
Mining Biomedical Texts for Pediatric Information
Tian Yun
1
, Deepti Garg
2
and Natalia Khuri
1 a
1
Department of Computer Science, Wake Forest University, 1834 Wake Forest Road, Winston-Salem, U.S.A.
2
Department of Computer Science, San Jos
´
e State University, One Washington Square, San Jos
´
e, U.S.A.
3
Department of Computer Science, Wake Forest University, 1834 Wake Forest Road, Winston-Salem, U.S.A.
Keywords:
Text Mining, Classification, Machine Learning, Support Vector Machine.
Abstract:
To perform a comprehensive and detailed analysis of the gaps in knowledge about drugs’ safety and effective-
ness in neonates, infants, children, and adolescents, large collections of complex and unstructured texts need
to be analyzed. In this work, machine learning algorithms have been used to implement classifiers of biomed-
ical texts and to extract information about safety and efficacy of drugs in pediatric populations. Models were
trained using approved drug product labels and computational experiments were conducted to evaluate the ac-
curacy of the models. A Support Vector Machine with a radial kernel had the best performance by classifying
short texts with an accuracy of 94% and an excellent precision. Results show that classifiers perform better
when trained using features comprising multiple words rather than single words. The proposed text classifier
may be used to mine other sources of biomedical information, such as research publications and electronic
health records.
1 INTRODUCTION
Data-driven modeling is prevalent in the development
of biomedical and bioinformatics methods, tools, and
approaches. These methods, tools, and approaches
rely on the availability of large data sets, acquired by
scientists in academia, industry and government or-
ganizations. The majority of the data sets are col-
lected in the medium and high throughput experi-
ments, which measure activities of interest. Alterna-
tively, the data may be also collected by mining of
published works and extracting the information of in-
terest. For example, data sets of side effects of drugs
may be extracted from the published works and used
to develop bioinformatics tools and software to pre-
dict novel associations and interactions.
Every marketed drug undergoes rigorous clinical
studies to assess its safety and efficacy in the target
population. However, “off-label” drug prescribing is
not uncommon, especially in the treatment of pedi-
atric patients. Here, “off-label” refers to a prescrip-
tion that differs from the approved use or dosage of a
drug. Such practice is legal, and it is informed by the
clinical experience of the physicians or by the avail-
ability of treatments. For example, drugs may be pre-
scribed to pediatric patients when no alternative treat-
a
https://orcid.org/0000-0001-9031-8124
ments exist (Ito, 2017). Even when pediatric treat-
ments are available, their formulation or dosage may
be less effective or less well tolerated than adult’s for-
mulation or dosage of a newer drug (Lowenthal and
Fiks, 2016).
The rates of the “off-label” prescribing in pedi-
atric patients range from 36% in inpatient settings to
97% in intensive care units (Hoon et al., 2019). Ad-
ditionally, drugs are most frequently prescribed “off-
label” to neonates and infants compared to other pedi-
atric age groups, and to girls compared to boys (Hoon
et al., 2019).
Due to an increased attention to the “off-label”
pediatric prescribing and due to several national leg-
islative actions (U.S. Congress. Best Pharmaceuticals
for Children Act Amending Section 505A of the Fed-
eral Food, Drug & Cosmetic Act (Public Law 107-
109). (2002), 2002; U.S. Congress. Pediatric Re-
search Equity Act amending Section 505B of the Fed-
eral Food, Drug & Cosmetic Act (Public Law 108-
155). (2003), 2003; FDASIA, 2012), the number of
pediatric clinical studies has been growing. Infor-
mation about these studies may be found in the li-
braries of biomedical publications, pediatric clinical
research networks (Fiks et al., 2016), electronic health
records, insurance claims, dedicated portals (Desh-
mukh and Khuri, 2018; U.S. Food and Drug Admin-
60
Yun, T., Garg, D. and Khuri, N.
Mining Biomedical Texts for Pediatric Information.
DOI: 10.5220/0010310200600071
In Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2021) - Volume 3: BIOINFORMATICS, pages 60-71
ISBN: 978-989-758-490-9
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

istration. New pediatric labeling information dataset,
2020), and even social media (Mulugeta et al., 2018).
However, retrieval of information about drugs’
use in pediatric patients is challenging due to the
unstructured format of biomedical texts and the di-
versity of terms that characterize pediatric popula-
tions. Pediatric populations encompass children from
birth to 17 years of age, and they are typically di-
vided into four age groups, namely, neonates, infants,
children and adolescents. However, drug regulations
do not prescribe the exact division but rather allow
drug developers to identify the appropriate pediatric
age cohort based on the scientific evidence, such as
the body weight, ability to swallow a specific drug
formulation, metabolism of drug enzymes, expres-
sion levels of drug membrane transporters, and so on.
Thus, it is challenging to search for pediatric infor-
mation in biomedical literature. For example, while
recent search for articles mentioning “pediatric” stud-
ies in the PubMed repository of biomedical litera-
ture returned 1,010 results, searching for studies in
“neonates” returned 426 additional publications.
Supervised machine learning (ML) algorithms
may improve or augment biomedical text mining. For
instance, a binary classifier may be trained to predict
whether a previously unseen text contains informa-
tion about drug’s safety and efficacy in pediatric pop-
ulations. Additionally, large collections of biomedi-
cal texts, could be rapidly screened to retrieve only
those texts that contain information relevant to pedi-
atric prescribing, such as drug’s efficacy, adverse re-
actions, dosage, and so on. Finally, automated text
mining of biomedical literature may assist drug devel-
opers and regulators in identifying unmet needs and
research gaps in pediatric drug development.
To train ML classifiers for use in text mining, a
large training data set of labeled texts is needed. How-
ever, there is a lack of labeled biomedical texts that
focus specifically on pediatric patients. To create la-
beled texts for use in classifier’s training, drug product
labels may be used. In the US, approved drug prod-
uct labels contain the most reliable information about
drugs’ safety and efficacy in pediatric populations.
These labels are reviewed and approved by the regu-
lators, and they are updated regularly. They are stored
in a special format called Structured Product Labeling
(SPL) (Structured product labeling, 2019). The SPL
format is approved by the Health Level Seven (HL7)
organization, which administers standards for storage,
retrieval and exchange of digital health information
between different medical systems and entities (HL7
Standards, 2019).
SPL is divided into several hyperlinked sections,
and each section is coded using an identifier called
the Logical Observation Identifiers Names and Code
(LOINC). For example, PEDIATRIC USE section is
coded with LOINC 34081-0. Each drug product is
described in its own SPL file, and on average, there
are about twice as many SPL files as there are ap-
proved drugs. The number of drug product labels
exceeds the number of approved drugs because there
may be several products associated with a single drug,
such as drug products from different manufacturers,
drug products in different dosage forms or routes of
administration.
Therefore, public availability of the SPL files
presents an opportunity to create a training data set.
Yet, the process of extracting tagged texts from drug
labels is onerous. Firstly, many older (prior to 2005)
drug labels do not contain LOINC identifiers. Sec-
ondly, pediatric information may be also included in
other sections of the SPL files. We propose to cir-
cumvent these challenges by using a semi-supervised
approach to design and implement a text mining
pipeline to accurately and rapidly identify if an un-
structured text is related to pediatric use or not. To
validate the proposed pipeline, we collected, cleaned,
preprocessed and transformed unstructured texts into
real-valued vectors containing the term frequency–
inverse document frequency (TFIDF) scores, and la-
beled them as pediatric or nonpediatric texts. The
accuracy of our ML classifiers was high, indicating
that our proposed approach is a viable first step in
the curation of unstructured texts. Additionally, we
showed that classification accuracy can be further im-
proved by the selection of most informative features.
To the best of our knowledge, our application of ML-
powered text mining to the retrieval of pediatric infor-
mation is a novel contribution.
The remainder of the article is organized as fol-
lows. Section 2 reviews relevant prior work. Our ap-
proach for the classification of pediatric texts in drug
labels is described in Section 3. Section 4 presents ex-
perimental results, which are placed in a broader con-
text in Section 5. We conclude the article and present
possible future directions in Section 6.
2 PRIOR WORK
Biomedical text mining is an active area of research
motivated by the opportunities for the extraction of
actionable insights from massive collections of un-
structured texts. Among these unstructured texts,
publicly available drug product labels provide sci-
entific summaries of nonclinical and clinical drug
studies, and they include information about drug’s
indications and contraindications, drug-drug interac-
Mining Biomedical Texts for Pediatric Information
61

tions, adverse effects, dosage, and so on. To date,
text mining of drug product labels has resulted in
the accurate extraction of drug indications (N
´
ev
´
eol
and Lu, 2010; Li et al., 2013; Fung et al., 2013;
Khare et al., 2014), adverse drug reactions (Bisgin
et al., 2011; Demner-Fushman et al., 2018a; Demner-
Fushman et al., 2018b; Pandey et al., 2019; Tiftikci
et al., 2019), pharmacogenomic biomarkers (Fang
et al., 2016; Mehta et al., 2020), patient-reported out-
comes (Gnanasakthy et al., 2019) and pregnancy drug
risks (Rodriguez and Fushman, 2015).
The majority of methods focus on named entity
recognition and relation extraction tasks, and make
extensive use of biomedical ontologies, controlled vo-
cabularies, and linguistic information. The reported
accuracy of some of these tools is about 80%, how-
ever most of the automated information extraction
tools are still far from delivering a gold standard with-
out human intervention. Manual curation is needed to
either filter the results or to aid with the extraction of
information. This is due to the fact that drug indi-
cations and adverse reactions are difficult to extract
because of co-existing conditions, characteristics of
patient cohorts, and so on. In addition to software
development, several data repositories have been cre-
ated from data extracted from biomedical texts, in-
cluding drug labels (Khare et al., 2014; Fang et al.,
2016; Kuhn et al., 2016). They provide easy access to
information about each drug, such as its ingredients,
dose forms, adverse reactions, and so on.
Despite the rich history of biomedical text mining
for the information about drugs’ safety and efficacy
in general population of patients, little attention has
been paid to mining information about drugs’ use in
special populations, such as pediatric and geriatric pa-
tients, pregnant and nursing women. Only two online
resources exist for querying drug labels about pedi-
atric use. First, US Food and Drug Administration
(FDA) maintains the Pediatric Labeling Information
Database (U.S. Food and Drug Administration. New
pediatric labeling information dataset, 2020). This
database is built from regulatory submissions, which
include drug product labels. This resource has very
limited search capabilities and is constructed manu-
ally, thus, lagging behind the updates of drug prod-
uct labels. For instance, although over 1,200 pediatric
studies have been submitted to the FDA in response to
pediatric regulations, only about 800 of these studies
are currently listed in the database.
To address the paucity of information about drugs’
safety and efficacy in pediatric populations, a second
online resource, PediatricDB, was built (Deshmukh
and Khuri, 2018). The data of this portal can be
queried using drug names, pediatric age group, ther-
apeutic category and so on. Similarly, the frequency
of updates in PediatricDB is lagging behind the up-
dates of SPL repository because of its reliance on the
manual data curation.
Our work differs from prior research. It addresses
the need for the automated retrieval of information
which may better inform prescribers, regulators, man-
ufacturers and patients. The output of our classifier
may also be used as an input to the existing tools, such
as an automated extraction of indications, drug reac-
tions, and so on. Next, we describe our approach in
details.
3 DATA AND METHODS
3.1 Text Mining Workflow
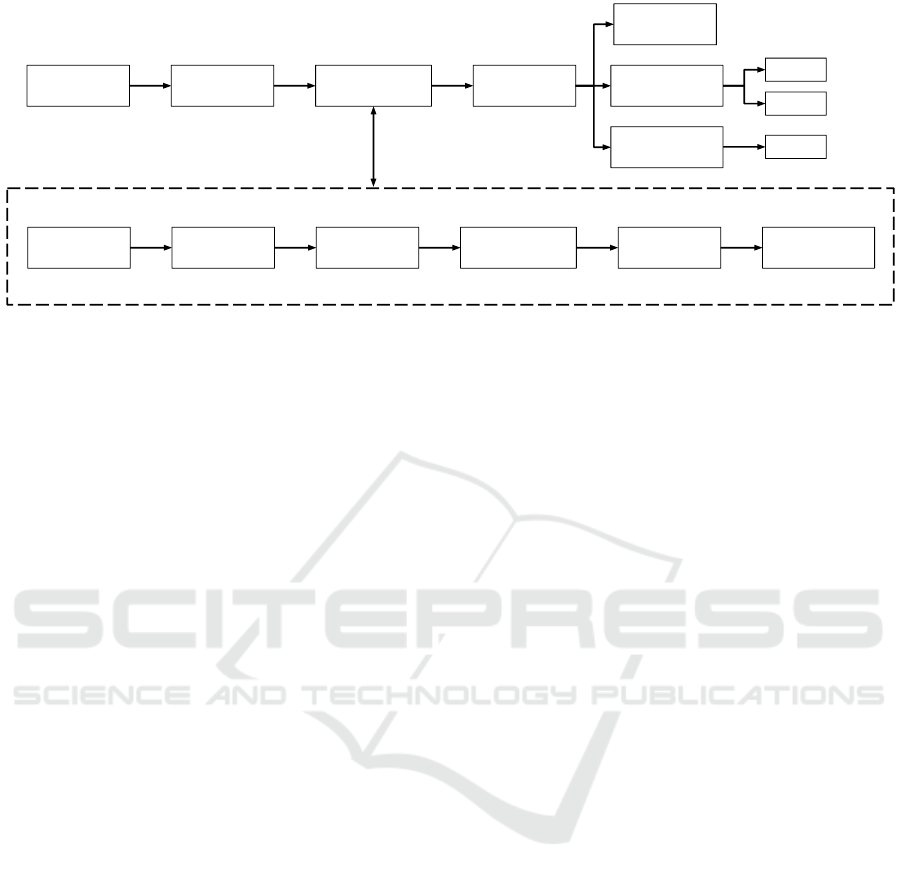
Our text mining workflow comprises five steps,
namely (1) data parsing, (2) data partitioning, (3) data
preprocessing, (4) data transformation, and (5) valida-
tion (Fig. 1). The workflow was executed on Google’s
cloud servers using CPUs. We experimented with
three ML classifiers and performed different valida-
tion experiments to assess their usability in different
real-life scenarios. First, we estimated classifiers’ per-
formance in a 10-fold cross-validation. Second, we
validated their performance retrospectively, by classi-
fying texts that were collected at the same time point
as the training data set but using a different data col-
lection protocol. Finally, we prospectively validated
the performance of classifiers on texts, which were
collected at a later time point.
3.2 Data Collection and Pre-pocessing
Weekly archives of approved human prescription
drugs were downloaded from the public repository
DailyMed (DailyMed, 2019) on August 31, 2019.
From the downloaded files, 500 SPL files were ran-
domly sub-sampled. Files without the Indication
And Usage section were filtered out, leaving 494 SPL
files for the downstream processing. Next, SPL files
were parsed using a custom SPL parser implemented
in the Python programming language, making use of
the lxml XML toolkit (lxml, 2019).
We constructed a training data set of pediatric
texts by extracting from 494 SPL files, all texts
tagged with LOINC 34081-0 (Pediatric Use sec-
tions). Next, these texts were removed from the SPL
files and two test sets were constructed as follows.
The first test data set (Test 1) comprised all texts,
which had keywords Pediatric Use in the docu-
ment tags. After Test 1 texts were removed from the
BIOINFORMATICS 2021 - 12th International Conference on Bioinformatics Models, Methods and Algorithms
62
Parse
SPL Files
Partition
Data
Preprocess
Data
Compute
TFIDF
Retrospective
Validation
Split
Sentences
Tokenize
Documents
Convert to
Lower Case
Remove Stop
Words
Preprocessed
Document
Cross
Validation
Test2
Test3
Test1
Prospective
Validation
Raw
Document
Figure 1: Text mining workflow. Shown are the five steps of building classifiers of pediatric and nonpediatric texts. The
details of the data preprocessing steps are shown in the dotted box.
494 files, the second test set (Test 2) was constructed.
It comprised the remaining pediatric texts which were
found using a case-insensitive pattern search in the re-
maining sections of the SPL files.
Out of 494 SPL files, 34 contained no pediatric
information, and these 34 files were segmented into
nonpediatric texts, as follows. First, statistical anal-
ysis of sentence lengths of pediatric texts was con-
ducted, and a Poisson distribution was fitted to that
data. Next, we sampled text lengths from the fitted
Poisson distribution and generated nonpediatric texts
of sampled lengths to be used for training and testing.
Sampling was done without replacement.
Finally, the last test set (Test 3) was constructed
from 31,565 SPL files, which were retrieved at a later
date (May 17, 2020). These SPL files were parsed
into pediatric and nonpediatric texts. Pediatric texts
were extracted using the same protocol as the train-
ing pediatric data set. Nonpediatric texts were con-
structed from the SPL files which did not contain any
tagged pediatric sections. We removed all texts from
Test 3 that overlapped with the training set, Test 1 or
Test 2. Both pediatric and nonpediatric texts in Test 3
were left in their original length, that is neither Pois-
son sampling nor text partitioning was done to create
Test 3.
3.3 Feature Engineering
Each text was encoded into a numeric feature vector
using a custom Python code and the TfidfVectorizer
function from the Scikit-Learn library (Scikit-Learn,
2019). Each document was first split into sentences
using period (.) as a delimiter, and regular expres-
sions were used to avoid splitting numeric values. We
then tokenized each sentence, such that hyphenated
words and numbers containing decimal places were
kept together as tokens. Each token was converted
to lowercase. A token was removed if it was a stop
word, such as and, the, a, is, and so on. Addition-
ally, we included the word pediatric in the list of stop
words.
Next, we constructed a dictionary of unique n-
grams using the training set; we included unigrams
(single words), bigrams (two consecutive words) and
trigrams (three consecutive words) in the dictionary.
The frequency of each n-gram in the documents was
computed, and n-grams with low (less than 0.05) and
high frequency (0.90) were removed from the dictio-
nary. This helped in removing words that do not con-
tribute toward the classification of pediatric and non-
pediatric texts, such as specific drug and manufacturer
names, and/or drug ingredients.
Third, the occurrence of each n-gram in every doc-
ument was determined, and these counts were nor-
malized using the TFIDF transformation. TFIDF
scores help identify important n-grams in a document,
with higher scores reflecting more important n-grams.
TFIDF values close or equal to zero are representative
of noninformative words. TF scores were sublinearly
smoothed by replacing TF with 1 + log(T F), and the
IDF normalization weights were smoothed by adding
one to document frequencies. Finally, L2 normaliza-
tion was applied. In the end, each text was encoded
by a vector of floating-point TFIDF scores ranging be-
tween 0 and 1.
Finally, we annotated every text using a binary
class label. Each pediatric text was labeled with a “1”
and each nonpediatric text with a “0”.
To visualize the data, principal component anal-
ysis (PCA) was performed using PCA function from
Scikit-Learn (Scikit-Learn, 2019). The first two prin-
cipal components were plotted to visualize pediatric
and nonpediatric texts in the training data set.
Texts in the test data sets were also transformed
into vectors of TFIDF scores, computed as described
Mining Biomedical Texts for Pediatric Information
63

above. Notably, to encode test data sets, we used the
dictionary that was built from the training texts. Thus,
all n-grams found in the test data sets, which were
not present in the training dictionary, were ignored
during the transformation of the test data sets. Texts
in the test data sets were labeled with a “1” to denote
a pediatric text or with a “0” to denote a nonpediatric
text.
3.4 Classifier Training and Validation
We selected three ML algorithms to train our classi-
fiers, namely, k-nearest Neighbors (kNN), Decision
Tree (D-Tree), and Support Vector Machines (SVM).
These algorithms were selected due to their desir-
able characteristics, such as their simplicity and in-
terpretability. All classifiers were trained using the
same training data set, and training was done with the
default parameters in the Scikit-Learn library (Scikit-
Learn, 2019).
D-Tree is a simple supervised ML classifier,
which distinguishes between the target classes by
learning binary decision rules. There exist various
variants of the algorithm, and in this work, we used
the Scikit-Learn (Scikit-Learn, 2019) library imple-
mentation of the Classification and Regression Trees
(CART) algorithm (Everitt, 2005).
kNN is another basic classification algorithm
which is based on the assumption that similar data
points are closer to each other than dissimilar data
points (Cover and Hart, 2006). To classify an unseen
text, its k-closest texts are identified, and the majority
class of the neighbors is used to assign the label to the
unseen text. We used the Euclidean distance function,
which is a commonly used distance metric in the im-
plementations of the kNN algorithm (Hu et al., 2016).
The kNN algorithm is easy to implement as it does not
need any assumptions about the underlying data dis-
tribution and we used the Scikit-Learn (Scikit-Learn,
2019) library implementation.
SVM is a supervised machine learning classifier
which learns a separating hyperplane between the two
classes (Chang and Lin, 2011). In a two-dimensional
space, this hyperplane corresponds to a line between
the two target classes in the training data set. The tun-
ing parameters for the SVM algorithm are the kernel,
regularization and gamma. Kernel defines the higher
dimension where the separating hyperplane is to be
computed. Regularization defines the extent of per-
missible misclassification. Gamma defines to what
extent the data points should be considered while
computing the separating line. The Sequential Mini-
mal Optimization algorithm (Fan et al., 2005), imple-
mented in the Scikit-Learn library, was used to train
SVM classifiers.
In the validation experiments, five metrics were
used to assess performance, namely, accuracy, pre-
cision, recall, F1 score, and area under the Receiver
Operating Characteristic (ROC) curve (AUC).
3.4.1 Experiment 1: Cross-validation
Cross-validation is a common resampling technique
used to evaluate and compare the performance of the
ML models (Refaeilzadeh et al., 2009). To implement
a cross-validation experiment, we split the training
data set into 10 folds and used 9 folds for training
and the remaining 1 part of the data for validation.
The training data set was shuffled prior to splitting,
and stratified partitioning was used to ensure that the
distribution of labels in each fold was similar to the
distribution of class labels in the original training data
set. Ten ROC curves were constructed for each clas-
sifier, and AUC scores were averaged across the 10
folds.
3.4.2 Experiment 2: Retrospective Validation
In the second experiment, we evaluated each classi-
fier by retrospective validation (Prospective and ret-
rospective cohort studies, 2019). In this experiment,
classifiers were trained on the entire training data set,
and the best models were used to predict the class la-
bels of the two test data sets, Test 1 and Test 2. Be-
cause the true class labels of these two data sets were
known, this experiment evaluated the generalizability
of each model. Moreover, this experiment examined
the accuracy of our models on the data set acquired
using a different data collection protocol.
3.4.3 Experiment 3: Prospective Validation
In the third experiment, the objective was to accu-
rately label pediatric and nonpediatric texts in a large
collection of texts, which were retrieved at a time dif-
ferent from the collection date of the training data set.
This experiment evaluated the feasibility of an auto-
mated text classification on a large unseen collection
of texts.
4 RESULTS
4.1 Construction of the Data Sets
ML classifiers automatically learn relationships be-
tween features of the data and their class labels. Be-
cause they learn these relationships from labeled data,
BIOINFORMATICS 2021 - 12th International Conference on Bioinformatics Models, Methods and Algorithms
64

it is important to collect, pre-process and annotate
training and testing data sets. We performed two data
collections, separated in time by approximately one
year. The first data collection was used to construct
the training set and two test sets, Test 1 and Test 2.
The second data collection was used to construct Test
3. The training data set comprised 407 pediatric and
1,524 nonpediatric texts ((Table 1). Test 1 and Test 2
data sets were smaller, with 20 to 33 pediatric texts,
and 34 nonpediatric texts. The number of pediatric
texts in Test 3 was 28,720 compared with 2,845 non-
pediatric texts.
Table 1: Number of texts in training and test data sets.
Pediatric Nonpediatric Total
Training Set 407 1524 1931
Test 1 Set 20 34 54
Test 2 Set 33 34 67
Test 3 Set 28,720 2,845 31,565
On average, pediatric texts in the training data set
comprised 5.98 sentences and about 97.05 words, ex-
cluding stop words. Texts with a single sentence were
over-represented, and the longest pediatric text con-
tained 47 sentences. In Test 1, the average number
of sentences and words was 7.65 and 140.05, respec-
tively, and one-sentence texts were over-represented.
In Test 2 data set, text lengths were 6.21 sentences
and 88.18 words, on average.
Thirty-four SPL files from the first collection did
not contain pediatric information. These files were
used to create nonpediatric texts. The composition of
these nonpediatric texts differed from the composition
of pediatric texts. More specifically, the texts com-
prised, on average, 138.65 sentences and ranged be-
tween 30 and 595 sentences. Additionally, the num-
ber of words in these nonpediatric texts ranged be-
tween 300 and 8,071. Therefore, we post-processed
nonpediatric texts to create a sufficient number of
nonpediatric texts for training and testing.
We made the sentence distribution of nonpediatric
texts follow the sentence distribution of pediatric seg-
ments. More specifically, a shifted Poisson distri-
bution with mean of 1 was fitted to the distribution
of sentence lengths in pediatric texts. Considering
that texts with length of 1 may not generate meaning-
ful tokens, we sampled from the Poisson distribution
with the mean of 2, and implemented a shift of 1 to
the right to make sure that sampling does not return
empty sentences with lengths of 0. For instance, if
the sampled value from the Poisson distribution was
2, then it would be shifted to 3. Thus, 3 consecutive
sentences would be sampled from the 34 nonpediatric
texts to generate shorter texts.
All texts in Test 3 were kept in their original
lengths. Notably, while there were fewer nonpedi-
atric texts, on average, they were longer than pediatric
texts.
4.2 Feature Engineering and Encoding
We constructed a common dictionary of all unique
unigrams, bigrams, and trigrams from the training
texts. In all, 115 such n-grams were extracted from
the training set, and they were used to compute
TFIDF scores for each text. The same dictionary of
115 n-grams was used to compute the TFIDF scores
of texts in Test 1, Test 2, and Test 3.
To better understand the data, we performed a
principal component analysis of the TFIDF scores in
the training data set. Even though the first two princi-
pal components only explained 10.67% variance of
the data, there was still a clear separation between
the pediatric data points and nonpediatric data points
(Fig. 2, left).
We observed the presence of a region where 38
pediatric and 831 nonpediatric texts overlapped. This
region is defined by the values of the first principal
component ranging from -0.1 to 0.1 (x-axis) and by
the values of the second principal component ranging
from -0.15 to 0.0 (y-axis). Notably, most nonpedi-
atric texts in Test 1 concentrated in this overlapping
region. The projection of Test 2 onto the principal
components of the training set was similar. Most pe-
diatric texts had scores between 0.2 and 0.6 for the
first principal component and between -0.1 and 0.2 for
the second principal component, respectively. Test 3
had a similar distribution.
4.3 Experiment 1: Cross-validation
Three ML algorithms were evaluated by cross-
validation, namely, kNN, D-Tree and SVM. In this
experiment, the SVM model showed the strongest
performance in the 10-fold cross-validation, achiev-
ing an average AUC of 0.98 (Fig. 3, right). Notably,
SVM performance was consistent across the 10 val-
idation experiments. Its standard deviation of AUC
scores was 0.01. Both, kNN and D-Tree classifiers,
had high accuracy as well. The average AUC scores
of the kNN and D-Tree classifiers were 0.96 and 0.94,
respectively (Fig. 3, left and middle). However, there
was significantly more variation in the AUC scores in
the different folds. The standard deviation of AUC
scores of kNN was 0.02, while that of D-Tree was
0.03.
Mining Biomedical Texts for Pediatric Information
65
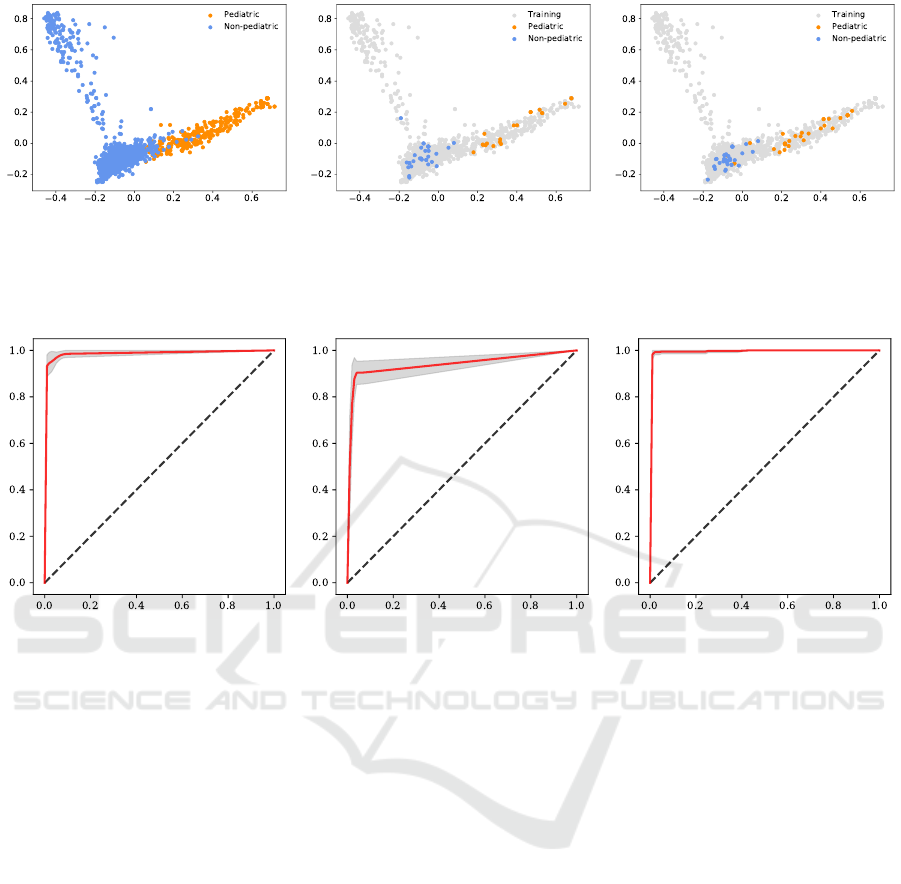
Figure 2: Visualization of the first two principal components of pediatric and nonpediatric texts. First principal component is
shown on the x-axis and the second principal component on the y-axis. Pediatric texts are denoted by the orange circles and
nonpediatric texts by the cornflower blue circles. Left: Training set. Middle: Test 1 projected onto the principal components
of the training set shown with grey circles. Right: Test 2 projected onto the principal components of the training set shown
with grey circles.
Figure 3: Receiver Operating Characteristic Curves of 10-fold cross-validation of three classifiers. Shown are ROC curves of
kNN, D-Tree, and SVM classifiers. True positive rate is plotted on the y-axis and false positive rate on the x-axis. Red line
denotes the average ROC curve from the 10-fold cross-validation, and the dotted line indicates the performance of a random
guess. The gray region is ±1 standard deviation of the ROC curves in the 10-fold cross-validation. Left: kNN. Middle:
D-Tree. Right: SVM.
4.4 Experiment 2: Retrospective
Validation
In this experiment, we trained the three classifiers us-
ing the entire training data set and evaluated their pre-
dictive performance using Test 1 and Test 2. The suc-
cess of this experiment is defined by how well classi-
fiers predict the labels for the test data sets.
All three classifiers, kNN, D-Tree and SVM, were
trained using their default parameters. For the kNN
classifier, Euclidean distance was used to compute the
distance between neighborhood points and the num-
ber of neighbors was set to k = 5. For the D-Tree clas-
sifier, criterion = Gini impurity was used for learning.
For the SVM, kernel = radial basis function, gamma
= ’scale’ and probability = True were used.
We did not carry out a grid search for the best pa-
rameters, since our main objective is to show the util-
ity of the ML predictors for the classification of pe-
diatric information, rather than building the most ac-
curate model via hyperparameter optimization. How-
ever, parameter tuning could be easily implemented
using a grid search in Python’s Scikit-Learn library.
Next, the trained classifiers were used to predict
labels of all texts in Test 1 and Test 2. Both, kNN and
D-Tree performed well; the kNN classifier reached
F1-score of 0.97 and AUC score of 0.98, while the
D-Tree classifier reached F1-score of 0.98 and AUC
score of 0.99 (Table 2). Out of the three classifiers,
SVM had the best performance, achieving an accu-
racy of 1.00 in Test 1. Considering how Test 1 and
Test 2 were constructed, this result was expected. Test
1 was built from pediatric sections that were very sim-
ilar in their writing style to those in the training set.
In predicting labels in Test 2, D-Tree outper-
formed kNN, and the D-Tree classifier reached F1-
score of 0.92 and AUC score of 0.92, while the kNN
classifier reached F1-score of 0.84 and AUC score
of 0.86. Again, SVM had the best performance. It
reached F1-score of 0.94 and AUC score of 0.94 (Ta-
ble 2). The decrease in the performance of all clas-
sifiers was expected because texts in Test 2 do not
BIOINFORMATICS 2021 - 12th International Conference on Bioinformatics Models, Methods and Algorithms
66

Table 2: Performance of classification models in three validation experiments. Test 1 reports 10-fold cross-validation experi-
ments. Test 2 and Test 3 refer to retrospective and prospective validation, respectively.
Metrics
Test 1 Test 2 Test 3
kNN D-Tree SVM kNN D-Tree SVM kNN D-Tree SVM
Accuracy 0.98 0.98 1.00 0.87 0.93 0.94 0.91 0.89 0.94
Precision 1.00 0.95 1.00 1.00 1.00 1.00 0.99 0.97 1.00
Recall 0.95 1.00 1.00 0.73 0.85 0.88 0.91 0.91 0.94
F1-score 0.97 0.98 1.00 0.84 0.92 0.94 0.95 0.94 0.97
ROC AUC 0.98 0.99 1.00 0.86 0.92 0.94 0.92 0.80 0.96
necessarily come from the PEDIATRIC USE sections
of the SPL files. Although they refer to pediatric in-
formation, these texts originated from other sections
of the drug labels and may contain auxiliary details.
Therefore, these texts are more challenging to classify
yet they are important to detect.
4.5 Experiment 3: Prospective
Validation
In the last experiment, we trained the three classi-
fiers using the entire training set, and predicted la-
bels for all texts in a large Test 3. This experiment
evaluates the scenario closer to the intended appli-
cation in a real setting. In practice, the size of the
data available for training is much smaller compared
to the number of texts to be classified. Thus, by
training with a smaller data set and by predicting la-
bels of a much larger data set, information could be
gained about classifier’s utility. Moreover, our train-
ing data set comprised TFIDF scores computed from
much shorter texts than those present in Test 3. In this
experiment, we observed that kNN outperformed D-
Tree in all metrics, where kNN reached F1 score of
0.95 and AUC score of 0.92, while D-Tree reached
F1 score of 0.94 and AUC score of 0.80. SVM also
classified the input data well, achieving the F1 score
of 0.97 and AUC score of 0.96 (Table 2).
4.6 Evaluation of Feature Importance
Dictionary, which was used for computing TFIDF
scores of texts, comprised 115 n-grams and consisted
of unigrams, bigrams and trigrams. In ML, it is de-
sirable to include the fewest number of the simplest
features to avoid overfitting and to increase the in-
terpretability of the results. Therefore, we examined
the effect of including bigrams and trigrams into the
set of features prior to computing the TFIDF scores.
More specifically, we evaluated if the inclusion of
the higher order n-grams improved classifier’s perfor-
mance. Therefore, we computed two feature sets, one
comprised 115 n-gram TFIDF scores and the second
contained 261 unigram TFIDF scores only. All three
classifiers, kNN, D-Tree, and SVM, were separately
trained with the n-gram TFIDF scores and unigram
TFIDF scores, and the entire training set was used to
build classifiers.
Results showed that with the inclusion of higher
order n-grams, classifiers performed better in most
cases, with the exception of the kNN classifier in Test
2. For instance, the SVM classifier trained with the
higher order n-grams outperformed the SVM trained
with unigrams in all performance metrics (Fig. 4). In
Test 2, the unigram kNN outperformed the n-gram
kNN with F1-score of 0.96 versus 0.84, and the n-
gram D-Tree reached F1-score of 0.92 and AUC score
of 0.92, while unigram D-Tree reached F1-score of
0.86 and AUC score of 0.88. In Test 3, the n-gram
kNN had F1-score of 0.95 and AUC score of 0.92
compared to the unigram kNN with F1 score of 0.96
and AUC score of 0.71. F1-score of the n-gram D-
Tree was 0.94 and AUC score was 0.80, while the un-
igram D-Tree had F1-score of 0.84 and AUC score of
0.74.
We observed that with the unigram TFIDF scores,
SVM was still the best-performing model. Interest-
ingly, kNN results were stronger than those of the
D-Tree classifier. Among the three classifiers, uni-
gram kNN had the best performance in Test 2. Over-
all, these results underscore the importance of using
multi-word tokens for the accurate classification of
pediatric and nonpediatric texts.
Finally, we carried out a feature selection process
using SelectKBest function from Scikit-Learn (Scikit-
Learn, 2019). Because SVM classifier outperformed
the other two methods in all three validation experi-
ments, we applied the feature selection process to the
SVM model only. Additionally, only Test 2 and Test 3
were used in this experiment, because they were more
challenging. Specifically, n-gram features were se-
lected and they were used to train the SVM model,
which was tested with Test 2 and Test 3 data sets. The
number of selected features, denoted as k, was deter-
mined, and the search was done with all possible val-
Mining Biomedical Texts for Pediatric Information
67
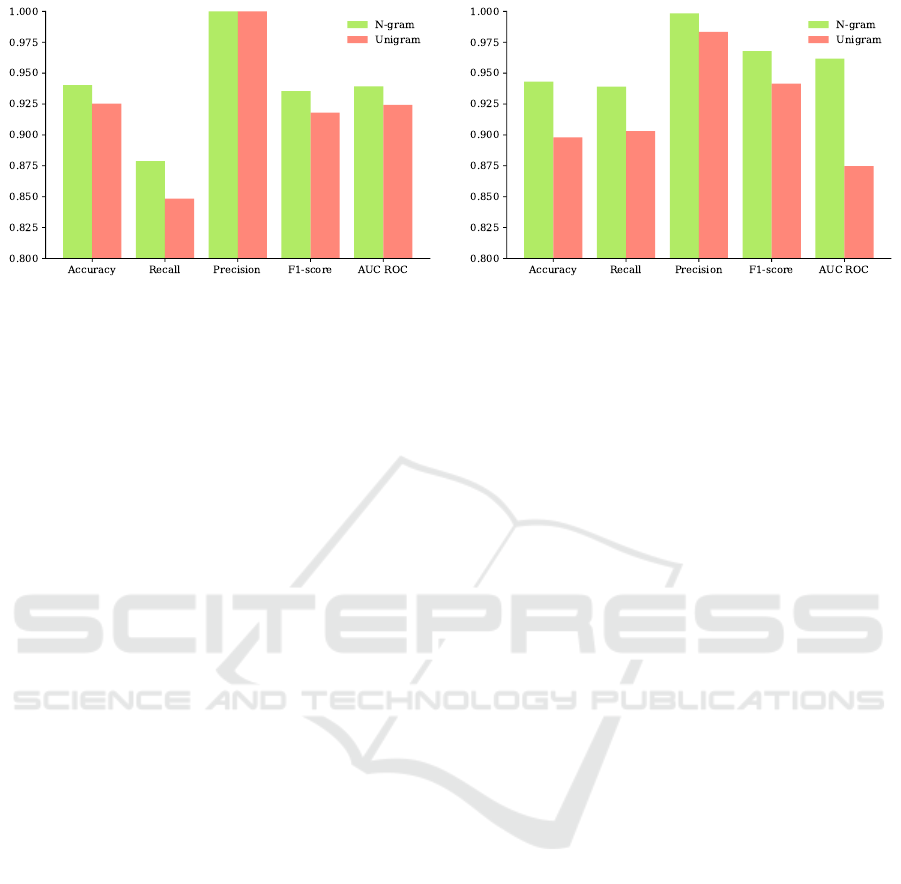
Figure 4: Performance of SVM models trained with n-gram TFIDF versus unigram TFIDF scores. Shown are performance
metrics of SVM models tested on Test 2 (Left) and Test 3 (Right). Green bars denote n-gram SVM model and red bars refer
to unigram SVM model.
ues of k, from 1 to 115. The ANOVA F-values were
computed for each feature and then sorted. The se-
lected k features were the ones with the top k ANOVA
F-values. Our results show that k = 31 is a reasonable
number of features to use (Fig. 5). In Test 2, SVM
achieved an accuracy of 0.94, precision of 1.0, recall
of 0.88, F1-score of 0.94, and AUC score of 0.94. In
Test 3, SVM reached F1-score of 0.96 and AUC score
of 0.96. By further increasing the number of selected
features, for instance to k = 32, the performance on
Test 3 began to decrease. Even though the perfor-
mance of SVM with k = 100 on Test 3 was slightly
better than that of SVM with k = 31, the dimension of
data needed to be increased from 31 to 100, which is
a big change.
5 DISCUSSION
Pediatric drug prescribing must rely on scientific ev-
idence about drugs’ safe and effective use in this
specific population of patients. Yet, drugs are often
prescribed to pediatric patients without this evidence
due to the challenges of pediatric drug development
and evaluation. Stimulated by the legislative actions,
more than 1,200 pediatric labeling changes have been
submitted to the US FDA since 2002 (Green et al.,
2019). Yet, these changes are not easily accessible
nor are they machine-readable. Thus, there is a lag in
how fast the data becomes available to the public.
This information gap is due to several reasons, in-
cluding the lack of the standard machine-readable for-
mat for disseminating such information. For instance,
information about pediatric drug use may appear in
several sections of drug product labels, or it may be
tagged differently altogether. Even when found in a
well tagged section of a drug label, pediatric infor-
mation may describe patients using diverse keywords
such as neonates, infants, children and adolescents or
using specific ages, such as 12, for example. This
makes keyword extraction challenging. Finally, while
drug developers and regulators often mine regulatory
and scientific data as well as data from the electronic
health records, insurance claims and so on, such data
are not freely and readily available to academic re-
searchers and consumer scientists.
To construct a repository of pediatric informa-
tion, text documents must undergo manual curation,
a time-intensive and labor-intensive process. In an ef-
fort to address the paucity of information about drugs’
safety and efficacy in pediatric populations, a differ-
ent approach is needed. We propose to expedite this
process by a high throughput text classification us-
ing ML algorithms. Our work aims to streamline data
analysis by identifying relevant pediatric texts in drug
labels, which are updated at a rate of 500 per day.
Tested under three different scenarios, ML predic-
tors showed encouraging results in differentiating be-
tween pediatric and nonpediatric information found
in SPL files. We selected simple yet interpretable ML
methods to construct text classifiers, namely, kNN, D-
Tree and SVM. Among these three methods, SVM
outperformed the other two in all validation experi-
ments (Table 2). These validation experiments ranged
from the 10-fold cross-validation to retrospective val-
idation using small test sets, to prospective validation
using a large collection of documents obtained at a
later time point.
More specifically, the SVM classifier trained with
the multi-word tokens achieved high accuracy of
94%, excellent precision (1.00) and high recall of
0.94% (Table 2). SVM classifiers execute very fast
and do not require expensive hardware for text pro-
cessing. Our results show that the number of multi-
word tokens can be reduced from 115 to 31 without
the loss in accuracy, making the process of classifica-
BIOINFORMATICS 2021 - 12th International Conference on Bioinformatics Models, Methods and Algorithms
68
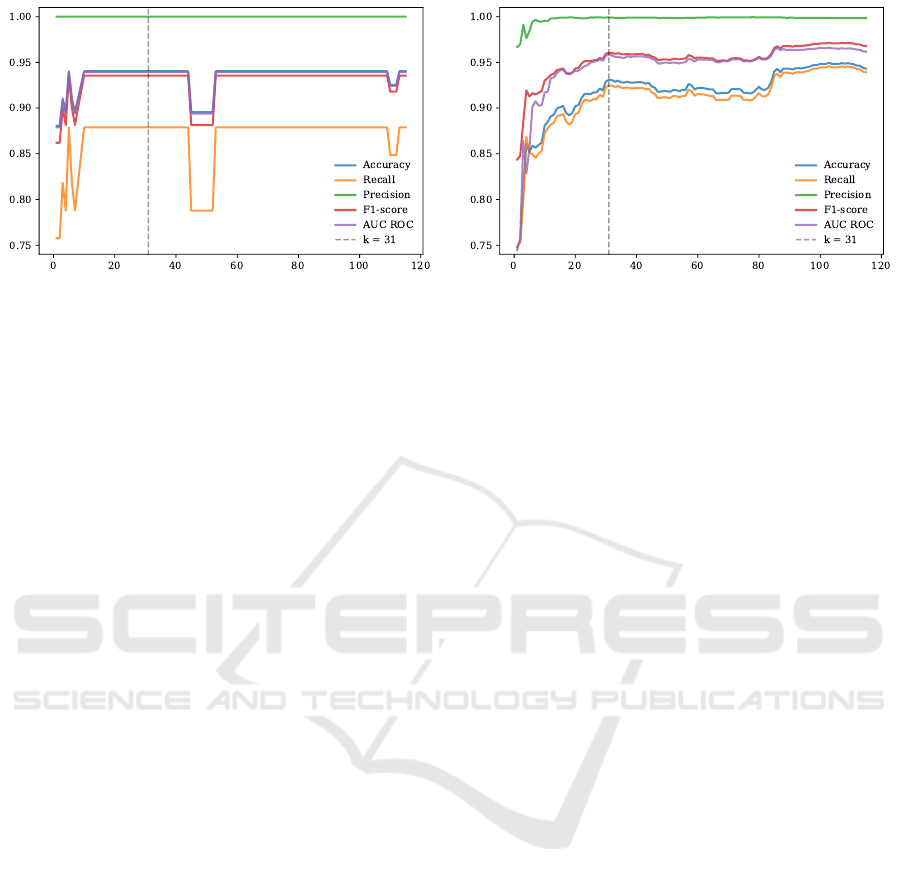
Figure 5: Feature Selection using SelectKBest function. X-axis represents the number of features selected, denoted as k.
Y-axis represents the values of performance metrics of SVM model trained with k features on Test 2 (Left) and Test 3 (Right).
The gray dotted line represents a selected number of features to use, k = 31.
tion even faster (Fig. 5).
ML classifiers have become ubiquitous and we in-
vestigated their potential in our specific application
domain. To overcome the paucity of a well-curated
training data set, we used texts extracted from 494
randomly sampled SPL files. Training data set was
constructed by extracting texts relevant to pediatric
use using a LOINC identifier. LOINC identifiers have
been adopted by the FDA as a standard way of for-
matting the drug product labels.
We decided to train classifiers with a smaller data
set, although current public SPL archives contain
thousands of files. The motivation behind this was
that, in a practical setting, the size of the labeled data
is significantly smaller than the size of the data that
needs to be classified. For instance, if the classifier
were to be applied to the data from the electronic
health records, the number of texts to be screened
would be far greater than the number of the training
texts.
Despite high classification accuracy, some issues
may arise from training with a small data set. These
issues include outliers, missing values, overfitting and
sampling bias. Overfitting was observed, it is seen
in the decreased classification performance between
cross-validated estimates and testing results. For ex-
ample, the AUC score of SVM classifiers dropped
from the 0.98 (cross-validation) to 0.96 (Test 3) and
0.94 (Test 2). Similar patterns were observed in test-
ing of the kNN and D-Tree classifiers. The estimated
AUC score of the kNN classifier decreased from 0.96
in cross-validation to 0.86 for Test 2 and 0.92 for Test
3. Likewise, there was a 0.02% decrease between
cross-validated and testing AUC scores for D-Tree in
Test 2, and even greater decrease of 0.16% in Test 3
(Fig. 3 and Table 2).
Overfitting is a common problem in ML, and it
can be somewhat remedied in several ways, rang-
ing from a manual data review to a consensus voting
on the predicted class. Avoiding extensive parameter
tuning and selecting the simplest, interpretable mod-
els are the two approaches which we selected to pur-
sue. To train the ML classifiers, all parameters were
set to their default values (Section 3), and we used
three algorithms that are simple to construct yet yield
explainable results. Although overfitting is seen with
all three methods, SVM model outperforms the other
two approaches, and it seems to be the most stable in
its performance. Moving forward, we expect to in-
clude texts from other biomedical texts in our training
data set, aiming to reduce the overfitting by increasing
the diversity of texts.
Some limitations exist in the current work. First,
classifiers were validated only with texts extracted
from the drug product labels. The vocabulary and the
semantics of these texts are tailored for the regulatory
submissions. However, it is desirable to apply our
method to any text, such as biomedical research liter-
ature, clinical trials data, and even social media. The
SVM model can be periodically retrained using ad-
ditional data sources, thus, increasing model’s appli-
cability domain. For instance, one could use the cur-
rent model to classify texts from other sources, then
review predicted labels manually and add newly la-
beled texts to the training set for the creation of a new
model.
Second, classifiers were trained using pediatric
texts extracted using LOINC identifier for PEDIATRIC
USE, which may introduce a bias into the derivation
of features, in form of the n-grams derived from these
texts. These LOINC identifiers were absent in older
SPL files or the SPL files may have been tagged dif-
ferently. It may be possible that the vocabulary and
the semantics of the older texts differ from those that
were submitted to the FDA more recently. This is in-
deed confirmed by our results, in the retrospective and
prospective validation. For instance, Test 2 was con-
structed by scanning through the entire SPL file with
Mining Biomedical Texts for Pediatric Information
69

a regular expression containing the word “pediatric”.
Thus, texts in Test 2 may have a different sentence
structure than those in the training set. There was also
a noticeable decrease in the accuracy of the trained
classifiers when they were tested with Test 3, which
may be explained by a much richer vocabulary found
in Test 3. More specifically, the number of unique n-
grams computed for Test 3 was 424 compared to only
115 found in the training set. On the other hand, all
classifiers performed strongly in Test 1, which resem-
bled training set very closely. This underscores the
importance of testing classifiers with a variety of the
test data sets, obtained from different sources, as has
been proposed in this work.
To address the potential concerns about the small
size of the training data set, we conducted the follow-
ing experiment. We trained all three classifiers using
Test 3 data set of 31,565 texts, and tested these clas-
sifiers using the data set comprising 1,931 texts. We
note that these two data sets do not overlap, that is
they do not have any texts in common. In this experi-
ment, all three classifiers were able to perfectly divide
pediatric and nonpediatric texts; all performance met-
rics were 1.00. We point out that these results may
not be representative of the future application of the
current method, as is shown by our prospective vali-
dation (Table 2).
6 CONCLUSION
Rapid and accurate data acquisition and collection is
a prerequisite step in the development of methods
and tools in bioinformatics and biomedical data sci-
ences. Often, data collection is done manually, requir-
ing scientists to read and annotate large libraries of
biomedical and life science publications. We demon-
strated a viable approach to expedite the data col-
lection process by combining tools from text mining
and machine learning. We applied our approach to
an important problem of identifying texts that con-
tain information relevant to the safety and efficacy of
drugs in pediatric patients. This vulnerable popula-
tion of patients is not included in the clinical stud-
ies of drugs, and remains exposed to the “off-label”
prescribing. Such exposure is due to the insufficient
evidence about drugs’ safety and efficacy in pediatric
patients, arising from the small size of pediatric study
groups, stratification of the pediatric age groups and
differences in the development and maturation of pe-
diatric patients (Mulugeta et al., 2018). Additionally,
existing computational tools are mostly targeted to-
wards the analyses of averaged and age-agnostic data
sets and molecular processes.
We designed, implemented and evaluated a text
processing pipeline based on machine learning and
showed that despite the diversity in formats, styles
and terms, our SVM classifier can accurately pre-
dict whether they contain the information relevant
for pediatric prescribing. The binary SVM classifier
achieved high accuracy of 0.98 in the 10-fold cross-
validation experiments, where it outperformed two
other ML classifiers, kNN and D-Tree. In two ad-
ditional validation experiments, the SVM model also
achieved high classification accuracy of 0.94, again
outperforming the other two predictors. Our experi-
mental results indicate that it is important to train clas-
sifiers with features derived from a dictionary of un-
igrams, bigrams and trigrams rather than from single
words. Although a more powerful machine learning,
such as deep learning, could be used instead of SVM,
we opted for the less complex and interpretable mod-
els. Future work will focus on the applications of the
trained SVM classifier in profiling of biomedical lit-
erature and clinical trials with the goal of extracting
new knowledge.
ACKNOWLEDGEMENTS
The authors thank Wendy Lee and Ching Seh Mike
Wu for helpful discussions.
REFERENCES
Bisgin, H., Liu, Z., Fang, H., Xu, X., and Tong, W. (2011).
Mining fda drug labels using an unsupervised learn-
ing technique-topic modeling. BMC bioinformatics,
12:S11.
Chang, C.-C. and Lin, C.-J. (2011). Libsvm: A library
for support vector machines. ACM Trans. Intell. Syst.
Technol., 2(3):27:1–27:27.
Cover, T. and Hart, P. (2006). Nearest neighbor pattern clas-
sification. IEEE Trans. Inf. Theor., 13(1):21–27.
DailyMed (2019). (Accessed on 8/31/2019).
Demner-Fushman, D., Fung, K. W., Do, P., Boyce, R. D.,
and Goodwin, T. R. (2018a). Overview of the tac
2018 drug-drug interaction extraction from drug la-
bels track. In Proceedings of the Text Analysis Con-
ference (TAC 2018).
Demner-Fushman, D., Shooshan, S. E., Rodriguez, L.,
Aronson, A. R., Lang, F., Rogers, W., Roberts, K.,
and Tonning, J. (2018b). A dataset of 200 structured
product labels annotated for adverse drug reactions.
Scientific data, 5:180001.
Deshmukh, S. and Khuri, N. (2018). Pediatricdb: Data ana-
lytics platform for pediatric healthcare. In 2018 Thir-
teenth International Conference on Digital Informa-
tion Management (ICDIM), pages 216–221. IEEE.
BIOINFORMATICS 2021 - 12th International Conference on Bioinformatics Models, Methods and Algorithms
70

Everitt, B. (2005). Classification and Regression Trees.
Chapman & Hall.
Fan, R.-E., Chen, P.-H., and Lin, C.-J. (2005). Working
set selection using second order information for train-
ing support vector machines. J. Mach. Learn. Res.,
6:1889–1918.
Fang, H., Harris, S. C., Liu, Z., Zhou, G., Zhang, G., Xu, J.,
Rosario, L., Howard, P. C., and Tong, W. (2016). Fda
drug labeling: Rich resources to facilitate precision
medicine, drug safety, and regulatory science. Drug
discovery today, 21(10):1566–1570.
FDASIA (2012). (Accessed on 5/21/2020).
Fiks, A. G., Scheindlin, B., and Shone, L. (2016). 30th
anniversary of pediatric research in office settings
(pros): An invitation to become engaged. Pediatrics,
138(3):e20161126.
Fung, K. W., Jao, C. S., and Demner-Fushman, D. (2013).
Extracting drug indication information from struc-
tured product labels using natural language process-
ing. Journal of the American Medical Informatics As-
sociation : JAMIA, 20(3):482–488.
Gnanasakthy, A., Barrett, A., Evans, E., D’Alessio, D., and
Romano, C. D. (2019). A review of patient-reported
outcomes labeling for oncology drugs approved by
the fda and the ema (2012-2016). Value in Health,
22(2):203–209.
Green, D. J., Sun, H., Burnham, J., Liu, X. I., van den
Anker, J., Temeck, J., Yao, L., McCune, S. K., and
Burckart, G. J. (2019). Surrogate endpoints in pedi-
atric studies submitted to the us fda. Clinical Phar-
macology & Therapeutics, 105(3):555–557.
HL7 Standards (2019). (Accessed on 3/14/2019).
Hoon, D., Taylor, M. T., Kapadia, P., Gerhard, T., Strom,
B. L., and Horton, D. B. (2019). Trends in off-label
drug use in ambulatory settings: 2006–2015. Pedi-
atrics, 144(4).
Hu, L.-Y., Huang, M.-W., Ke, S.-W., and Tsai, C.-F.
(2016). The distance function effect on k-nearest
neighbor classification for medical datasets. Springer-
Plus, 5(1):1304–1304.
Ito, S. (2017). Drugs for children. Clinical Pharmacology
& Therapeutics, 101(6):704–706.
Khare, R., Wei, C.-H., and Lu, Z. (2014). Automatic ex-
traction of drug indications from fda drug labels. In
AMIA Annual Symposium Proceedings, volume 2014,
page 787. American Medical Informatics Association.
Kuhn, M., Letunic, I., Jensen, L. J., and Bork, P. (2016).
The sider database of drugs and side effects. Nucleic
acids research, 44(D1):D1075–D1079.
Li, Q., Deleger, L., Lingren, T., Zhai, H., Kaiser, M.,
Stoutenborough, L., Jegga, A., Cohen, K., and Solti,
I. (2013). Mining fda drug labels for medical condi-
tions. BMC medical informatics and decision making,
13:53.
Lowenthal, E. and Fiks, A. G. (2016). Protecting children
through research. Pediatrics, 138(4):e20162150.
lxml (2019). (Accessed on 2/18/2019).
Mehta, D., Uber, R., Ingle, T., Li, C., Liu, Z., Thakkar,
S., Ning, B., Wu, L., Yang, J., Harris, S., et al.
(2020). Study of pharmacogenomic information in
fda-approved drug labeling to facilitate application of
precision medicine. Drug Discovery Today.
Mulugeta, L. Y., Yao, L., Mould, D., Jacobs, B., Florian, J.,
Smith, B., Sinha, V., and Barrett, J. S. (2018). Lever-
aging big data in pediatric development programs:
Proceedings from the 2016 american college of clini-
cal pharmacology annual meeting symposium. Clini-
cal Pharmacology & Therapeutics, 104(1):81–87.
N
´
ev
´
eol, A. and Lu, Z. (2010). Automatic integration of
drug indications from multiple health resources. In
Proceedings of the 1st ACM International Health In-
formatics Symposium, IHI’10, pages 666–673, New
York, NY, USA. Association for Computing Machin-
ery.
Pandey, A., Kreimeyer, K., Foster, M., Dang, O., Ly, T.,
Wang, W., Forshee, R., and Botsis, T. (2019). Ad-
verse event extraction from structured product labels
using the event-based text-mining of health electronic
records (ether) system. Health informatics journal,
25(4):1232–1243.
Prospective and retrospective cohort studies (2019). (Ac-
cessed on 11/23/2019).
Refaeilzadeh, P., Tang, L., and Liu, H. (2009). Cross-
validation. Encyclopedia of Database Systems, pages
532–538.
Rodriguez, L. M. and Fushman, D. D. (2015). Automatic
classification of structured product labels for preg-
nancy risk drug categories, a machine learning ap-
proach. In AMIA Annual Symposium Proceedings,
volume 2015, page 1093. American Medical Infor-
matics Association.
Scikit-Learn (2019). (Accessed on 4/19/2019).
Structured product labeling (2019). (Accessed on
3/15/2019).
Tiftikci, M.,
¨
Ozg
¨
ur, A., He, Y., and Hur, J. (2019). Machine
learning-based identification and rule-based normal-
ization of adverse drug reactions in drug labels. BMC
bioinformatics, 20(21):1–9.
U.S. Congress. Best Pharmaceuticals for Children Act
Amending Section 505A of the Federal Food, Drug &
Cosmetic Act (Public Law 107-109). (2002) (2002).
(Accessed on 5/4/2020).
U.S. Congress. Pediatric Research Equity Act amending
Section 505B of the Federal Food, Drug & Cosmetic
Act (Public Law 108-155). (2003) (2003). (Accessed
on 5/4/2020).
U.S. Food and Drug Administration. New pediatric labeling
information dataset (2020). (Accessed on 5/20/2020).
Mining Biomedical Texts for Pediatric Information
71
