
Segmentation of Agricultural Images using Vegetation Indices
Jean Fabr
´
ıcio Batista Santos, Jocival Dantas Dias Junior, Andr
´
e Ricardo Backes
and Maur
´
ıcio Cunha Escarpinati
School of Computer Science, Federal University of Uberl
ˆ
andia, Brazil
Keywords:
Precision Agriculture, Plant Segmentation, Vegetation Indices.
Abstract:
Identifying and segmenting plants from the background in agricultural images is of great importance for pre-
cision agriculture. It serves as a basis for several tasks such as identification of planting lines, identification
of weed plants, agricultural automation, among others. Given this importance, in this paper, we evaluated
the application of five vegetation indices for RGB images together with two binarization techniques for the
plant/background segmentation process. The results showed promising performance in all evaluated indices.
It was also possible to identify a relationship between the performance obtained in each index and the capture
conditions in each dataset.
1 INTRODUCTION
The diversity of land areas, types of soils and plants,
products, and machinery makes the management of
a simple planting area a complex task. To facilitate
this task, in the last few decades Precision Agricul-
ture (PA) has emerged as a new way to manage agri-
cultural resources. Literature defines PA as the use of
different technologies (such as artificial intelligence,
internet of things, data analysis, and image process-
ing) with the main objective of optimizing results, re-
ducing costs, and creating a more sustainable produc-
tion chain.
A simple example of PA applied to a plantation
is the use of an Unmanned Aerial Vehicle (UAV) to
acquire images of the plantation. Depending on the
sensors present in the UAV, it is possible to obtain a
map of the area with many types of information, such
as land topology and vegetation distribution. There is
a wide range of sensors that can be used with a UAV.
The most common are cameras that capture color pat-
terns in RGB format. However, other bands, such as
infrared and ultraviolet, can be also used. RGB cam-
eras are widely used in image processing because they
are the standard that most devices use, such as smart-
phones, allowing image processing algorithms to run
even in mobile applications (Riehle et al., 2020). By
using specific algorithms, it is possible to extract from
these maps high-level information from the area, such
as the location of crop lines, plant count, and the pres-
ence of sowing failures. These informations enable
the farm to improve the use of the resources and al-
lows the use of other operations such as robots, trac-
tors, and autonomous pruning (Bargoti and Under-
wood, 2016).
Identifying and separating plants from the soil is
an important task because it allows the monitoring of
plant growth, health, and the identification of pests
and weeds. Mathematical equations applied to the
RGB channels of the images result in different in-
dices that highlight certain wavelengths, such as the
green levels of the image, thus facilitating the sep-
aration of the plant from background pixels (Riehle
et al., 2020). However, an index that highlights green
pixels is not enough to separate weed crops, and other
types of vegetation index may be needed for this prob-
lem. The use of vegetation indices has some advan-
tages such as low computational cost in comparison
with other segmentation techniques or machine learn-
ing approaches, easy implementation, and handling.
Nevertheless, they are sensitive to brightness varia-
tion, presence of shadows, and manual definition of
the threshold (Riehle et al., 2020).
In literature, there are a wide variety of RGB in-
dices that can be used for image segmentation pur-
poses. In the present work, we aimed to evaluate some
of these vegetation indices in a plant/soil segmenta-
tion task when combined with a clustering algorithm
and a simple threshold method to obtain the differ-
ent regions of interest. We compared the performance
of each vegetation index in the dataset presented in
(Riehle et al., 2020), which presents images with pre-
defined masks of the plant/soil regions.
The remaining of the paper is structured as fol-
506
Santos, J., Dias Junior, J., Backes, A. and Escarpinati, M.
Segmentation of Agricultural Images using Vegetation Indices.
DOI: 10.5220/0010325005060511
In Proceedings of the 16th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2021) - Volume 4: VISAPP, pages
506-511
ISBN: 978-989-758-488-6
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

lows: Section 2 presents a review of recent paper pub-
lished on this topic and that motivated our research.
In Section 3 we describe the vegetation indices used
in this work as also the datasets used for evaluation.
Section 4 describes how experiments were performed
while Section 5 presents the results obtained by each
vegetation index. Finally, Section 6 concludes the pa-
per.
2 RELATED WORK
Computing a vegetation index is a common pre-
processing technique used for plant/soil segmentation
problems. It increases the contrast between vegeta-
tion and soil, generating a grayscale image that high-
lights particular regions of interest. For example, the
Excess Green Index (ExG) and the Excess Red Index
(ExR) aim to increase the contract of the Green and
Red levels, respectively, highlighting the plant from
other elements, such as soil and waste. And when
combined, these indices can generate even more effi-
cient results in segmentation, such as ExGR (Riehle
et al., 2020), which is calculated as follows:
ExG = 2G − R − B (1)
ExR = 1.4R − B (2)
ExGR = ExG − ExR (3)
ExGR index results in an image where pixels
range from positive (plant) to negative (soil and
residues) values so that the image is easily segmented
using a fixed threshold at zero value, without the need
to use an automatic threshold method, such as Otsu
(Otsu, 1979).
Another way to use these indices is to generate
masks from the indexed image. While the ExG index
highlights the pixels of the green channel of the im-
age, the ExR index highlights the red channel. These
two indices can be used to produce a mask for, re-
spectively, vegetation and background, that combined
by a logical AND operation enable us to extract the
vegetation of the original image (Riehle et al., 2020).
Other methods can be used to segment
plant/background without the need for manual
parameterization (e.g., threshold or color selection).
Some approaches use Naive Bayes to classify the
pixels belonging to the plant and background (Abbasi
and Fahlgren, 2016). First, the image is converted
from RGB to HSV color model. Then, the color
distribution of vegetation and soil are approximated
by probability distribution functions, which allows
the use of the Naive Bayes method to segment the
plants.
More recently, deep learning approaches, such
as Deep Convolutional Neural Networks (DCNN’s),
have been proposed as an alternative for plant/soil
segmentation problems. This is explained due to their
great success in many classification and segmentation
problems in different areas. They have been proven to
be very efficient in identifying objects and as a generic
solution for various types of soils, terrains, and ex-
posure to sunlight and shading. The network archi-
tecture varies from problem to problem as, for exam-
ple, the number of convolutional and pooling layers
(Zhuang et al., 2018). Depending on the application,
the input can be images generated from the vegetation
indices or simple RGB images. Performance analy-
sis, however, shows that vegetation indices generate
information loss and that using RGB images as input
usually generates better results (Zhuang et al., 2018).
For segmentation purposes, the basic structure of a
DCNN is composed of an encoder and a decoder. The
encoder is trained to compress and to extract image
features by using several convolutional layers, orga-
nized hierarchically, where each layer corresponds to
a different semantic level. In the sequence, the de-
coder reconstructs the input image based on the model
compressed by the encoder, thus resulting in an image
labeled with different regions of interest.
3 MATERIALS AND METHODS
3.1 Vegetation Indices
Given its usage in several precision agriculture works,
we evaluated the following plant/background indices
for segmentation: Modified Green Red Vegetation
(4) (Bendig et al., 2015), Green Leaf Index (Equa-
tion 5) (Louhaichi et al., 2001), Modified Photo-
chemical Reflectance Index (Equation 6) (Yang et al.,
2018), Red Green Blue Vegetation Index (Equation 7)
(Bendig et al., 2015), Excess of Green (Equation 8)
(Woebbecke et al., 1995) and Vegetativen (Equation
9) (Hague et al., 2006):
MGRV =
G
2
− R
2
G
2
+ R
2
(4)
GLI =
2G − R − B
2G + R + B
(5)
MPRI =
G − R
G + R
(6)
RGBV I =
G − (B ∗ R)
(G
2
) + (B ∗ R)
(7)
Segmentation of Agricultural Images using Vegetation Indices
507

ExG = 2G − R − B (8)
V EG =
G
R
a
∗ B
b
(9)
* a = 0.667 and b = (1-a)
While ExG, GLI and VEG highlight pixels in the
green channel, MGRVI, MPRI, and RGBVI highlight
more than one channel of the image. For example,
MGRVI enhances the information of both green and
red channels, while MPRI emphasizes the reflectance
produced by chlorophyll present in leaves. These in-
dices act directly with the ability of the soil and plants
to reflect, respectively, red and green shades. Figure 1
shows an example of the MGRVI index obtained for
an image.
Figure 1: Example of the MGRVI index obtained for an
image.
3.2 Dataset
To evaluate the proposed method we considered four
datasets presented in (Riehle et al., 2020). Each
dataset contains 50 images and it was obtained us-
ing a different camera, resulting in a total of 200 im-
ages. The cameras used for the first two sets were
“GoPro HERO6 BLACK” and “Parrot SEQUOIA +”
which originated the datasets, respectively, GP and
SE. These images were obtained by an autonomous
caterpillar robot “Phoenix”. The other data set called
K2 was obtained by the robot “TALOS” using a “Mi-
crosoft Kinect v2” camera and the fourth dataset was
made available by the University of Bonn using a “JAI
AD-130GE” camera. For all camera datasets images
have 2280 × 2256 pixels size in RGB format. The
datasets contain images of different plants (maize and
sugar beet) at various growth stages and vegetation
coverages.
3.3 Evaluation
We used five metrics to assess the performance of
the proposed segmentation approach: Dice coefficient
(Equation 10), Jaccard index (Equation 11), Precision
(Equation 12), Sensitivity (Equation 13) and Speci-
ficity (Equation 14). In these equation, A and B are,
respectively, the proposed and the expert’s segmenta-
tion images, T P is the number of true positives, FN
is the false negative and T N is the true negative.
Dice =
2|A ∩ B|
|A| + |B|
(10)
Jaccard =
|A ∩ B|
|A ∪ B|
(11)
Precision =
T P
T P + FN
(12)
Sensitivity =
T P
T P + FN
(13)
Speci f icity =
T N
T N + FP
(14)
4 EXPERIMENTS
In our experiments, we aimed to segment images in
each dataset in order to extract the regions contain-
ing plants from the background image. To accom-
plish that we computed all selected vegetation indices
for each image in the datasets. In the sequence, we
used the K-means method to group the pixels into
two classes (K = 2), i.e., Plant and Background. We
initialized the centroids with the highest and lowest
value present in the image for each index. This was
performed to avoid any randomness and to guarantee
the reproducibility of the algorithm. By associating
the clusters obtained from K-means with the original
pixel position, we obtain a binary image representing
the two clusters, i.e., a segmented image represent-
ing regions of plant and background. Additionally,
we used an opening morphological operation, with a
15 × 15 structuring element in square format, to re-
move small noises in the segmented image.
The selected 15 × 15 kernel size of the structur-
ing element was defined manually and gradually. We
started with a 5 × 5 kernel size and we increased its
size as it kept reducing the presence of small noises
and improved the result. The decision to use a square
kernel was not based on any particular need, but due
to the fact this kernel is commonly used in morpho-
logical opening and closing operations.
VISAPP 2021 - 16th International Conference on Computer Vision Theory and Applications
508
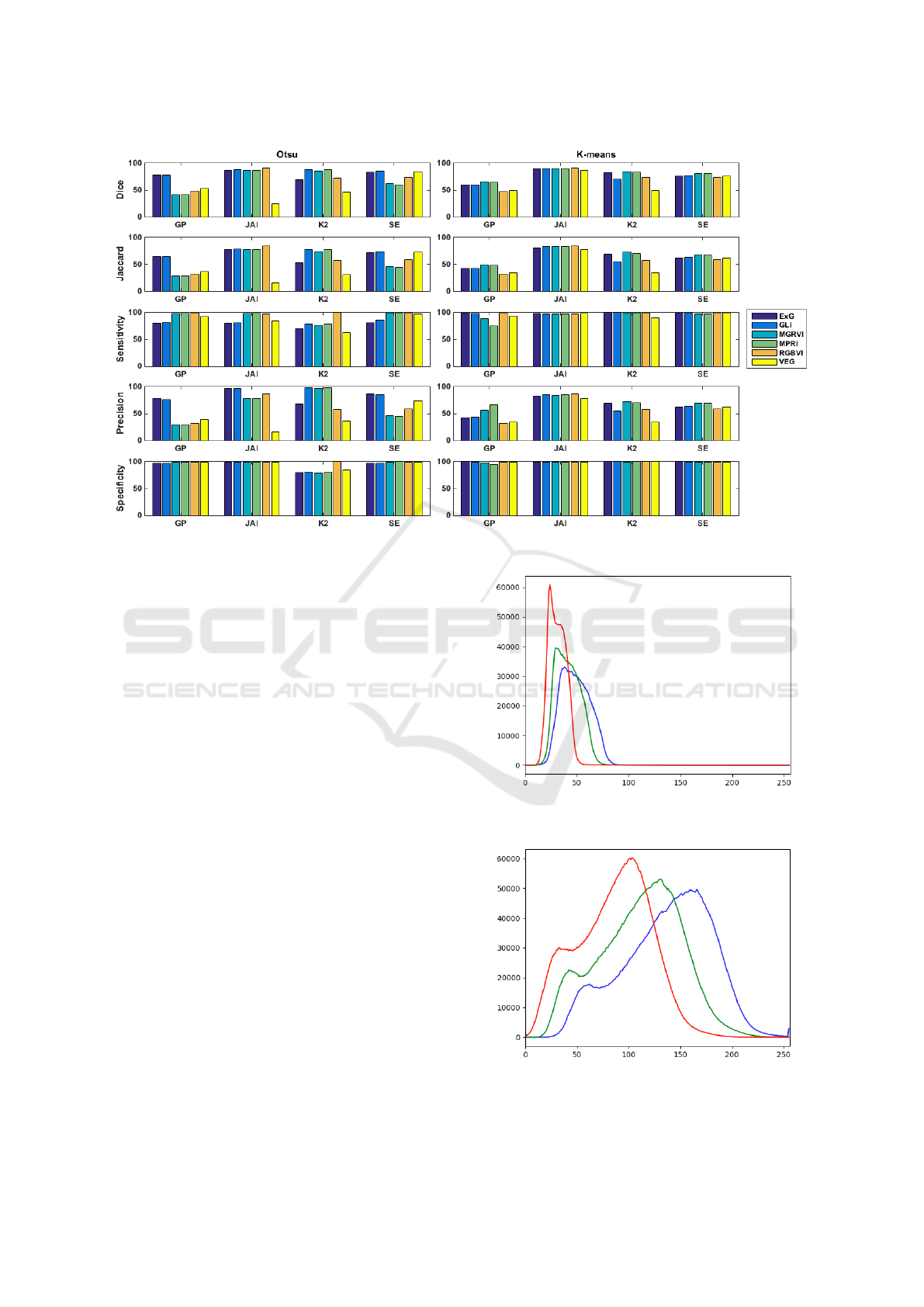
Figure 2: Comparison results for Otsu and K-means method for each vegetation index in all datasets.
Alternatively, we also evaluated the performance
of a simple automatic threshold approach to segment
the vegetation index images. In this case, we replaced
the K-means method in the approach previously de-
scribed by the Otsu method.
5 RESULTS
In this section, the results obtained by the experiments
conducted on this work were presented. Each vegeta-
tion index was applied to each dataset as explained
in Section 3. Figure 2 shows the results obtained in
each metric by each vegetation index in all datasets,
for both K-means and the Otsu method.
In general, the best results are obtained in the JAI
dataset. Images in this dataset present small varia-
tion in luminosity and brightness, increasing the ef-
fectiveness of segmentation, which is corroborated
by Dice coefficients ranging from between 86.82% to
91.31%. We also notice that, for this dataset, the K-
means approach is slightly superior in comparison to
Otsu. Moreover, this dataset presents a small varia-
tion in green shades present in the plants and the soil
color pattern is very homogeneous (Figure 3). On the
other hand, the GP dataset presented, in general, the
worst results. This is explained due to the large varia-
tion of luminosity present in the images, as shown in
Figure 4. A superficial analysis of the images in this
dataset shows there exists a variation in luminosity
caused by the local climate, as also an increase in the
Figure 3: Example of histogram of an image from JAI
dataset.
Figure 4: Example of histogram of an image from GP
dataset.
Segmentation of Agricultural Images using Vegetation Indices
509

Figure 5: Noise reduction using morphological opening over the result of the K-means approach. Left: Binary image obtained
from segmentation. Right: Image after morphological opening.
image exposure due to differences in the reflection of
the sunlight by the soil. Even for images where there
was small light variation, as in the SE dataset, the dif-
ference in the reflection of light on the ground still
influences the results.
When comparing Otsu and K-means, we notice
that K-means tend to obtain higher results. For
the same dataset, Otsu presents higher oscillation in
their results depending on the vegetation index used.
Meanwhile, K-means, on average, presents similar re-
sults for different vegetation indices. This indicates
that the process of clustering can detect more effi-
ciently different regions of interest (soil and plant)
than thresholding. This is partially explained by the
fact that K-means uses a distance metric to compute
the clusters and iteratively moves the centroid for the
positions that best separate the different classes of ob-
jects.
We must also emphasize the importance of mor-
phological opening, as shown in Figure 5. As one
can notice, this operation diminishes the level of noise
(e.g., loose pixels and other small objects) resulting
from the segmentation process.
Among all compared indices, MGRVI presented
the best results for the Dice coefficient, ranging from
65.0% to 90.0%. This is a normalization of GRVI
whose vegetation and soil reflectance pattern is rela-
tively easy to interpret, where the vegetation reflects
the green band more than the red, while the soil re-
flects the red band more than the green (Motohka
et al., 2010). Figure 6 shows the best segmentation
obtained using K-means for each dataset.
6 CONCLUSIONS
In this paper, we presented a study of different veg-
etation indices to segment plants/soil in images. We
evaluated two approaches to segment the images from
four different datasets: K-means and Otsu. Results
demonstrated the superiority of the K-means method
to segment these images when applied in combina-
tion with the MGRVI index. The evaluation carried
out by this work gives us an overview of the behav-
ior of each vegetation index for different types of
images (acquired at different conditions and equip-
ment) and their limitations, making it clear that a sin-
gle index may not be satisfactory to segment plants
from the background. In future work, we intend
to combine different vegetation indices to obtain a
more robust result for different datasets, to evaluate
whether the use of other color model (e.g., HSV)
combined with vegetation indices can improve the re-
sult obtained, and evaluate different automatic thresh-
old methods. We also intend to study the application
of vegetation indices that use multispectral images for
the plant/background segmentation process.
ACKNOWLEDGEMENTS
Andr
´
e R. Backes gratefully acknowledges the fi-
nancial support of CNPq (Grant #301715/2018-1).
This study was financed in part by the Coordenac¸
˜
ao
de Aperfeic¸oamento de Pessoal de N
´
ıvel Superior -
Brazil (CAPES) - Finance Code 001.
VISAPP 2021 - 16th International Conference on Computer Vision Theory and Applications
510
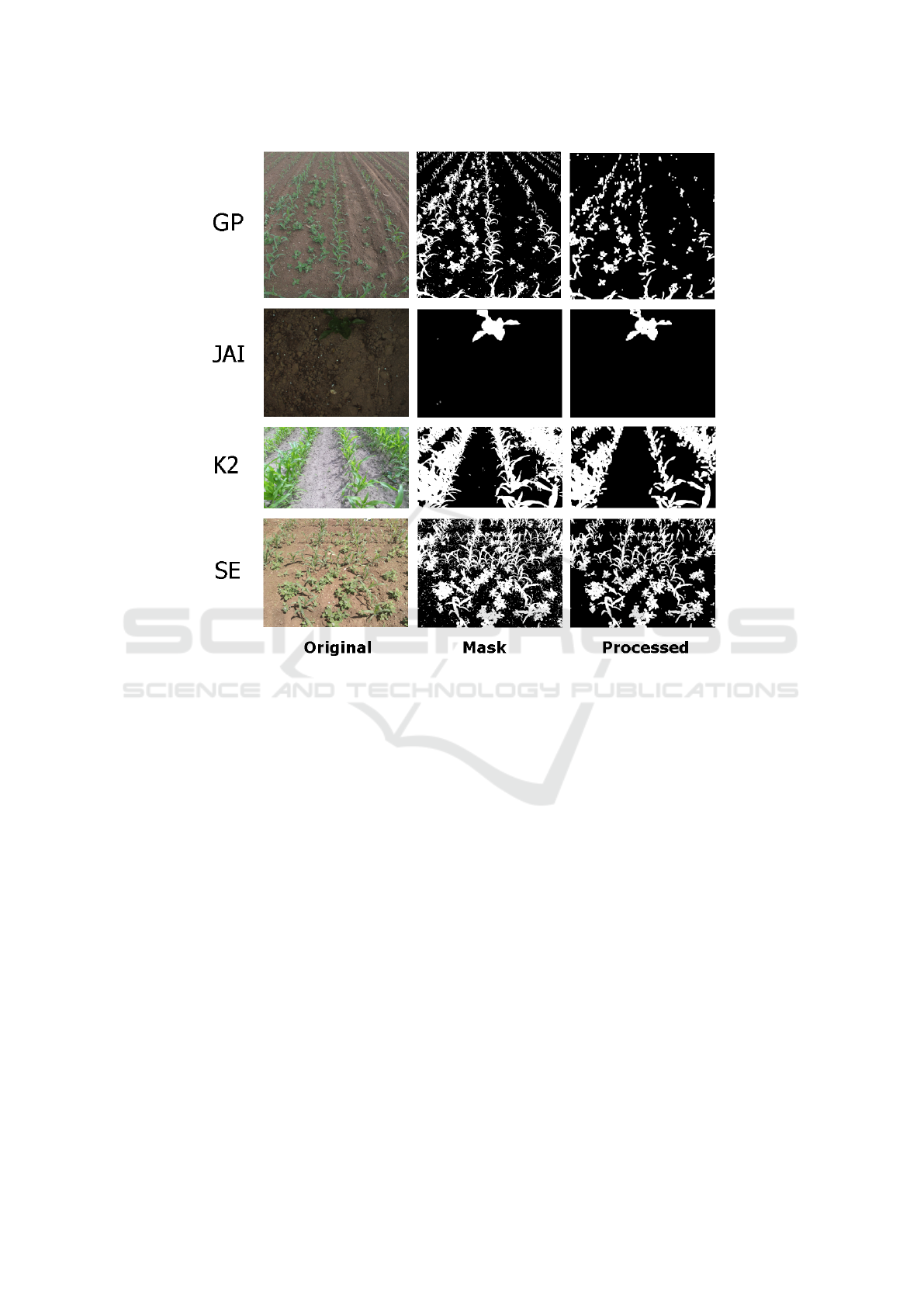
Figure 6: Best results obtained for each dataset. For datasets GP and K2, best results is obtained using MGRVI index. For
JAI dataset, best result is obtained using RGBVI, while MPRI index performs best in SE dataset.
REFERENCES
Abbasi, A. and Fahlgren, N. (2016). Na
¨
ıve bayes pixel-level
plant segmentation. In 2016 IEEE Western New York
Image and Signal Processing Workshop (WNYISPW),
pages 1–4.
Bargoti, S. and Underwood, J. P. (2016). Image segmen-
tation for fruit detection and yield estimation in apple
orchards. CoRR, abs/1610.08120.
Bendig, J., Yu, K., Aasen, H., Bolten, A., Bennertz, S.,
Broscheit, J., Gnyp, M. L., and Bareth, G. (2015).
Combining uav-based plant height from crop surface
models, visible, and near infrared vegetation indices
for biomass monitoring in barley. International Jour-
nal of Applied Earth Observation and Geoinforma-
tion, 39:79 – 87.
Hague, T., Tillett, N., and Wheeler, H. (2006). Automated
crop and weed monitoring in widely spaced cereals.
Precision Agriculture, 7:21–32.
Louhaichi, M., Borman, M., and Johnson, D. (2001). Spa-
tially located platform and aerial photography for doc-
umentation of grazing impacts on wheat. Geocarto
International, 16.
Motohka, T., Nasahara, K., Hiroyuki, O., and Satoshi, T.
(2010). Applicability of green-red vegetation index
for remote sensing of vegetation phenology. Remote
Sensing, 2.
Otsu, N. (1979). A threshold selection method from gray-
level histograms. IEEE Transactions on Systems,
Man, and Cybernetics, 9(1):62–66.
Riehle, D., Reiser, D., and Griepentrog, H. W. (2020).
Robust index-based semantic plant/background seg-
mentation for rgb- images. Comput. Electron. Agric,
169:105201.
Woebbecke, D., Meyer, G., Bargen, K., and Mortensen, D.
(1995). Color indices for weed identification under
various soil, residue, and lighting conditions. Trans-
actions of the ASAE, 38:259–269.
Yang, S., Wang, L., Shi, C., and Lu, Y. (2018). Evalu-
ating the relationship between the photochemical re-
flectance index and light use efficiency in a mangrove
forest with spartina alterniflora invasion. International
Journal of Applied Earth Observation and Geoinfor-
mation, 73:778 – 785.
Zhuang, S., Wang, P., and Jiang, B. (2018). Segmentation
of green vegetation in the field using deep neural net-
works*. In 2018 13th World Congress on Intelligent
Control and Automation (WCICA), pages 509–514.
Segmentation of Agricultural Images using Vegetation Indices
511
