
Computer-aided Abnormality Detection in Chest Radiographs in a
Clinical Setting via Domain-adaptation
∗,†
Abhishek K. Dubey
a
, Michael T. Young, Christopher Stanley, Dalton Lunga and Jacob Hinkle
Oak Ridge National Laboratory, Oak Ridge, TN, U.S.A.
Keywords:
Computer-aided Diagnosis of Lung Conditions, Domain-shift Detection and Removal, Chest Radiographs.
Abstract:
Deep learning (DL) models are being deployed at medical centers to aid radiologists for diagnosis of lung
conditions from chest radiographs. Such models are often trained on a large volume of publicly available
labeled radiographs. These pre-trained DL models’ ability to generalize in clinical settings is poor because
of the changes in data distributions between publicly available and privately held radiographs. In chest ra-
diographs, the heterogeneity in distributions arises from the diverse conditions in X-ray equipment and their
configurations used for generating the images. In the machine learning community, the challenges posed by
the heterogeneity in the data generation source is known as domain shift, which is a mode shift in the genera-
tive model. In this work, we introduce a domain-shift detection and removal method to overcome this problem.
Our experimental results show the proposed method’s effectiveness in deploying a pre-trained DL model for
abnormality detection in chest radiographs in a clinical setting.
1 INTRODUCTION
Chest radiography is one of the most ubiquitous di-
agnostic modalities for cardiothoracic and pulmonary
abnormalities in the clinical setting. A timely diag-
nostic based on the radiographs is a critical step in the
clinical workflow. However, many healthcare centers
often suffer either from a heavy workload or shortage
of experienced radiologists. Deployment of a reliable
abnormality detection system would be advantageous
in both scenarios. Deep learning (DL) based abnor-
mality detection systems are an emerging technology,
which is yet to be successfully deployed in clinical
settings. The domain shift encountered in privately
a
https://orcid.org/0000-0001-8052-7416
∗
Biomedical Science, Engineering, and Computing Group,
Oak Ridge National Laboratory, Oak Ridge, USA
†
This manuscript has been authored by UT-Battelle, LLC
under Contract No. DE-AC05-00OR22725 with the U.S.
Department of Energy. The United States Government re-
tains and the publisher, by accepting the article for publi-
cation, acknowledges that the United States Government
retains a non-exclusive, paid-up, irrevocable, world-wide
license to publish or reproduce the published form of the
manuscript, or allow others to do so, for United States Gov-
ernment purposes. The Department of Energy will pro-
vide public access to these results of federally sponsored
research in accordance with the DOE Public Access Plan
(http://energy.gov/downloads/doe-public-access-plan).
held datasets due to heterogeneity in data generation
sources continue to be a prime impediment when de-
ploying pre-trained DL models. In this work, we in-
troduce a domain-shift detection and removal method
to deploy pre-trained DL models in clinical settings.
Domain-shift in this context is formally defined as
the changes in the marginal probability density p(x)
between privately held chest radiographs and publicly
available radiographs. The goal of domain-shift de-
tection is to quantify the changes in the marginal p(x).
While training a model on a public labeled data source
{x
i
, y
i
}
n
i=0
, the best hope is to learn the conditional
probability p(y|x) that is stable or varies smoothly
with the marginal p(x). Even if the conditional is sta-
ble, learned models may suffer from model misspec-
ification, i.e., the learned model may not perfectly
capture the functional relationship between x and y
and the approximate solution may become sensitive to
changes in p(x). The goal of domain-shift removal is
to find a transformation B of the data that minimizes
the difference between marginal distributions of the
transformed samples B(x) of privately-held data and
public data to reduce the effect of sensitivity on pre-
diction.
Domain separation (Bousmalis et al., 2016) pro-
vides a competing pioneering technique to handle
domain-shift. Domain separation aims to separate
the feature representation between publicly available
Dubey, A., Young, M., Stanley, C., Lunga, D. and Hinkle, J.
Computer-aided Abnormality Detection in Chest Radiographs in a Clinical Setting via Domain-adaptation.
DOI: 10.5220/0010302500650072
In Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2021) - Volume 2: BIOIMAGING, pages 65-72
ISBN: 978-989-758-490-9
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
65
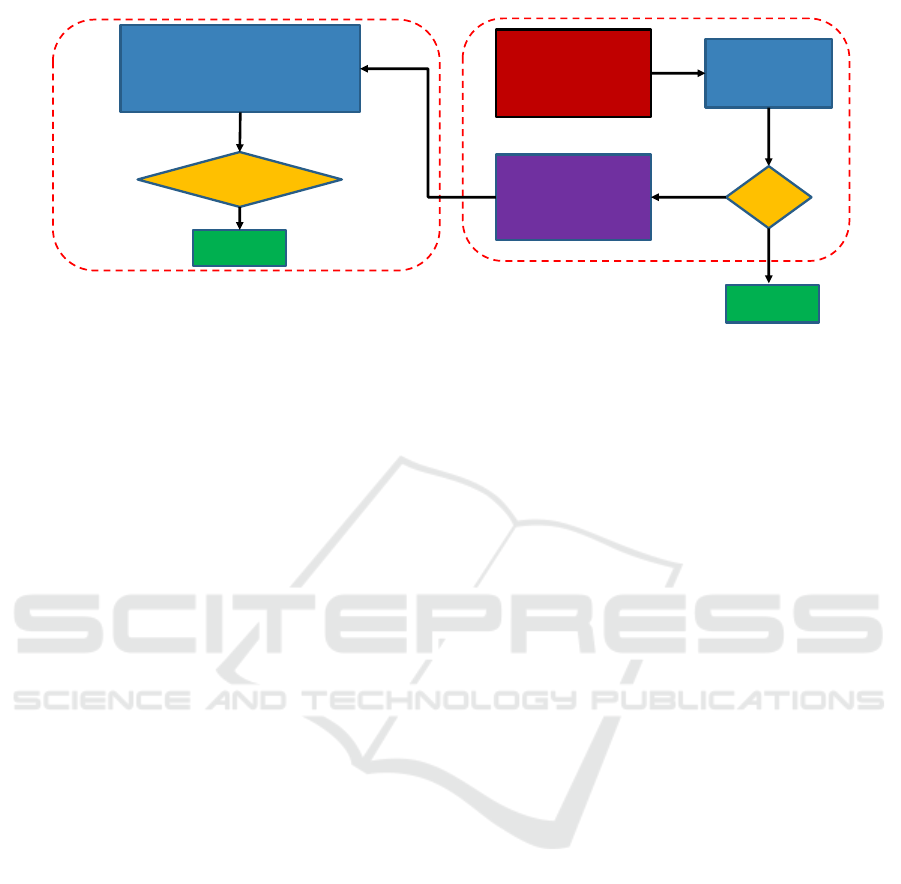
Download pretrained DL
model (trained on public
dataset)
Perform B-test for
domain shift
detection
Shift
?
Domain shift removal
from privately held
dataset
No
Yes
No
adaptation
Domain shift detection and removal
Performance evaluation with and
without domain shift removal on
privately held dataset
Improved
Performance?
Success
Evaluation
Clinical workflow
METHOD, 3
Figure 1: Clinical workflow for computer-aided abnormality detection in chest radiographs.
and privately held radiographs into domain-invariant
and domain-specific features. Domain-shift removal
and predictive modeling are tightly coupled in the do-
main separation technique, which requires non-trivial
changes in predictive models to overcome domain-
shift. This paper introduces a novel workflow for
deploying the state-of-the-art pre-trained DL model
for abnormality detection in clinical settings with-
out requiring any changes in network architecture
to overcome domain-shift. Figure 1 shows the pro-
posed workflow. In this workflow, we first char-
acterize domain-shift between samples of privately
held and public chest radiographs. In particular, we
show that the two sources differ in the distribution
of high-frequency components such as noise and tex-
ture, which we characterize by the density of wavelet
scattering transform of radiographs. Then we learn
a generative adversarial network to map samples of
a privately held dataset to match their style distribu-
tion to that of public chest radiographs. To evaluate
the workflow, we assess the pre-trained DL model’s
performance on privately held radiographs for abnor-
mality detection with and without the domain-shift re-
moval step.
2 RELATED WORK
In this context, one approach is to learn a transfor-
mation that embeds data into domain invariant fea-
ture space, which has domain generalization ability to
previously unseen domains. Domain invariant com-
ponent analysis (DICA) (Muandet et al., 2013) is
among such methods in the literature. DICA assumes
that data samples come from various unknown dis-
tributions and it estimates the distributional variance
from the data sources. DICA then finds the orthog-
onal transform B onto a low-dimensional subspace
that minimizes the distributional variance while pre-
serving the functional relationship between samples
and class-labels. However, such methods require data
samples coming from various unknown distributions
to estimate the distributional variance.
Another method in this category includes domain
invariant variational autoencoder (DIVA) (Ilse et al.,
2019). DIVA extends the variational autoencoder
framework by disentangling latent representations for
a domain label (z
d
), a class label (z
y
) and any resid-
ual variations in the inputs (z
r
). This work claimed
to learn a domain-invariant representation using semi-
supervised training utilizing the labeled and unlabeled
data from both domains. They used three separate en-
coders q
φ
z
d
(z
d
|x), q
φ
z
y
(z
y
|x) and q(φ
z
r
)(z
r
|x) and an
additional parameterized neural network p
θ(x|z
d
,z
y
,z
r
)
as a decoder. Their work looks promising for the
domain-adaptation task in general. However, their
network architecture has non-trivial differences from
the existing state-of-the-art architecture developed for
the abnormality detection in chest radiographs.
In this work, we propose a workflow to facili-
tate the use of state-of-the-art DL architecture via
domain-shift removal from the privately held radio-
graphs. The domain-shift removal problem broadly
falls in the computer vision community under the un-
paired image-to-image translation category. We iden-
tify the changes in noise and texture characteristics of
radiographs as the main difference between the data
sources. We use CycleGAN (Zhu et al., 2017) for
removing these differences through image-to-image
translation. CycleGAN improves upon generative ad-
versarial networks by exploiting cycle consistency
property (Dubey et al., 2018; Iliopoulos et al., 2019;
Dubey, 2018) in the forward and backward translation
maps to avoid mode collapse in the process of image-
to-image translation.
BIOIMAGING 2021 - 8th International Conference on Bioimaging
66

3 METHOD
3.1 Domain-shift Detection
The goal of domain-shift detection is to identify the
shift in the marginal probability density p(x) be-
tween two domains X and Y given training samples
{x
i
}
N
i=1
and {y
i
}
N
i=1
, where x
i
∈ X and y
i
∈ Y. We
denote the true marginal probability density of two
datasets as x ∼ p
x
(x) and y ∼ p
y
(y). We formulate the
domain-shift detection as a hypothesis testing prob-
lem, whether to accept the null hypothesis that there
is no domain-shift H
0
: p
x
= p
y
or to accept the al-
ternative hypothesis that there is a domain-shift H
1
:
p
x
6= p
y
. The hypothesis testing often suffers from the
curse of dimensionality in high-dimensional data set-
tings in estimating test statistic. In this work, we use
a kernel two-sample test initially proposed by (Gret-
ton et al., 2012), which addressed the problem posed
by high-dimensional data settings by introducing the
maximum mean discrepancy (MMD) as test statistic.
The MMD is a distance-measure between probability
densities and is defined as the largest difference in ex-
pectations between the two probability distributions
over functions in the unit ball of a suitable reproduc-
ing kernel Hilbert space (RKHS). The MMD can be
empirically estimated between the probability density
p
x
and p
y
by the squared distance between their mean
embeddings in the RKHS as
η
k
(p
x
, p
y
) = kµ
k
(p
x
) −µ
k
(p
y
)k
2
H
k
, (1)
where µ
k
(p
x
) and µ
k
(p
y
) are mean embedding of p
x
and p
y
, and H
k
is an RKHS with reproducing kernel
k. In this work, we use the B-test statistic as an MMD
estimate proposed by (Zaremba et al., 2013). The B-
test statistics is an MMD estimate obtained by averag-
ing the
ˆ
η
k
(i), where each
ˆ
η
k
(i) is the empirical MMD
based on a subsample of size B. The asymptotic distri-
bution for
ˆ
η
k
under H
0
and H
1
are shown to be Gaus-
sian in (Zaremba et al., 2013). Following (Zaremba
et al., 2013), we set the subsample size B to
√
n to ob-
tain a consistent estimator. A user-defined threshold
α, which denotes the test level, is used to determine
whether the test statistic is sufficiently large as to ac-
cept the alternative hypothesis H
1
, that is a shift in the
marginal distributions p
x
and p
y
.
In this work, we use a fixed convolutional neural
network called Wavelet scattering transform in com-
position with the radial basis function as the kernel
function. The Wavelet scattering transform is used
to extract the features that are invariant to transla-
tion and Lipschitz stable to deformation. The higher-
order wavelet scattering transform is shown to char-
acterize the noise and texture in the signal by (Bruna
and Mallat, 2013). We use the scattering transform
to capture this high-frequency component of the ra-
diographs essentially, which is the characteristic dif-
ference between the domains. Then we use the radial
basis kernel to map the scattering coefficient to the
kernel space to find the B-test statistics.
Next we identify the out-of-distribution (OOD)
samples that require domain-shift removal. For this
purpose, we empirically estimate the density of the
samples from the source and target domain in a low-
dimensional subspace spanned by the principal com-
ponents of the scattering coefficients. We identify the
samples that are in the non-overlapping region be-
tween the source and target domains as potential can-
didates for the domain-shift removal.
3.2 Domain-shift Removal
The goal of the domain-shift removal is to learn a
mapping G : X → Y from the privately held dataset
domain X to publicly available dataset domain Y . We
use the state-of-the-art method, CycleGAN (Zhu et al.,
2017), to perform this task. CycleGAN additionally
learns the reverse mapping F : Y → X and two adver-
sarial discriminators D
X
and D
Y
in conjunction with
F from the unpaired samples from X and Y as shown
in Figure 2. CycleGAN enforces inverse consistency
conditions, F ◦G = G ◦F = , between the two maps,
where is an identity map. Additionally, the discrim-
inator D
X
is learned to distinguish between the real
images {x ∈ X } and translated images {F(y), y ∈Y }
and similarly D
Y
is learned to discriminate between
{y ∈Y } and {G(x), x ∈X}.
4 EXPERIMENTAL SETUP
This section describes two chest radiograph datasets,
presents their noise and texture characterization, and
describes the experimental set-up for abnormality de-
tection.
4.1 Dataset Description
We present abnormality detection results on MIMIC-
CXR dataset. MIMIC-CXR is a publicly available chest
radiograph in Digital Imaging and Communications
in Medicine (DICOM) format. The diagnosis labels
are derived from the radiology reports associated with
these images. The dataset contains radiographs as-
sociated with 227, 827 patients collected at the Beth
Israel Deaconess Medical Center between 2011 and
2016. The dataset is de-identified to satisfy the Health
Computer-aided Abnormality Detection in Chest Radiographs in a Clinical Setting via Domain-adaptation
67
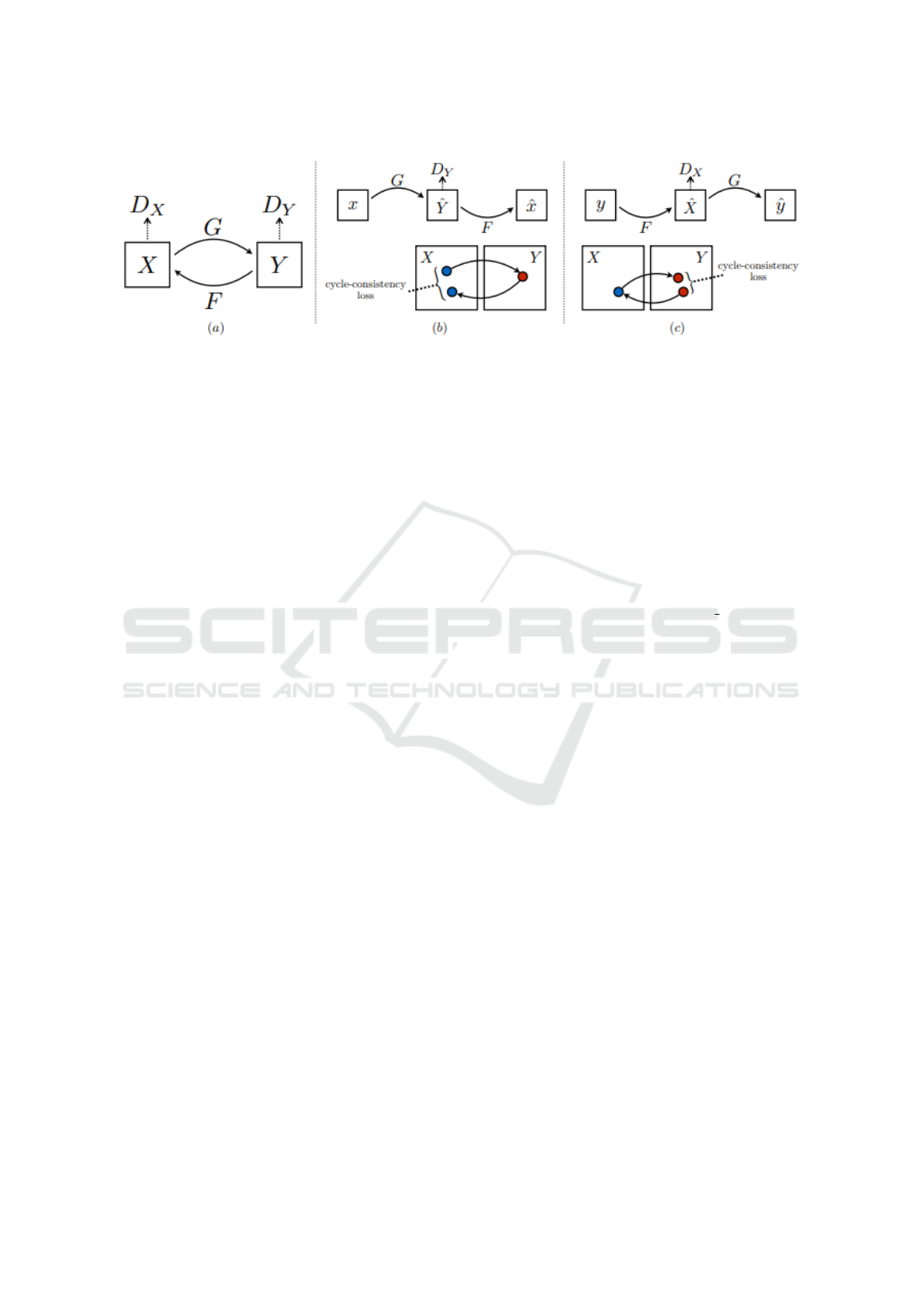
Figure 2: CycleGAN contains mappings between two domains G : X →Y and F : Y → X , and one discriminator for each
domain, D
X
and D
Y
. The purpose of including discriminators is to encourage the generators G and F to generate samples that
can not be indistinguished with the available real samples from the two domains. Additionally, CycleGAN introduced cycle
consistency losses to enforce forward and backward cyclic consistency between the generators, i.e., x →G(x) →F(G(x)) ≈x,
and y → F(y) →G(F(y)) ≈y. We have included this figure into this manuscript from the CycleGAN paper (Zhu et al., 2017).
Insurance Portability and Accountability Act require-
ments, and protected health information are removed.
We converted the DICOM file format (16-bit depth
raw format) to JPEG file format (8-bit depth raw for-
mat) using the pydicom library and downsample the
radiographs to 256 ×256 pixels for further analysis.
We normalized the dynamic range of the images to
[0, 255] by the following steps: (i) subtracting the im-
age pixel values with the lowest pixel value in the im-
age, (ii) dividing the image pixel values by the high-
est pixel value and multiplying pixel values by 255
in the image, (iii) truncating and converting the result
to an unsigned integer. Finally, we stored the radio-
graphs in the compressed JPEG format with a quality
value of 95. We did not perform any filtering or pre-
processing of the images before storing them in JPEG
format.
We used a pre-trained DenseNet121 (Tang
et al., 2020) for the abnormality detection, which
was trained on another publicly available dataset,
ChestXray14 (Wang et al., 2017), released by the
National Institute of Health. This dataset provides
112, 120 radiographs from 30, 805 patients in PNG
format at 1024 ×1024 resolution. The dataset was
rigorously screened to remove all personally iden-
tifiable information. The ChestXray14 radiographs
have the same dynamic range of [0, 255]. We down-
sample the radiographs to 256 ×256 pixels to make
it consistent with MIMIC-CXR. We use ChestXray14
to learn an image-to-image translation model be-
tween the samples of MIMIC-CXR and ChestXray14,
which we use for removing the domain-shift from
out-of-the-distribution samples of MIMIC-CXR. We
did not perform any filtering or pre-processing to
ChestXray14 radiographs before using it for MIMIC-
CXR to ChestXray14 translation.
4.2 Domain-shift Characterization
We characterized the noise and texture of MIMIC-CXR
and ChestXray14 by computing the distribution of
wavelet scattering transforms of the datasets. We used
Scattering2D method from Kymatio package (An-
dreux et al., 2020) for computing the scattering trans-
form. We computed up to the second-order of the
scattering coefficients by setting max order=2. We
set the filter parameters J=4 and L=8 while maintain-
ing other parameters to default values. We summed
the scattering coefficients over the image domain to
obtain a translational invariant feature. This way,
we extracted 417 coefficients for every image in the
two datasets. We whitened the coefficients and re-
duced their dimensionality using the principal com-
ponent analysis and estimated data distribution in re-
duced space. We estimated the distribution by binning
the coefficient space [−4, 4] × [−4, 4] into 50 × 50
bins and counting the samples in every bin for both
datasets. Figure 4 shows the count of samples of
MIMIC-CXR and ChestXray14 in the binned area. A
domain-shift between MIMIC-CXR and ChestXray14
is evident from the figure.
We performed a two-sample test (B-
test) (Zaremba et al., 2013) on the extracted
wavelet scattering coefficients with radial basis
function kernel. With the kernel scale parameter
γ=1, we get a p-value=0 for the two-sample kernel
test, indicating an overwhelming support for the
hypothesis that the two datasets come from different
distributions. Figure 3 shows the supporting statistics
in the B-test for the null and alternative hypotheses.
BIOIMAGING 2021 - 8th International Conference on Bioimaging
68
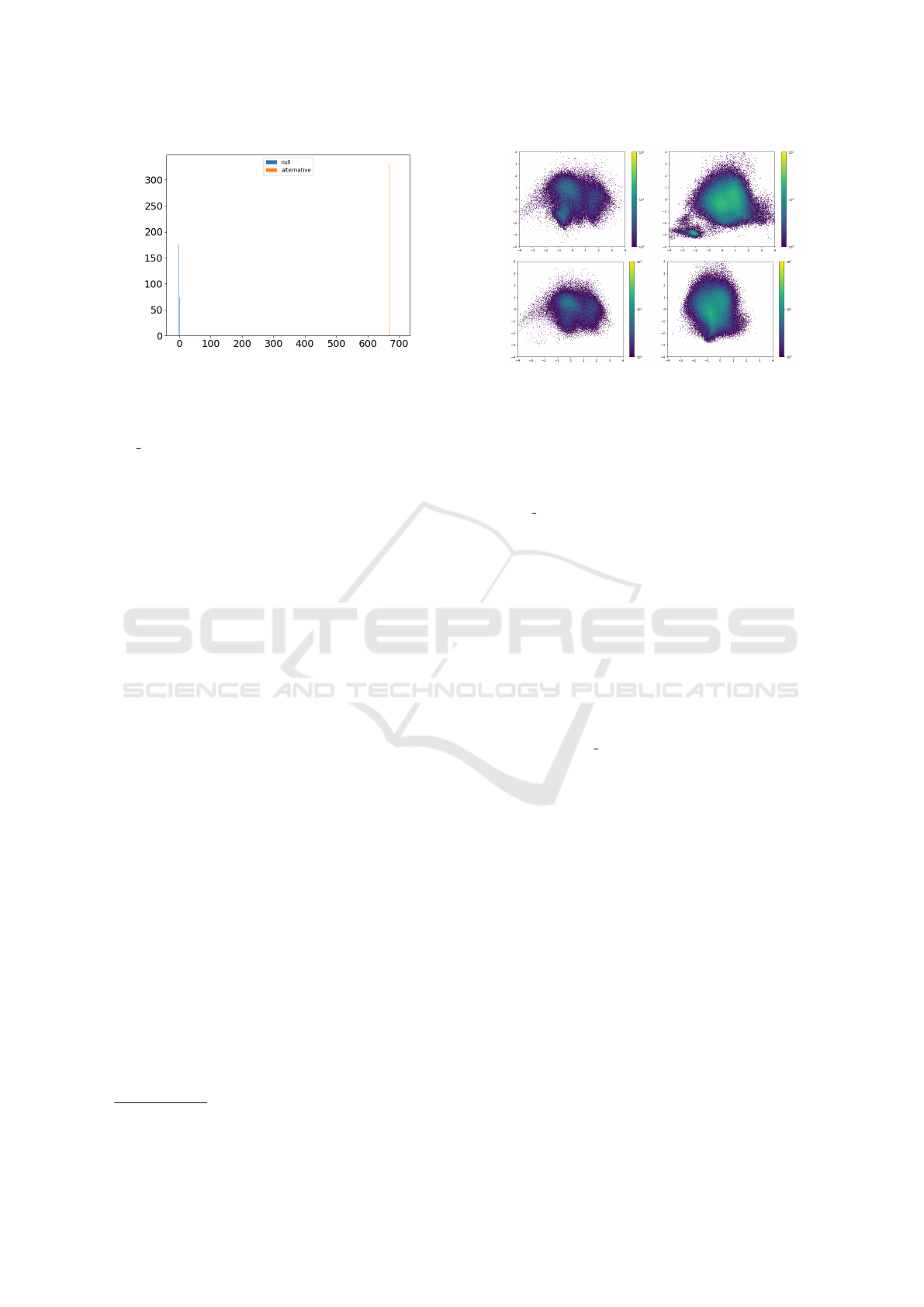
Figure 3: Empirical MMD distributions under null (H
0
) and
alternative (H
1
) hypothesis between the ChestXray14 and
MIMIC-CXR data sources. We used scattering transform
in composition with the radial basis function (RBF) as ker-
nel function in order to find the B-test statistics. We set
max order=2 to compute up to the second-order of the scat-
tering coefficients and set other filter parameters J = 4 and
L = 8. We summed the scattering coefficients over the im-
age domain to obtain a translational invariant 417 features.
We set the RBF scale parameter γ = 1.
4.3 Evaluation Measures
We assess the pre-trained DenseNet121’s perfor-
mance on the abnormality detection in chest radio-
graphs on MIMIC-CXR. We compare the area under
the receiver operating characteristic curve (AUC), ac-
curacy, precision, sensitivity, specificity, positive pre-
dictive value (PPV), and negative predictive value
(NPV) scores of the pre-trained model with and with-
out domain-shift removal. We show the class activa-
tion maps for some selected examples to aid in the
interpretation of DenseNet121 results. We compare
the class activation maps of the selected radiographs
with and without domain-shift removal to study the
model’s sensitivity to noise and texture characteristics
in the radiographs.
5 RESULTS
5.1 Domain Adaptation by CycleGAN
We present the distributions of wavelet scattering co-
efficients of ChestXray14 and MIMIC-CXR dataset be-
fore and after the domain-adaptation by CycleGAN
in Figure 4. We used the PyTorch implementation
1
of CycleGAN. We used default network architecture
and default training and testing parameters. Table 1
includes some notable parameters. We trained Cy-
cleGAN for 14 epochs with all 112, 120 ChestXray14
1
https://github.com/junyanz/CycleGAN
No adaptation
CycleGAN
(a) ChestXray14 (b) MIMIC-CXR
Figure 4: Density plot of two PCA modes of wavelet scat-
tering coefficients (WSCs) of two datasets are displayed.
Top row shows the original distributions of the scatter-
ing coefficients, whereas bottom row shows the distribu-
tions after the image-to-image translation by CycleGAN.
The WSCs are computed using Scattering2D implemen-
tation from Kymatio package with parameter setting J=4,
L=8, max order=2 while keeping the other parameters to
default values. We summed the scattering coefficients over
the image domain to obtain translational invarient features.
We also whitened the features before extracting two PCA
components.
and selected 243, 332 MIMIC-CXR radiographs. The
MIMIC-CXR radiographs with Posterior-Anterior (PA)
and Anterior-Posterior (AP) views were selected to
keep them consistent with ChestXray14 source. We
used the image-to-image translation maps learned by
CycleGAN to adapt the samples of ChestXray14 and
MIMIC-CXR to the other domain. Figure 5 reports the
training loss of CycleGAN averaged over 100 mini-
batchs with batch size=1 during entire training pro-
cess. Density plots in the figure 4 shows that Cycle-
GAN performs well in domain adaptation and success-
fully eliminates most of the non-overlapping areas be-
tween the domains.
5.2 Abnormality Detection
We present the AUC, accuracy, precision, sen-
sitivity, PPV, and NPV score of the pre-trained
DenseNet121 (Tang et al., 2020) on the abnormality
detection task on MIMIC-CXR with and without the
domain-shift removal. DenseNet121 was trained by
the Authors of (Tang et al., 2020) on ChestXray14
dataset using images with a 256 ×256 resolution and
was shown to achieve a new state-of-the-art perfor-
mance on the abnormality detection binary task. We
downloaded their pre-trained model and tested their
model’s accuracy on MIMIC-CXR with and without
the domain-shift removal.
We derived the labels for the abnormality task de-
Computer-aided Abnormality Detection in Chest Radiographs in a Clinical Setting via Domain-adaptation
69
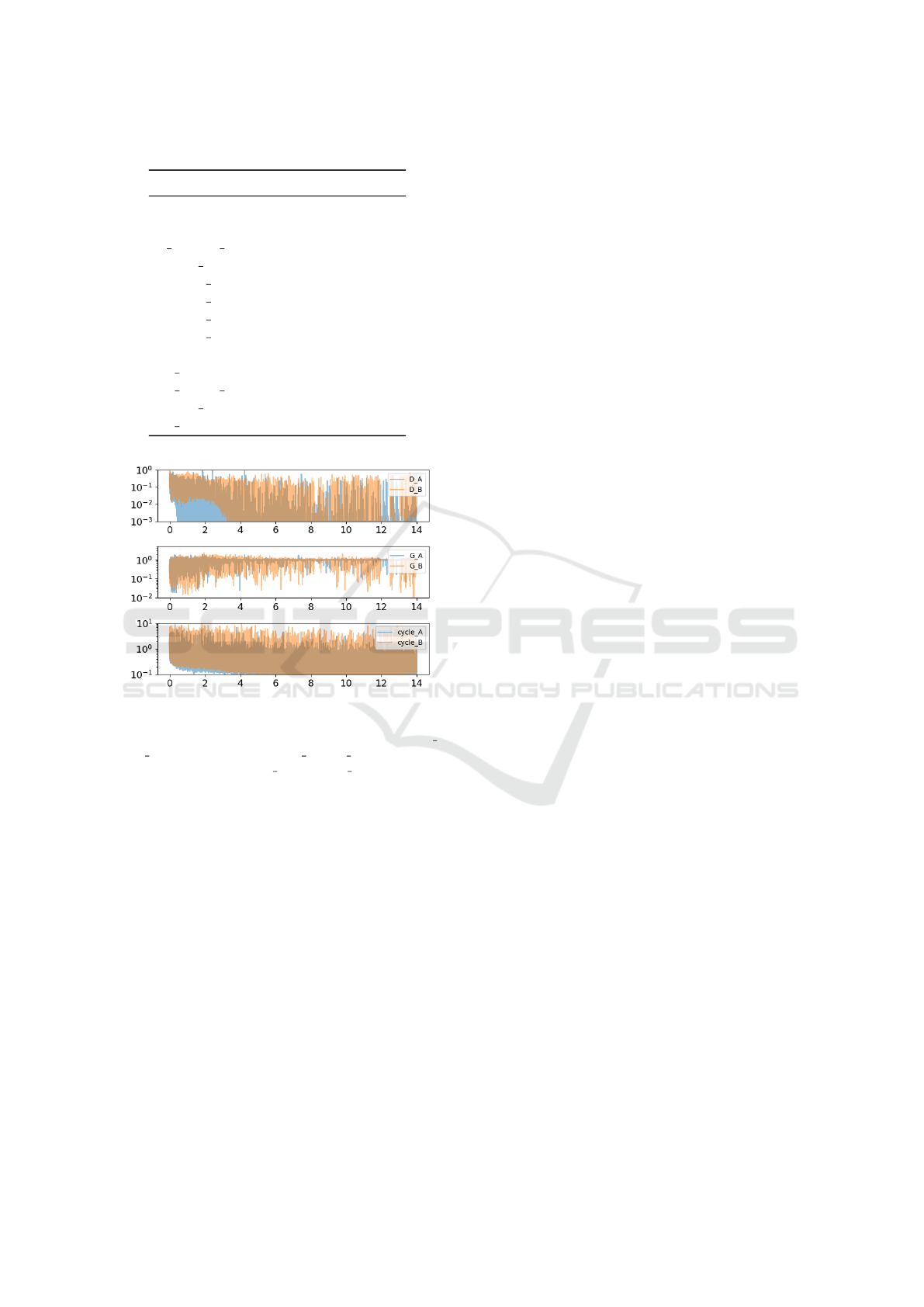
Table 1: CycleGAN architecture and training parameters.
Parameter Value
netG resnet-9blocks
netD basic
n layers D 3
input nc 3
output nc 3
lambda A 10
lambda B 10
lambda identity 0.5
lr 0.0002
lr policy linear
lr decay iter 50
batch size 1
no dropout true
Figure 5: Six training loss of CycleGAN, trained between
the samples of ChestXray14 and MIMIC-CXR, are displayed
for 14 epochs. We denote two discriminator losses by D A
and D B, two generator losses by G A and G B, and two cycle-
consistency losses by cycle A and cycle B.
tection for the MIMIC-CXR dataset. We set the abnor-
mality label to 1 when any of the following 6 con-
ditions are detected by both Chexpert (Irvin et al.,
2019) and NegBio (Peng et al., 2018): Cardiomegaly,
Consolidation, Edema, Pleural Effusion, Pneumonia,
Pneumothorax. We set the abnormality label to 0
when both Chexpert and NegBio report No Find-
ing. We exclude all other cases from the MIMIC-CXR
test cohort. This screening process yields a total of
193, 974 labeled radiographs, including 81, 847 nor-
mal radiographs and 112, 127 radiographs with abnor-
mality.
We present our experimental findings in Table 2.
The pre-trained model performs much lower on the
full MIMIC-CXR than on the small ChestXray14 test
cohort of 1, 344. With the abstention of 50%, the pre-
trained model achieves an accuracy of 90% on MIMIC-
CXR. To calculate the model’s performance with ab-
stention, we calculate the model’s confidence in ab-
normality detection by calculating |p−0.5|, where p
is the abnormality detection score returned by the pre-
trained model. We ranked the predictions based on
the model’s confidence, and we kept the top 50% pre-
dictions. Next, we report the pre-trained model’s per-
formance on the out-of-distribution (OOD) test sets.
We used the original MIMIC-CXR samples that lie in
the region [−4,−1]×[−4,−2] in Figure 4 as the OOD
test set. We report an improvement of 3% accuracy
on the OOD test set with domain-shift removal.
5.3 Grad-CAM to Study Sensitivity to
Domain-shift
We show the class activation maps computed by the
Grad-CAM (Selvaraju et al., 2017) for some selected
examples to aid interpretation of DenseNet121 re-
sults. For each examples, we show the original ra-
diographs of ChestXray14 and MIMIC-CXR, adapted
radiographs by the CycleGAN to the other domain, a
heatmap overlaid on the images indicating the predic-
tion of abnormal regions by the Grad-CAM. Examples
suggest that the DenseNet121 model is potentially fo-
cusing on clinically meaningful abnormal regions of
the chest radiographs for the classification task, how-
ever it is sensitive to the noise and texture of the input
radiographs, as seen in Figure 7.
6 DISCUSSION
We hypothesized a distributional difference in noise
and texture characteristics between the data sources
due to diverse conditions in X-ray equipment and
their configurations for generating the images. We
based our hypothesis on the recent findings (Pooch
et al., 2019; Yao et al., 2019), which implicitly
showed the existence of some characteristic differ-
ences between these data sources. This work has de-
veloped an explicit method to show the characteristic
differences between the data sources. The density-
plot of the high-order wavelet scattering transform
of these radiographs confirms our hypothesis. We
exploited the unpaired image-to-image translation
method, CycleGAN, to remove this shift and ex-
perimentally validated its effectiveness in domain-
shift removal. Our findings also should be applica-
ble to discerning unique, private features from com-
mon, public ones, which could facilitate more tar-
geted privacy-aware DL approaches to best balance
privacy-utility. We also showed that the state-of-the-
art model for abnormality detection is susceptible to
BIOIMAGING 2021 - 8th International Conference on Bioimaging
70
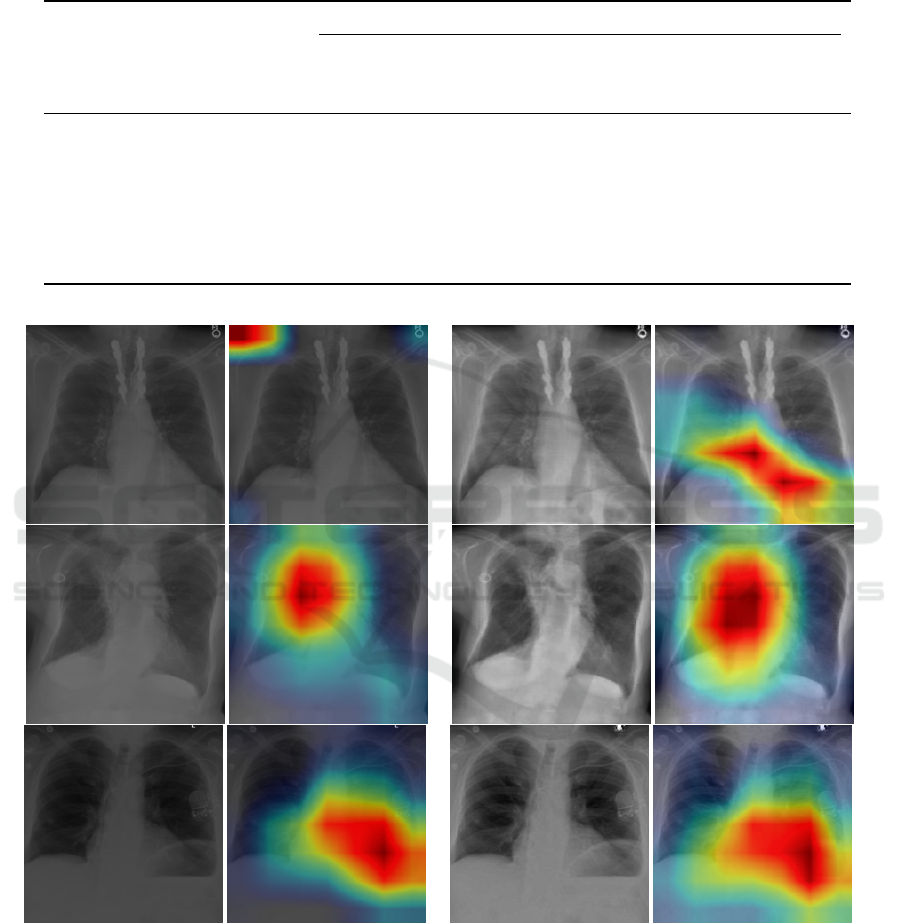
Table 2: Classification evaluation scores of pre-trained DenseNet121 on the out-of-distribution (OOD) MIMIC-CXR radio-
graphs are compared to the CycleGAN’s mapped OOD radiographs’ scores in the last two columns. The second column
includes DenseNet121’s performance on a small ChestXray14 test cohort reported by a previous study (Tang et al., 2020).
The third and fourth column contains DenseNet121’s evaluation scores on full and 50% abstained MIMIC-CXR datasets. The
evaluation scores with abstention are computed on the top 50% of predictions, ranked based on the model’s confidence.
Metric
ChestXray14 MIMIC-CXR
Hold out
(1344)
Full (193974)
Abstention
(96987)
OOD (5800)
No adaptation Cycle-GAN
AUC 0.98 0.79 0.87 0.73 0.75
Accuracy 0.95 0.79 0.90 0.71 0.74
Precision 0.90 0.83 0.89 0.83 0.85
Sensitivity 0.97 0.82 0.96 0.60 0.63
Specificity 0.93 0.76 0.79 0.85 0.87
PPV 0.90 0.83 0.89 0.83 0.85
NPV 0.95 0.75 0.91 0.64 0.66
(a) Original MIMIC-CXR (b) Style-adjusted MIMIC-CXR
Figure 6: Left group is the original MIMIC-CXR radiographs and the class activation maps of abnormal regions in the radio-
graph found by Grad-CAM. Right group is the translated MIMIC-CXR radiographs obtained by applying CycleGAN’s translation
map to the MIMIC-CXR radiographs and the class activation maps of abnormal regions in the radiograph found by Grad-CAM.
model misspecification and is sensitive to input distri-
bution changes when trained on a single data source.
This finding is consistent with the literature work
for other diagnostic tasks (Pooch et al., 2019; Yao
et al., 2019). We have decoupled the domain-shift re-
moval and model construction due to the applicability
of such a decoupled method to various downstream
tasks. However, a problem-specific coupled solution
Computer-aided Abnormality Detection in Chest Radiographs in a Clinical Setting via Domain-adaptation
71
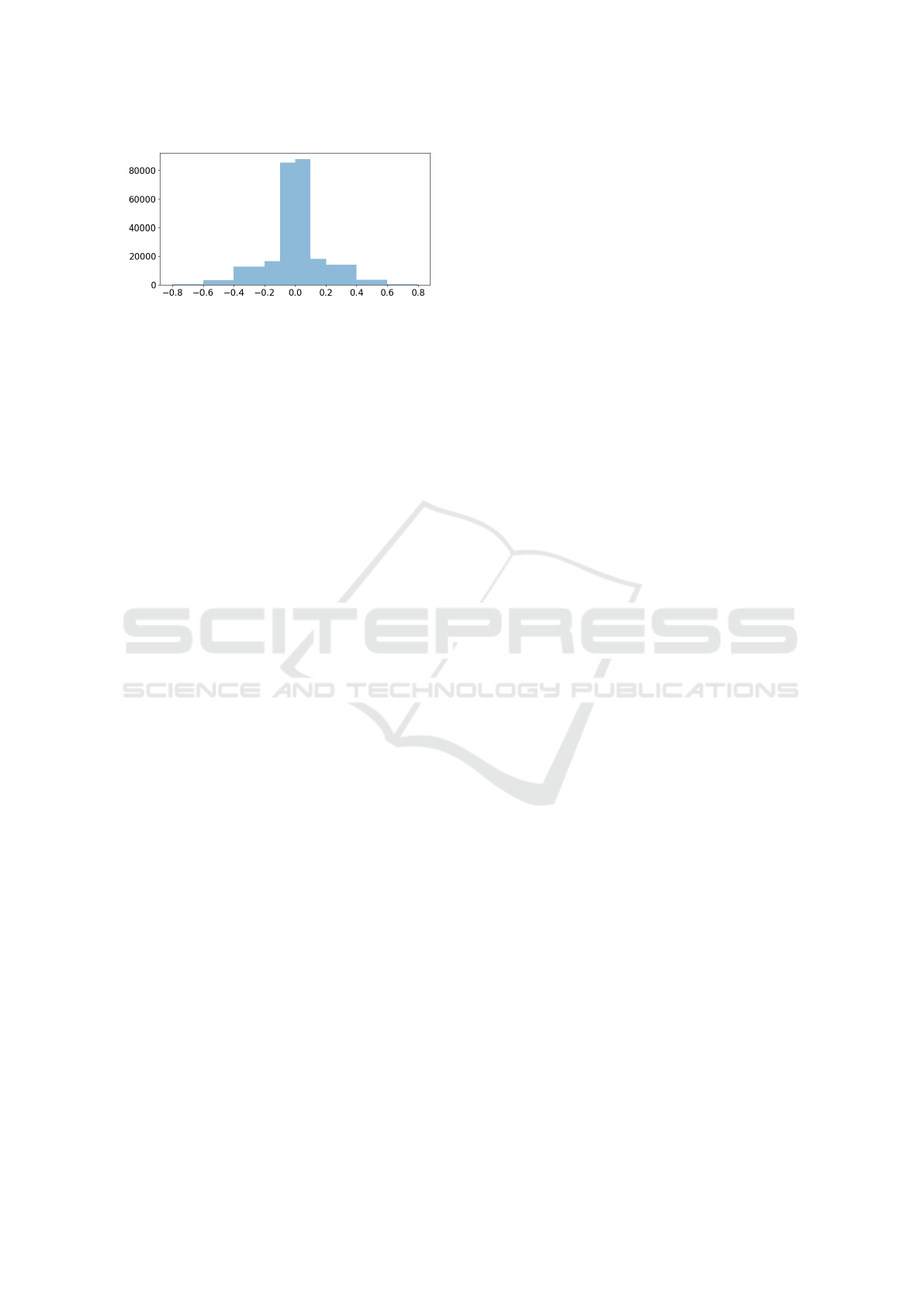
Figure 7: Differences in the abnormality prediction scores
obtained by DenseNet121 on MIMIC-CXR and style-
adjusted MIMIC-CXR by CycleGAN is binned into non-
overlapping intervals, and the counts in every interval are
displayed.
to abnormality detection with adversarial training is
also possible, which is out-of-scope of this paper. Our
main contribution in this work is the introduction of
distribution of high-frequency components to char-
acterize the data sources and relating it to the diffi-
culty of pre-trained models to generalize on unseen
domains. In this work, we have introduced a frame-
work for domain-shift detection and removal to over-
come this problem.
ACKNOWLEDGEMENTS
This research is sponsored in whole or in part by the
AI Initiative (LOIS 9613) and Privacy research (LOIS
9831) as part of the Laboratory Directed Research and
Development Program of Oak Ridge National Labo-
ratory.
REFERENCES
Andreux, M., Angles, T., Exarchakis, G., Leonarduzzi, R.,
Rochette, G., Thiry, L., Zarka, J., Mallat, S., And
´
en,
J., Belilovsky, E., et al. (2020). Kymatio: Scattering
transforms in python. Journal of Machine Learning
Research, 21(60):1–6.
Bousmalis, K., Trigeorgis, G., Silberman, N., Krishnan, D.,
and Erhan, D. (2016). Domain separation networks. In
Advances in neural information processing systems,
pages 343–351.
Bruna, J. and Mallat, S. (2013). Invariant scattering convo-
lution networks. IEEE transactions on pattern analy-
sis and machine intelligence, 35(8):1872–1886.
Dubey, A. (2018). Symmetric completion of deformable reg-
istration via bi-residual inversion. PhD thesis, PhD
thesis). Duke University, Durham, NC, USA.
Dubey, A., Iliopoulos, A.-S., Sun, X., Yin, F.-F., and Ren, L.
(2018). Iterative inversion of deformation vector fields
with feedback control. Medical physics, 45(7):3147–
3160.
Gretton, A., Borgwardt, K. M., Rasch, M. J., Sch
¨
olkopf, B.,
and Smola, A. (2012). A kernel two-sample test. Jour-
nal of Machine Learning Research, 13(Mar):723–773.
Iliopoulos, A.-S., Dubey, A., and Sun, X. (2019). “idvf“:
Iterative inversion of deformation vector field with
adaptive bi-residual feedback control. Journal of
Open Source Software, 4(35):1076.
Ilse, M., Tomczak, J. M., Louizos, C., and Welling, M.
(2019). Diva: Domain invariant variational autoen-
coders. arXiv preprint arXiv:1905.10427.
Irvin, J., Rajpurkar, P., Ko, M., Yu, Y., Ciurea-Ilcus, S.,
Chute, C., Marklund, H., Haghgoo, B., Ball, R., Sh-
panskaya, K., et al. (2019). Chexpert: A large chest
radiograph dataset with uncertainty labels and expert
comparison. In Proceedings of the AAAI Conference
on Artificial Intelligence, volume 33, pages 590–597.
Muandet, K., Balduzzi, D., and Sch
¨
olkopf, B. (2013). Do-
main generalization via invariant feature representa-
tion. In International Conference on Machine Learn-
ing, pages 10–18.
Peng, Y., Wang, X., Lu, L., Bagheri, M., Summers, R.,
and Lu, Z. (2018). Negbio: a high-performance tool
for negation and uncertainty detection in radiology re-
ports. AMIA Summits on Translational Science Pro-
ceedings, 2018:188.
Pooch, E. H., Ballester, P. L., and Barros, R. C. (2019).
Can we trust deep learning models diagnosis? the im-
pact of domain shift in chest radiograph classification.
arXiv preprint arXiv:1909.01940.
Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R.,
Parikh, D., and Batra, D. (2017). Grad-cam: Visual
explanations from deep networks via gradient-based
localization. In Proceedings of the IEEE international
conference on computer vision, pages 618–626.
Tang, Y.-X., Tang, Y.-B., Peng, Y., Yan, K., Bagheri, M.,
Redd, B. A., Brandon, C. J., Lu, Z., Han, M., Xiao,
J., et al. (2020). Automated abnormality classification
of chest radiographs using deep convolutional neural
networks. NPJ Digital Medicine, 3(1):1–8.
Wang, X., Peng, Y., Lu, L., Lu, Z., Bagheri, M., and Sum-
mers, R. M. (2017). Chestx-ray8: Hospital-scale chest
x-ray database and benchmarks on weakly-supervised
classification and localization of common thorax dis-
eases. In Proceedings of the IEEE conference on
computer vision and pattern recognition, pages 2097–
2106.
Yao, L., Prosky, J., Covington, B., and Lyman, K.
(2019). A strong baseline for domain adaptation and
generalization in medical imaging. arXiv preprint
arXiv:1904.01638.
Zaremba, W., Gretton, A., and Blaschko, M. (2013). B-test:
A non-parametric, low variance kernel two-sample
test. In Advances in neural information processing
systems, pages 755–763.
Zhu, J.-Y., Park, T., Isola, P., and Efros, A. A. (2017).
Unpaired image-to-image translation using cycle-
consistent adversarial networks. In Proceedings of
the IEEE international conference on computer vi-
sion, pages 2223–2232.
BIOIMAGING 2021 - 8th International Conference on Bioimaging
72
