEblock + Supervision
39.23 64.99 73.34 59.19
Dropout (Same percentage as the proposed method) 36.44 65.50 75.09 59.01
Proposed method
40.53 67.58 76.75 61.62
Table 2: IoU of the Drosophila dataset.
menbrane[%] nuclear[%] background[%] Synapus[%] mIoU[%]
U-Net + SEblock 91.80 76.87 76.89 50.46 73.98
U-Net + SEblock without deep layers
92.32 78.24 76.93 51.31 74.70
U-Net + SEblock + Supervision 92.39 77.77 78.24 52.21 75.15
Dropout (Same percentage as the proposed method)
92.40 77.98 78.70 50.23 74.83
Proposed method 92.93 78.71 78.02 58.14 76.95
We use 35 images for training, 5 images for valida-
tion, and 10 images for test.
The second dataset includes 20 Drosophila feather
images (Stephan Gerhard et al.,2013). The size of
the image is 1024 × 1024 pixels and consists of four
classes; cell membrane, mitochondria, synapse, and
background. Training and inference were performed
by cropping 16 images of 256 × 256 pixels from one
image without overlap due to GPU memory size. In-
tersection over Union (IoU) and Mean IoU were used
as evaluation measure for both datasets.
4.2 Network
The proposed method introduces a module that ran-
domly enhances the features at the final layer of en-
coder during only training. We call it “Feature Ran-
dom Enhancement Module”. Fig. 4 shows the U-
Net used in this paper. As shown in Fig. 4, the pro-
posed module was implemented on a standard U-Net
with SE block. Some feature maps are selected from
512 feature maps at the farthest layer from the loss
function which is shown as the bottom right in Fig. 4
at training phase, and the value in the feature map is
multiplied by X.
4.3 Results
In all experiments, we trained all methods till 2000
epochs in which the learning converges sufficiently,
and evaluation is done when the highest mIoU ac-
curacy is obtained for validation images. We used
softmax cross entropy. The hyperparameters B and
X were searched 50 times using the Tree-structured
Parzen Estimator algorithm (TPE) which seem to be
a sufficient number.
For comparison, U-Net with only SE block is eval-
uated. This is the baseline. U-Net with SE block with-
out yellow layer in Fig.1 (the yellow dotted square in
Fig. 4) is evaluated to present the problem that deep
layers are difficult to train. The problem is that the
model without those deep layers achieved higher ac-
curacy than that with those deep layers.
We also evaluate the U-net with SE block which
uses SuperVision instead of the proposed module in
order to show the effectiveness of Feature Enhance-
ment module. SuperVision uses 1x1 convolution
to change the number of channel to the number of
classes at the end of encoder, and resize it to the size
of input image and softmax cross entropy loss is com-
puted. When we use SuperVision, we must optimize
two losses; the first loss is standard softmax cross en-
tropy loss at the final layer and the second loss is for
supervision. In general, the balancing weight for two
losses should be optimized.
Loss=(1- λ)*Loss.1+λ*Loss.2 (1)
where is λ the balancing weight. The parameter is
also optimized by TPE. The search was performed
15 times to find the appropriate parameter. Since
λ is a single parameter, the number of searches is
smaller than that of the two parameters B and X in
our method.
First, we show the experimental results on the
mouse cell dataset. Table 1 shows the results when
we use B = 162 and X = 632 which gives the highest
mean IoU for validation set. Table 1 shows that the
accuracy of our method is improved in all classes in
comparison with the conventional models. We con-
firmed 2.12% improvement from the baseline. In ad-
dition, the accuracy of the proposed method is better
than U-Net + SENet without deep layers. This result
shows our method can train deep layers effectively. In
addition, the accuracy of U-Net + SENet without deep
VISAPP 2021 - 16th International Conference on Computer Vision Theory and Applications
524
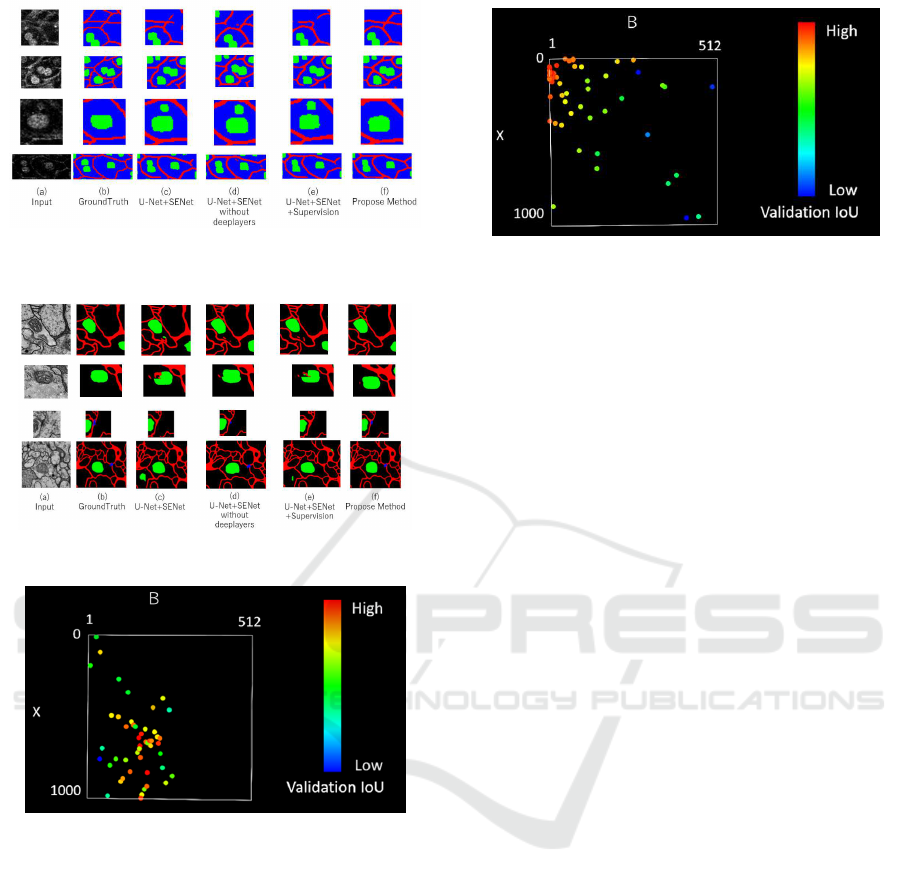
Figure 5: Segmentation results on transgenic mouse cell im-
ages.
Figure 6: Segmentation results on the Drosophila dataset.
Figure 7: TPE algorithm of Transgenic mouse cell.
layers is better than U-Net + SENet. It shows that the
deep layers of the U-Net + SENet used in the experi-
ments is far from the loss and not updated well. The
accuracy of mean IoU is not improved by U-Net with
SuperVision (λ = 0.3257 determined by TPE) even if
loss is computed at the end of encoder that our mod-
ule is used. When we use dropout with the same per-
centage as the proposed method (Dropout rate = B/the
number off filters = 162/512), it did not improve the
accuracy.
Figure 5 shows the qualitative results. In Fig. 5,
(a) is input image, (b) is ground truth, (c),(d) and
(e) are the results by the conventional U-Net with SE
block, U-Net with SE block without deep layers and
U-Net with SE block + SuperVision, respectively, and
(f) is the result by the proposed method. We see that
cell image is blurred and it is difficult for not experts
Figure 8: TPE algorithm of Drosophila feather.
to assign class labels. This is because cells are killed
by strong light and images are captured with low illu-
minace.
In conventional method (d), there are many non-
and over-detected cell membrane or nucleus. In addi-
tion, in conventionalmethod (e), there are many unde-
tected membranes. However, in the proposed method
(f), more accurate segmentation results are obtained.
This is because the proposed module enables to ex-
tract features from areas where training has not been
done successfully in conventional methods. In addi-
tion, the method using SuperVision gave lower accu-
racy than the proposed method. We consider that the
loss from the middle layer does not always give an
intermediate representation for good segmentation re-
sult.
Next, we show the experimental results on the
Drosophila dataset. Table 2 shows the results when
we use B = 8 and X = 250 when the highest mIoU is
obtained for validation set. Table 2 shows that the ac-
curacy of the proposed method is higher than that of
the U-Net with SE block, and the mean IoU was im-
proved 2.97% by baseline. Furthermore, the accuracy
was improved in comparison with the conventional
models. In addition, the accuracy of U-Net + SENet
without deep layers is better than U-Net + SENet.
This result is the same as mouse cell data set. It rein-
forces the theory that the deep layers of the U-Net +
SENet used in the experiments are not updated well.
The results by U-Net with SuperVision (λ = 0.2781
determined by TPE) and Dropout with the same per-
centage as the proposed method (Dropout rate = B/the
number of filters = 8/512) reinforce the theory.
Figure 6 shows qualitative results. In Fig. 6, (a) is
the input image, (b) is ground truth, (c),(d) and (e)
are the results by the U-Net with SE block, U-Net
with SE block without deep layers and U-Net with
SE block and SuperVision, and (f) is the results by
the proposed method. In the Drosophila dataset, the
image seems to contain enough information but it is
difficult for ordinary people to assign correct class la-
bels to each pixel. However, we confirmed that the
Cell Image Segmentation by Feature Random Enhancement Module
525
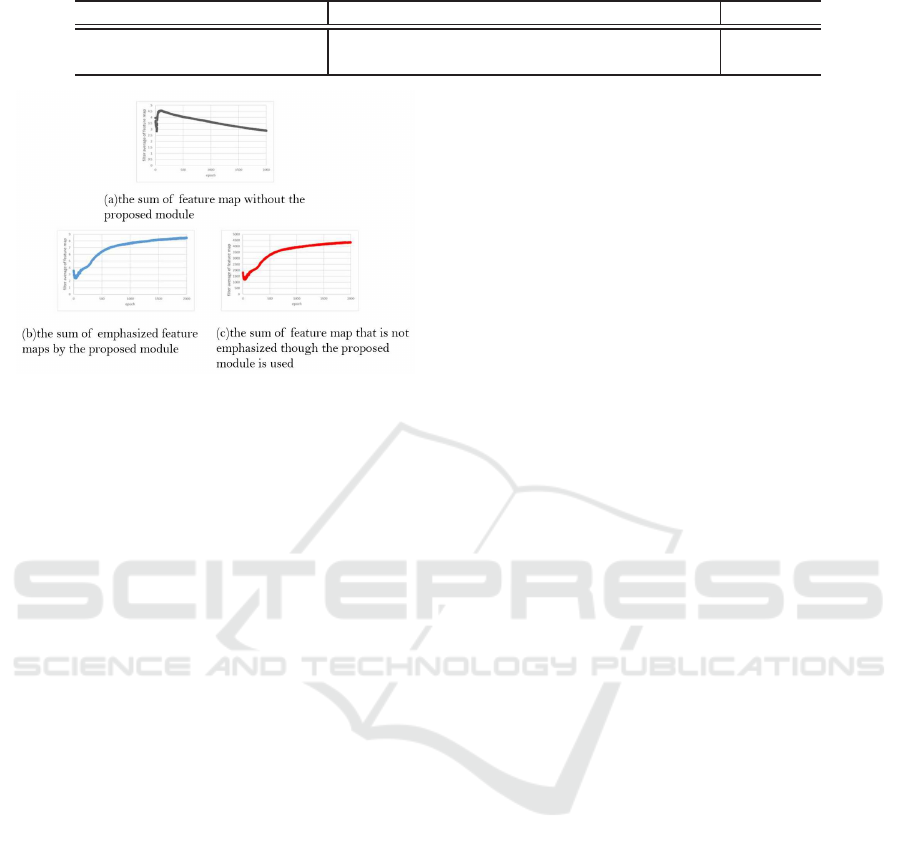
Table 3: For additional experiment,IoU of Transgenic mouse cell dataset.
menbrane[%] nuclear[%] background[%] mIoU[%]
U-Net + SEblock 37.78 65.75 74.96 59.50
Proposed method(additional experiment) 40.43 67.34 73.71 60.49
Figure 9: Sum of feature map with/without Feature En-
hancement module.
proposed method (f) performs better segmentation for
cell membrane and nucleus.
Figure 7 and 8 show the results of hyperparame-
ter search using the TPE algorithm. The vertical and
horizontal axes show the hyperparameters B and X
in the proposed module. Red points show high mean
IoU for validation set, and the blue points show low
accuracy. We see that the TPE algorithm focuses on
searching for places with high accuracy. Of course,
optimal B and X depend on the dataset. However, we
can find good hyperparameters by TPE.
4.4 Additional Experiments
The proposed method emphasizes some feature maps
randomly at each epoch to prevent over-fitting. How-
ever, when we apply 10,000 times enhancement to the
fixed ten filters during training, IoU accuracy was im-
proved by about 1% as shown in Table 3. This shows
that the effect can be seen even if the emphasis is not
performed randomly. Thus, we observed that the sum
of the values in the feature map that ReLU function is
adopted after convolution because we want to confirm
the behavior of the enhanced and unenhanced filters.
Figure 9 (a) shows the sum of feature map with-
out the proposed module, (b) shows the sum of em-
phasized feature map by the proposed module. In (c),
we used the proposed module but we computed the
sum of non-emphasized feature map for the compar-
ison with (b). In (a), the sum of feature map gradu-
ally decreases and the feature maps at the end of en-
coder are not used effectively. On the other hand, in
(b) and (c), the sum of feature map increased through
training. This means that the feature maps at the end
of encoder have large value automatically and those
features are used to obtain segmentation results. The
proposed method emphasizes some feature maps ran-
domly at each epoch to prevent over-fitting. There-
fore, from the change of the value of (c) compared
with (a), we see that the proposed module also has an
effect on the feature maps that are not emphasized.
5 CONCLUSION
In this paper, we introduced the Feature Random En-
hancement Module which is enhanced feature map
randomly during only training. We succeeded in im-
proving the accuracy on cell image segmentation. We
could propose the method for improving accuracy
though the amount of computation during inference
does not change.
A future work is to establish a method for deriv-
ing the parameters of the proposed module. Although
TPE seems to be effective for parameter search from
the results, it requires training for each parameter until
the accuracy converges. Therefore, the computational
cost for inference is fast but training takes longer time.
Thus, we would like to study whether those parame-
ters can be determined faster without convergence.
ACKNOWLEDGEMENT
This work is partially supported by MEXT/JSPS
KAKENHI Grant Number 18K111382 and
20H05427.
REFERENCES
Ciresan, D.C., Gambardella, L.M., Giusti, A., Schmid-
huber, J.: Deep neural networks segment neu-
ronal membranes in electron microscopy images. Ad-
vances in Neural Information Processing Systems.
pp.2852–2860 2012.
Jiawei Zhang, Yuzhen Jin, Jilan Xu, Xiaowei Xu, Yanchun
Zhang. MDU-Net: Multi-scale Densely Connected
U-Net for biomedical image segmentation. arXiv
preprint arXiv:1812.00352 .2018.
Qiangguo Jin, Zhaopeng Meng, Tuan D. Pham, Qi Chen,
Leyi Wei, Ran Su. DUNet: A deformable network for
retinal vessel segmentation. Knowledge-Based Sys-
tems 178 149-162. 2019.
VISAPP 2021 - 16th International Conference on Computer Vision Theory and Applications
526

Qiangguo Jin, Zhaopeng Meng, Changming Sun, Leyi Wei,
Ran Su. RA-UNet: A hybrid deep attention-aware net-
work to extract liver and tumor in CT scans. arXiv
preprint arXiv:1811.01328. 2018.
Y. Bengio, P. Simard, and P. Frasconi. Learning long-
term dependencies with gradient descent is difficult.
IEEE Transactions on Neural Networks, Vol.5(2),
pp.157–166.1994.
Kaiming He, Xiangyu Zhang, Shaoqing Ren, Jian Sun.
Deep residual learning for image recognition. Con-
ference on Computer Vision and Pattern Recognition.
2016.
Philipp Fischer, and Thomas Brox. U-Net: Convolutional
Networks for Biomedical Image Segmentation. Med-
ical Image Computing and Computer-Assisted Inter-
vention, Springer, LNCS, Vol.9351: 234-241, 2015.
Hengshuang Zhao, Jianping Shi, Xiaojuan Qi, Xiaogang
Wang, Jiaya Jia. Pyramid scene parsing network. Con-
ference on Computer Vision and Pattern Recognition
2017.
Hengshuang Zhao, Jianping Shi, Xiaojuan Qi, Xiaogang
Wang, Jiaya Jia. Pyramid scene parsing network. Con-
ference on Computer Vision and Pattern Recognition.
2017.
Chen-Yu Lee, Saining Xie, Patrick Gallagher, Zhengyou
Zhang, Zhuowen Tu. Deeply-supervised nets. Artifi-
cial intelligence and statistics. 2015.
Zongwei Zhou, Md Mahfuzur Rahman Siddiquee, Nima
Tajbakhsh, Jianming Liang. ”Unet++: A nested u-net
architecture for medical image segmentation.” Deep
Learning in Medical Image Analysis and Multimodal
Learning for Clinical Decision Support. Springer,
pp.3-11,2018.
Jie Hu, Li Shen, and Gang Sun. Squeeze-and-excitation net-
works. Conference on Computer Vision and Pattern
Recognition. 2018.
Ashish Vaswani, Noam Shazeer, NikiParmar, Jakob Uszko-
reit, Llion Jones, Aidan N. Gomez, Lukasz Kaiser, Il-
lia Polosukhin. Attention Is All You Need. Advances
in Neural Information Processing Systems. 2017.
Yulun Zhang, Kunpeng Li, Kai Li, Lichen Wang, Bineng
Zhong, Yun Fu. Image super-resolution using very
deep residual channel attention networks. In: Proceed-
ings of the European Conference on Computer Vision.
pp. 286-301, 2018.
Sanghyun Woo, Jongchan Park, Joon-Young Lee, In So
Kweon. Cbam: Convolutional block attention mod-
ule. In: Proceedings of the European Conference on
Computer Vision. pp. 3-19, 2018.
Yanting Hu, Jie Li, Yuanfei Huang, Xinbo Gao. Channel-
wise and Spatial Feature Modulation Network for
Single Image Super-Resolution. arXiv:1809.11130,
2018.
Ozan Oktay, Jo Schlemper, Loic Le Folgoc, Matthew
Lee, Mattias Heinrich, Kazunari Misawa, Kensaku
Mori, Steven McDonagh, Nils Y Hammerla, Bern-
hard Kainz, Ben Glocker, Daniel Rueckert. Attention
U-Net: learning where to look for the pancreas. Inter-
national Conference on Medical Imaging with Deep
Learning . 2018.
Nitish Srivastava, Geoffrey Hinton, Alex Krizhevsky, Ilya
Sutskever, Ruslan Salakhutdinov. Dropout: a simple
way to prevent neural networks from overfitting. The
Journal of Machine Learning Research, Vol.15, No.1,
pp.1929-1958, 2014.
J. Bergstra, R. Bardenet, Yoshua Bengio, B. & K´egl. Algo-
rithms for hyper-parameter optimization. Advances in
neural information processing systems. 2011.
A. Imanishi, T. Murata, M. Sato, K. Hotta, I. Imayoshi,
M. Matsuda, and K. Terai, “A Novel Morphological
Marker for the Analysis of Molecular Activities at
the Single-cell Level,” Cell Structure and Function,
Vol.43, No.2, pp.129-140, 2018.
Stephan Gerhard, Jan Funke, Julien Martel, AlbertCardona,
Richard Fetter. Segmented anisotropic ssTEM dataset
of neural tissue. figshare. Retrieved 16:09, (GMT)
Nov 20, 2013.
Cell Image Segmentation by Feature Random Enhancement Module
527
