
Segmented ECG Bio Identification using Fréchet Mean Distance
and Feature Matrices of Fiducial QRS Features
Abdullah Biran
1
and Aleksandar Jeremic
2
1
Department of Biomedical Engineering, McMaster University, Hamilton, Canada
2
Department of Electrical Engineering, McMaster University, Hamilton, Canada
Keywords: Biomedical Signal Processing, Biometrics, Electrocardiogram, QRS Segmentation, Short Time Fourier
Transform, Feature Lags.
Abstract: In this paper, we present a new segmented based method for human identification using Fréchet distances and
the characteristics of the lag-feature matrices of six fiducial based QRS features. We examined the
applicability of our methodology on 124 ECG records of 62 subjects from the publicly available ECG ID data
base. Our experiments show that the Fréchet distance can identify majority of the subjects (44 individuals)
using the feature matrix of QRS segment lagged by one beat with an identification accuracy ranging from
80% to 100%. Our preliminary results indicate that identifying humans using segmented approaches can be
potentially useful.
1 INTRODUCTION
One of the potential applications of wearable sensing
devices is human identification using biometric
signals. Historically, biometric has been using
fingerprinting and more recently iris scans. In
addition to identification accuracy issues some of the
currently used techniques can be either spoofed
and/or stolen. To this purpose, the identification using
in vivo signals has gained considerable research
interest in recent years. One of the most promising
techniques is based on the electrocardiography (ECG)
measurements. In addition to its in vivo
characteristics, ECG has robust advantages including
universality, hidden nature and simple acquisition
(Joao, S. Cardoso, & Lourenco, 2018). Generally,
ECG based human identification is done in two
different ways: fiducial points based and non-fiducial
feature based (Joao, S. Cardoso, & Lourenco, 2018).
The fiducial techniques relay on locating heartbeat
waveform points, onsets and offsets followed by
extracting different temporal, morphological and
amplitude features. The non-fiducial methods are
based on extracting features without relying on
segmentation algorithms such as using
autocorrelation and wavelet coefficients algorithms
(Chun Chi , Peng Tzu, & Pie Lun, 2019).
Many methods have been applied for personal
identification using ECG. Adrian et al. have
proposed a method using wavelet-based distance
measure for human identification (D.C. Chan, M.
Hamdy, Badre, & Badee, 2008). Saiful et al.
developed another method using the heartbeat
morphology features (Islam, Alajlan, Bazi, & S.
Hichri, 2012). Lin et al presented an algorithm by
applying non-linear Lyapunov exponents, root mean
square (RMS) and support vector machine (SVM)
(Lin, Chen, Lin, Yang, & Chiang, 2014).
Furthermore, Gutta and Cheng proposed discrete
cosine transform (DCT) and autocorrelation
techniques for extracting non-fiducial ECG features
(Gutta & Cheng, 2016). In contrast, Arteaga-Falconi
et al presented a numerical algorithm to extract
fiducial based time and amplitude features (Arteaga-
Falconi, Al Osman, & El Saddik, 2016). Moreover,
Biran et al. have developed segmented and non-
segmented techniques based on Short Time Fourier
Transform (STFT), Euclidian distance and Fréchet
distance to test the feasibility of using ECG as
biometric modality (Biran & Jeremic, Non-
Segmented ECG bio-Identification using Short Time
Fourier Transform and Frecht Mean Distance, 2020).
Liu et al. developed a multi scale autoregressive
model method (MSARM) for personal identification
using ECG (Liu, et al., 2018). Moreover, Sidek et al.
investigated the feasibility of ECG signal as biometric
Biran, A. and Jeremic, A.
Segmented ECG Bio Identification using Fréchet Mean Distance and Feature Matrices of Fiducial QRS Features.
DOI: 10.5220/0010262302230227
In Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2021) - Volume 4: BIOSIGNALS, pages 223-227
ISBN: 978-989-758-490-9
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
223
modality in abnormal cardiac conditions (Sidek,
Khalil, & F. Jelinek, 2014). Furthermore, Odinaka et
al. proposed a multibiometric identification system
based on combining both the electrical originating
signal, the ECG, and the laser Doppler vibrometry
(LDV) (Odinaka, A. O’Sullivan, J. Sirevaag, & W.
Rohrbaugh, 2015).
In this study, we present a fiducial based method
using electrocardiogram (ECG) by creating four
feature matrices of QRS segments. We segment the
ECG signal using our results in (Biran & Jeremic,
Automatic QRS Detection and Segmentaion using
Short Time Fourier Transfrom and Feature Fusion,
2020). We randomize the training/testing procedures
and calculate feature matrices using four different
lags. We identify the subjects using the Fréchet
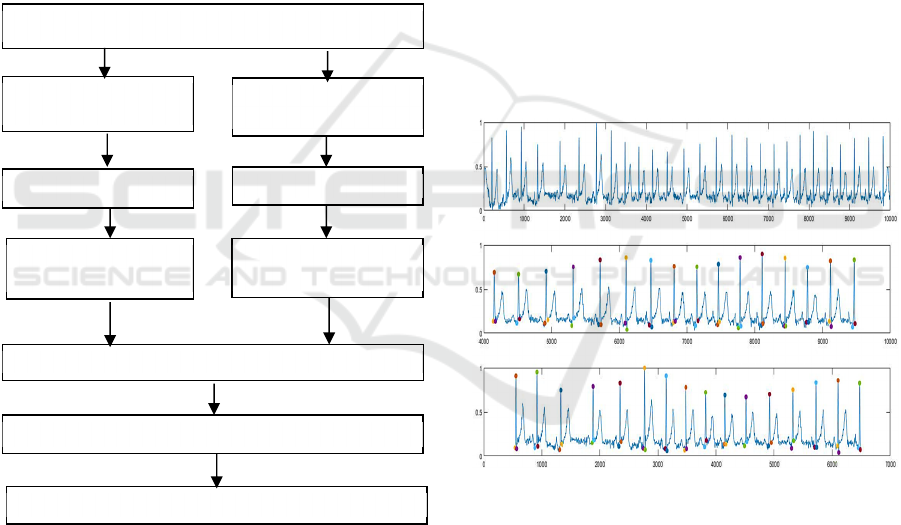
distance of feature matrices. In Figure 1 we illustrate
the outline of the methodology.
Figure 1: Block Diagram.
2 PROPOSED METHODOLOGY
2.1 ECG Database
In this study, we use filtered ECGs of 62 subjects
from the publicly available ECG ID database. The
ECG signals were measured using single lead at 500
sampling frequency and for a duration of 20 seconds.
The data record for each subject consists of two
measurements obtained at different times/days. In the
remainder of the paper, we use the normalized data as
it is commonly used procedure in many of the
machine learning based algorithms in order to
account for possible variability in signal range. In this
stage, we apply our previous work techniques on
automatically segmenting QRS peaks using
windowed STFT and QRS feature fusion. The
previously proposed method correctly and accurately
locates and segments the Q, R and S peaks. (Biran &
Jeremic, Automatic QRS Detection and Segmentaion
using Short Time Fourier Transfrom and Feature
Fusion, 2020).
Following majority of the methods that are used
for individual identification, we create two sets of
ECG data which are used for training \ referencing
and testing. To illustrate, we randomize the process
of selecting the QRS peaks in order to evaluate the
performance of the proposed algorithm. We
arbitrarily set the number of selected beats for both
test and reference ECG to 16. In Fig. 2 we show an
example of the randomization in the training and test
data selection.
Figure 2: An example of randomly selecting the training\
referencing and testing QRS waves.
2.2 QRS Feature Extraction
Using the results from our previous work we extract
the following features after aligning the
corresponding QRS segments: R-Q peak interval
( 𝑃
, S-R peak interval 𝑃
, R-Q amplitude
difference 𝑃
, R-S amplitude difference 𝑃
, QRS
wave distances 𝑃
, and slope distances of the QRS
wave𝑃
. Note that since the last two features are
calculated using the difference between the waves the
number of the feature measurements will be smaller
than the number of available heart beats.
Automatic QRS Segmentation using Windowed STFT
Randomly Selecting 16
training QRS Waves
Randomly Selecting 16
test QRS Waves
QRS Feature Extraction
QRS Feature Extraction
Calculating feature
matrices.
Calculating feature
matrices
Applying Fréchet Mean distances
Finding the Fréchet based decisions
Calculating the identification accuracies
BIOSIGNALS 2021 - 14th International Conference on Bio-inspired Systems and Signal Processing
224

In order to perform the identification task, we propose
to calculate the feature matrices using the training set
and then to identify unknown subject by calculating
the distances between the testing data and training
data and select a subject with the smallest distance.
Let 𝐹
∊𝑅
and 𝐹
∊𝑅
be the feature
vectors for any two QRS waves, then we define the
cross-feature ( 𝐶𝐹 ) matrix between any two QRS
waves using equation (1).
𝐶𝐹 𝐹
𝐹
(1)
By utilizing equation (1), multiple cross-feature
matrices can be calculated by applying particular lags
between QRS waves. Accordingly, we apply
equation (2) to calculate the QRS feature matrices
from both the training and testing data sets.
𝐶𝐹
∑
(2)
Where 𝐿 the lag coefficient, 𝐹 is the QRS feature
vector and 𝑛1 to 𝑐𝐿 is the index of QRS wave
where c is the number of segments.
Since we are using 4 different lags and we have two
set of QRS data (training and testing), by applying
equation (2), we define four training matrices and
four examining matrices of QRS features for each
subject.
2.3 Identification based on Fréchet
Distance
In this study, we apply the findings of Joromi’s work;
specifically, the Fréchet distance of covariance
matrices using Riemannian distances (Jahromi, 2014
). Technically, for any cross-feature matrix, we use
equation (3) to define the feature matrix
𝑅 𝐶𝐹
𝐶𝐹
(3)
Using equation (3) we first calculate the feature
matrix using training set and label them 𝐸
where L
is the lag coefficient and 𝑗1,2,….,𝑁 is the subject
number. Similarly, 𝑇
corresponds to lag l matrix
calculated using the test data. Then, we use the
following Fréchet distances between 𝑇
and 𝐸
𝐹𝑀𝐷1
𝑇𝑟
𝑇
𝑇𝑟𝐸
2𝑇𝑟𝑇
𝐸
𝑇
(4)
𝐹𝑀𝐷2
𝑇𝑟
𝑇
𝑇𝑟𝐸
2𝑇𝑟𝑇
𝐸
(5)
where 𝑇𝑟 is trace of the matrix.
3 RESULTS
We have applied our algorithm on 124 ECGs of 62
subjects from the ECG ID database. We test the
performance of the proposed algorithm in terms of the
average personal identification accuracy, which is
computed using equation (6)
𝐴
∑
𝐹𝐵𝐷
100
(6)
where is 𝐾 is the total number of experiments.
Consequently, to obtain the average performance,
we repeated the training and testing 10 times. Then
we use equation (13) to find the average personal
identification accuracy. As a result, Table 1 shows
that majority of the subjects (44 from record 1 and 43
from record 2 using FMD2) are correctly identified
with an average identification accuracy ranging from
80% to 100% at lag coefficient L = 1. In comparison,
40 subjects are correctly identified in the same
accuracy range by using FMD1. This shows that
FMD2 identifier has significantly better performance
compared to FMD1 as seen in Table 1, Figure 3 and
Figure 4. In addition, these findings indicate that
personal QRS features for most subjects remain stable
over one lag. However, for larger lag values, the
identification accuracy is fluctuating and is highly
dependent on the personal QRS properties. Figures 5
and 6 show examples of the fluctuating in the average
personal identification accuracy over the remaining
three lag coefficients.
4 CONCLUSIONS
In this paper, we proposed an automated fiducial
algorithm based on Fréchet distance and features of
QRS waves to identify subjects using their ECG. Our
preliminary results indicate that by measuring the
FMD2 distance between randomly created cross
feature matrices calculated by one beat lag, majority
of the individuals can be identified with high
accuracy range. Furthermore, applying different lag
coefficients can support the identification process
when the personal QRS feature remain stable over
time. Most importantly, our results indicate that the
ECG based human identification using fiducial
algorithms can achieve acceptable accuracy levels
Segmented ECG Bio Identification using Fréchet Mean Distance and Feature Matrices of Fiducial QRS Features
225
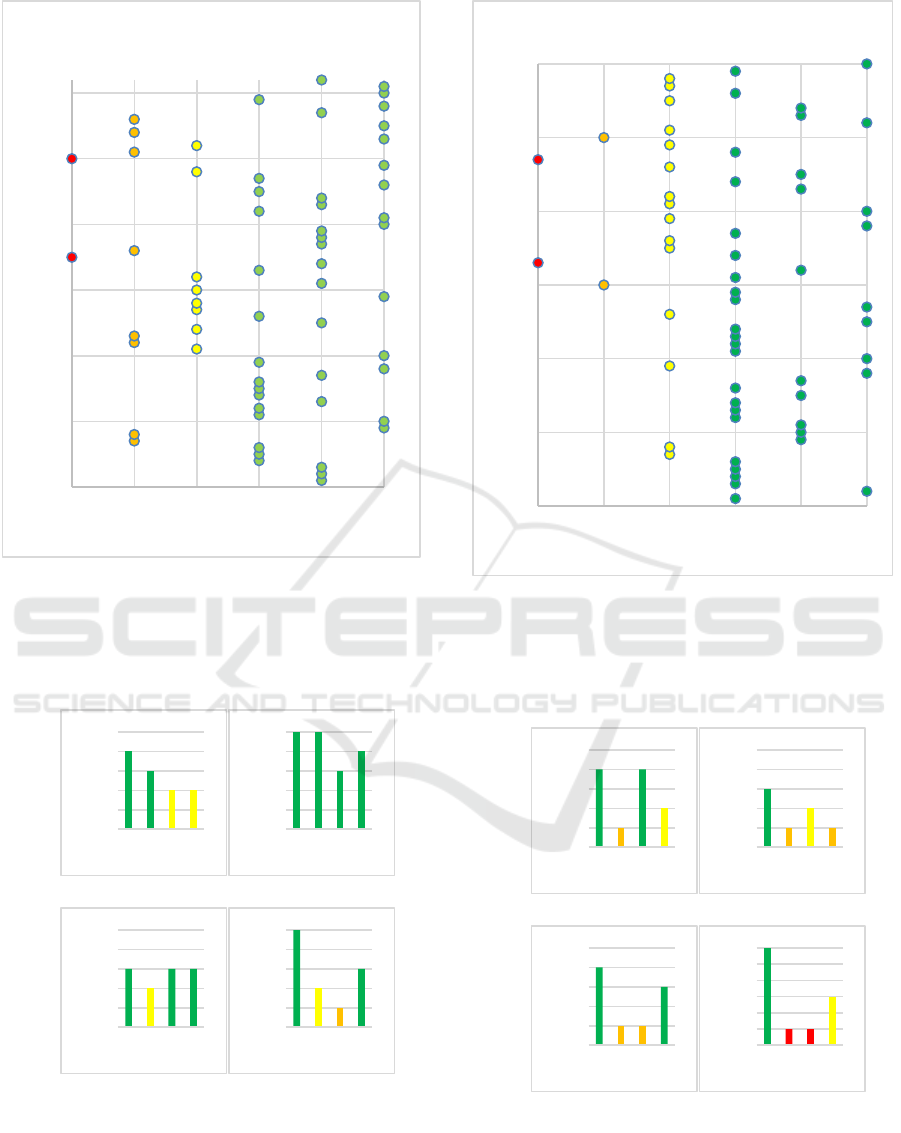
Figure 3: The average personal accuracy for all subject after
10 experiments using FMD2 and lag coefficient L =1 of
randomly selected QRS waves from record 1. We can see
that majority of the subjects (44 subjects) have an
identification accuracy ranging from (80 % to 100%).
Figure 5: The personal identification accuracy (Y-axis) of
different subjects using FMD2 over 4 lag coefficients (X-
axis) after 10 experiments. The top two are from record 1
and the bottom two are from record 2.
Figure 4: The Average Personal Accuracy for All Subject
after 10 Experiments using FMD2 and Lag Coefficient L
=1 of Randomly Selected QRS Waves from Record 2. We
See Can That Majority of the Subjects (43 Subjects) Have
an Identification Accuracy Ranging from (80 % to 100%).
Figure 6: The personal identification accuracy (Y-axis) of
different subjects using FMD1 over 4 lag coefficients (X-
axis) after 10 experiments. The top two are from record 1
and the bottom two are from record 2.
0
10
20
30
40
50
60
50 60 70 80 90 100
SubjectNumber
PersonalIdentificationAccuracy(100%)
FMD2basedIdentificationusingQRSfromrecord
1atL=1
0
10
20
30
40
50
60
50 60 70 80 90 100
Subject Number
PersonalIdentificationAccuracy(100%)
FMD2basedIdentificationusingQRSfromrecord
2atL=
1
Subject 62 Subject 10
Subject 4 Subject 18
50
60
70
80
90
100
1234
50
60
70
80
90
100
1234
50
60
70
80
90
100
1234
50
60
70
80
90
100
1234
Subject 17 Subject 38
Subject 1 Subject 19
50
60
70
80
90
100
1234
50
60
70
80
90
100
1234
50
60
70
80
90
100
1234
40
50
60
70
80
90
100
1234
BIOSIGNALS 2021 - 14th International Conference on Bio-inspired Systems and Signal Processing
226

Table 1: The total number of subjects identified per
accuracy range over all the distance measurements and data
records.
Accuracy 50% 60% 70% 80% 90% 100%
FMD2,
record 1
2 8 8 15 15 14
FMD2,
record 2
2 2 15 23 11 9
FMD1,
record 1
2 6 14 20 8 12
FMD1,
record 2
5 6 11 11 10 19
REFERENCES
Arteaga-Falconi, J., Al Osman, H., & El Saddik, A. (2016,
March). ECG Authentication for Mobile Devices. IEEE
Transactions onInstrumentation and Mesurement, 65,
591-600.
Biran, A., & Jeremic, A. (2020). Automatic QRS Detection
and Segmentaion using Short Time Fourier Transfrom
and Feature Fusion. 33rd Canadian Conference on
Electrical and Computer Engineering. London.
Biran, A., & Jeremic, A. (2020). ECG Based Human
Identification using Short Time Fourier Transform and
Histograms of Fiducial QRS Features. 13th
International Joint Conf. on Biomedical Engineering
Systems and Technologies, (pp. 324-329). Malta.
Biran, A., & Jeremic, A. (2020). Non-Segmented ECG bio-
Identification using Short Time Fourier Transform and
Frecht Mean Distance. 42nd Annual International
Conference of the IEEE Engineering in Medicine and
Biology Society, (pp. 5506-5509). Montreal.
Chun Chi , W., Peng Tzu, C., & Pie Lun, H. (2019, May).
Cancelable Biometric Recognition With ECGs:
Subspace-Based Approaches. IEEE Transactions on
Forensics and Security, 14, 1323-1336.
D.C. Chan, A., M. Hamdy, M., Badre, A., & Badee, V.
(2008, February). Wavelet Distance Measure for
Person IdentificationUsing Electrocardiograms. IEEE
Transactions on Instrumentation, 57, 248-253.
David, C., Silva, H., Gamboa, H., Fred, A., & Figueiredo,
M. (2013, February). Novel fiducial and non-fiducial
approaches to electrocardiogram-based biometric sys-
tems. Institution of Engineering Technology, 2, 64-75.
Gutta, S., & Cheng, Q. (2016, March). Joint Feature
Extraction and Classifier Design for ECG-Based
Biometric Recognition. IEEEJournal of Biomedical
and Health Informatics, 20, 460-468.
Islam, S., Alajlan, N., Bazi, Y., & S. Hichri, H. (2012,
May). HBS: A Novel Biometric Feature Based
onHeartbeat Morphology. IEEEtransactions on
Information Technology in Biomedicine, 16, 445-453.
Jahromi, M. (2014 ). Frechet Means with Respect to the
Riemannian Distances: Evaluations and Applications .
McMaster University .
Joao, R. P., S. Cardoso, J., & Lourenco, A. (2018, June ).
Evolution, Current challenges, and Future Possibilities
in ECG Biometrics. IEEE Access, 6, 34746-34776.
Li, H., & Jeremic, A. (2017). Semi-supervised Distributed
Clustering for Bioinformatics - Comparison Study., 4,
pp. 259-264.
Lin, S.-L., Chen, C.-K., Lin, C.-L., Yang, W.-C., & Chiang,
C.-T. (2014). Individual identification based on chaotic
electrocardiogram signals during muscular exercise.
The Institution of Engineering.
Liu, J., Yin, L., He, C., Wen, B., Hong, X., & Li, Y. (2018).
A Multiscale Autoregressive Model-Based Electro-
cardiogram Identification Method. IEEE Access,
18251-18263.
Odinaka, I., A. O’Sullivan, J., J. Sirevaag, E., & W.
Rohrbaugh, J. (2015, January). Cardiovascular
Biometrics: Combining Mechanical and Electrical
Signals. IEEE Transactions on Information on
Forensics and Security, 10, 16-27.
Odinaka, I., Lai, P.-H., D. Kaplan, A., A. O’Sullivan, J., J.
Sirevaag, E., & W. Rohrbaugh, J. (2012, December).
ECG Biometric Recognition: A Comparative Analysis.
IEEE Transactions on Information on Forensics and
Security, 7, 1812-1824.
Sidek, K., Khalil, I., & F. Jelinek, H. (2014, November).
ECG Biometric with Abnormal Cardiac Conditions in
Remote Monitoring System. ECG Biometric with
Abnormal Cardiac Conditions, 44, 1498-1509.
V. Oppenheim, A., & W. Schafer, R. (1989). Discrete-Time
Signal Processing. New Jersey: Prentie-Hall, Inc.
Zhang , Q., Zhou, D., & Zeng, X. (2017). HeartID: A
Multiresolution Convolutional Neural Network for
ECG-Based Biometric Human Identification in Smart
Health Applications. IEEE Access, 5, 11805-11816.
Segmented ECG Bio Identification using Fréchet Mean Distance and Feature Matrices of Fiducial QRS Features
227
