
Feature Space Reduction for Human Activity Recognition
based on Multi-channel Biosignals
Yale Hartmann, Hui Liu and Tanja Schultz
Cognitive Systems Lab, University of Bremen, Germany
Keywords:
Human Activity Recognition, Biosignals, Multi-channel Signal Processing, Feature Space Reduction,
Stacking.
Abstract:
In this paper, we study the effect of Feature Space Reduction for the task of Human Activity Recognition
(HAR). For this purpose, we investigate a Linear Discriminant Analysis (LDA) trained with Hidden Markov
Models (HMMs) force-aligned targets. HAR is a typical application of machine learning, which includes find-
ing a lower-dimensional representation of sequential data to address the curse of dimensionality. This paper
uses three datasets (CSL19, UniMiB, and CSL18), which contain data recordings from humans performing
more than 16 everyday activities. Data were recorded with wearable sensors integrated into two devices, a
knee bandage and a smartphone. First, early-fusion baselines are trained, utilizing an HMM-based approach
with Gaussian Mixture Models to model the emission probabilities. Then, recognizers with feature space
reduction based on stacking combined with an LDA are evaluated and compared against the baseline. Ex-
perimental results show that feature space reduction improves balanced accuracy by ten percentage points on
the UniMiB and seven points on the CSL18 datasets while remaining the same on the CSL19 dataset. The
best recognizers achieve 93.7 ± 1.4% (CSL19), 69.5 ± 8.1% (UniMiB), and 70.6 ± 6.0% (CSL18) balanced
accuracy in a leave-one-person-out cross-validation.
1 INTRODUCTION
The curse of dimensionality haunts all machine learn-
ing problems. Each feature dimension adds exponen-
tially to the data required to train a machine learn-
ing algorithm effectively, and various techniques have
been used and evaluated in a Human Activity Recog-
nition (HAR) context to address this explosion in the
required data. Feature selection using filter meth-
ods like minimum Redundancy Maximum Relevance
(mRMR), analysis of variance (ANOVA), and wrap-
per methods like forward selection, have been stud-
ied. They are known to successfully increase recog-
nition performance in many fields including HAR and
Automatic Speech Recognition (ASR) tasks (Bulling
et al., 2014) (Weiner and Schultz, 2018) (Capela
et al., 2015) (Suto et al., 2016). Feature space trans-
formation methods like Linear Discriminant Analy-
sis (LDA), Principal Component Analysis (PCA), and
Autoencoders (AE) have also been applied to HAR
with success (Almaslukh et al., 2018) (Mezghani
et al., 2013) (Li et al., 2018) (Hu and Zahorian, 2010).
However, most of these methods are designed for
single vector-based machine learning problems, not
sequential data, often found in HAR tasks. Com-
plex everyday activities, for example, are rarely deter-
mined properly by a snapshot of the human moving
around while performing this activity. Sitting down
and standing up look the same in a still image. Be-
side context, HAR tasks also require modeling and
transformation techniques adequate for sequences of
different lengths. Feature selection is possible with
a sequential classifier in a wrapper method like for-
ward selection, and transformation can be done using
AE. Alternatively, transformation is also possible by
re-labeling the sequential data to a vector-based prob-
lem using Hidden Markov Model (HMM) force align-
ments and assigning each vector of the sequence to a
distinct state. These alignment-based reductions have
been applied in ASR tasks (Haeb-Umbach and Ney,
1992) (Siohan, 1995).
Our previous work adapted this alignment-based
technique for an HAR task and showed improvements
in recognition accuracy (Hartmann et al., 2020). This
work evaluates the reduction technique on two larger
datasets, details improvements, and compares the
achieved performances with the state of the art.
Hartmann, Y., Liu, H. and Schultz, T.
Feature Space Reduction for Human Activity Recognition based on Multi-channel Biosignals.
DOI: 10.5220/0010260802150222
In Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2021) - Volume 4: BIOSIGNALS, pages 215-222
ISBN: 978-989-758-490-9
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All r ights reserved
215
2 DATASETS
In this paper, three datasets, CSL18, CSL19, and
UniMiB, are used. They provide segmented and an-
notated sequential human activity data recorded with
wearable sensors or smartphones. The following sec-
tion provides a brief overview of the three datasets.
2.1 CSL18 and CSL19
The CSL18-18A-21S-4P (CSL18) in-house four-
person dataset recorded 20 biosignals and distin-
guishes 18 different activities. It contains 40mins of
semi-automatic annotated data recorded from sensors
attached to a knee bandage and was recorded at the
Cognitive Systems Lab (CSL) in a controlled labora-
tory environment.
The CSL19-22A-17S-20P (CSL19) in-house
dataset is a follow up to the CSL18 dataset. The
22-activity dataset contains six hours of segmented
and annotated data from 20 participants. In contrast
to CSL18, the two microphones and one goniometer
channel are dropped as they do not contain useful
information. The activities (listed in Table 1) are
comprised of everyday and sports activities chosen
for their relevance to gonarthrosis and personal
monitoring of knee straining behavior. The sensors
are placed on a knee bandage, as gonarthrosis patients
wear them for support and pain relief.
The sensors include four electromyographic sen-
sors (EMG) to capture muscle activity, as well as a
goniometer and two 3D-inertial measurement units
(IMU) consisting of a triaxial accelerometer and gy-
roscope to capture motion. 17 biosignals with EMG
sensors sampled at 1000Hz and IMUs and goniometer
sampled at 100Hz are captured.
We chose the biosignalsplux Research Kit
1
as a
recording device. One PLUX hub can process signals
from 8 channels (each up to 16 bits) simultaneously.
Therefore, three synchronized hubs are used during
the entire session. For more detail on the setup and
segmentation, please refer to (Liu and Schultz, 2018)
and (Liu and Schultz, 2019).
2.2 UniMiB-SHAR
The 17-activity UniMiB-SHAR (UniMiB) dataset has
been recorded at the University Milano Bicocca and
published as a benchmark dataset (Micucci et al.,
2017). The dataset focuses on everyday activities as
well as different types of falls listed in Table 2. It con-
tains nine different Activities of Daily Living (ADLs)
and eight different falls from 30 subjects. A triaxial
1
biosignalsplux.com/products/kits/researcher.html
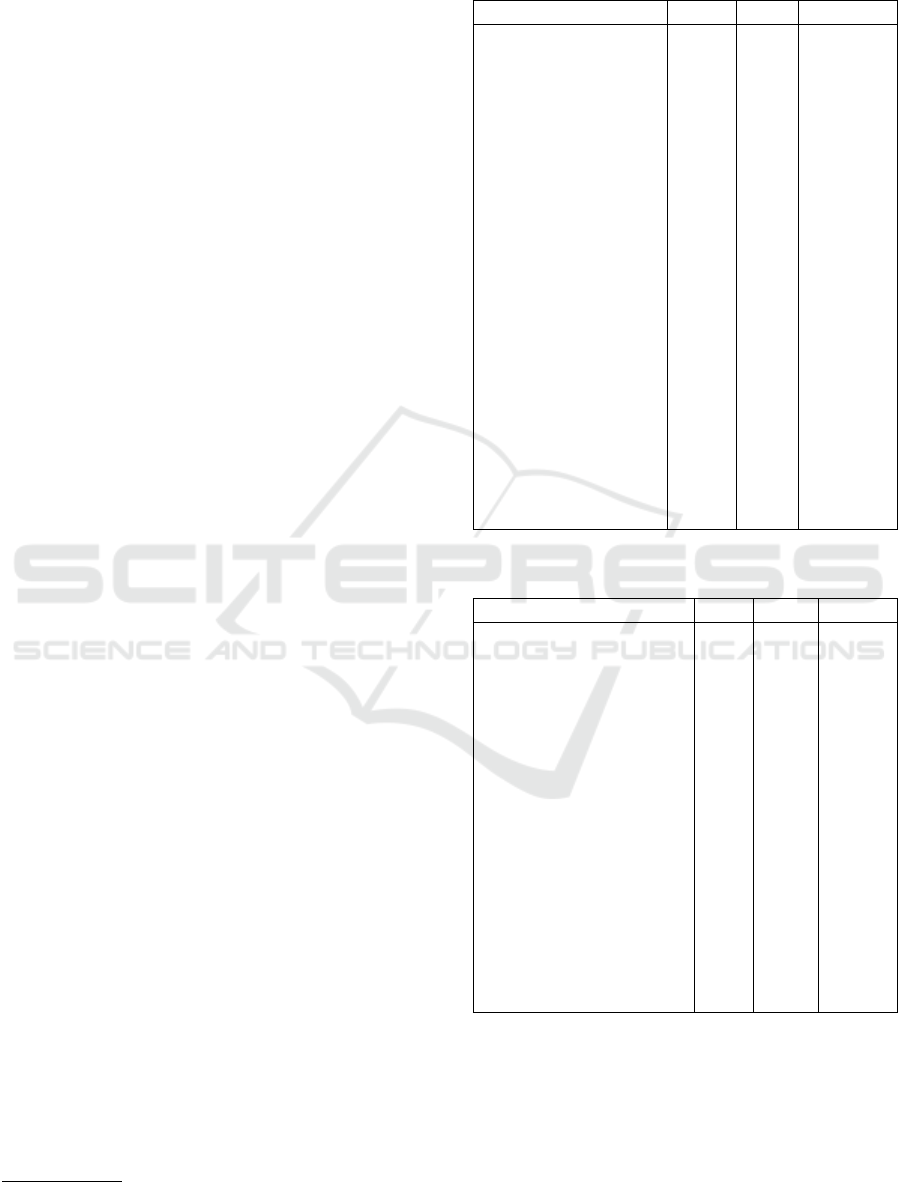
Table 1: Overview of recorded activities in CSL19 dataset.
Durations given in seconds.
Target Min. Occ. Total
Walk 3.138 400 1702.076
Walk upstairs 3.788 365 1736.030
Walk downstairs 3.068 364 1568.212
Walk 90
◦
-curve left 2.898 398 1725.124
Walk 90
◦
-curve right 3.228 393 1749.744
Spin left left-first 0.958 380 633.080
Spin left right-first 0.968 420 767.409
Spin right left-first 0.799 401 745.087
Spin right right-first 1.168 400 685.610
V-Cut left left-first 0.808 399 722.482
V-Cut left right-first 1.018 378 709.474
V-Cut right left-first 0.839 400 718.664
V-Cut right right-first 1.208 378 695.446
Shuffle left 1.738 380 1097.810
Shuffle right 2.088 374 1089.572
Run 2.318 400 1260.279
Jump one leg 0.829 379 639.363
Jump two legs 0.868 380 739.360
Sit 0.818 389 646.282
Stand 0.808 405 663.870
Sit down 1.128 389 746.222
Stand up 1.048 389 705.052
Table 2: Overview of recorded activities in UniMiB dataset.
Durations given in seconds.
Target Dur. Occ. Total
FallingBack 3.0 526 1578.0
FallingBackSC 3.0 434 1302.0
FallingForw 3.0 529 1587.0
FallingLeft 3.0 534 1602.0
FallingRight 3.0 511 1533.0
FallingWithPS 3.0 484 1452.0
GoingDownS 3.0 1324 3972.0
GoingUpS 3.0 921 2763.0
HittingObstacle 3.0 661 1983.0
Jumping 3.0 746 2238.0
LyingDownFS 3.0 296 888.0
Running 3.0 1985 5955.0
SittingDown 3.0 200 600.0
StandingUpFL 3.0 216 648.0
StandingUpFS 3.0 153 459.0
Syncope 3.0 513 1539.0
Walking 3.0 1738 5214.0
smartphone accelerometer sampled at 50Hz was used
for recording, and the gravitational constant was re-
moved post-recording. The smartphone was placed in
equal parts in the left and right subjects’ pocket dur-
ing recording. The data is automatically segmented
into three-second windows around a magnitude peak.
BIOSIGNALS 2021 - 14th International Conference on Bio-inspired Systems and Signal Processing
216
3 APPROACH
The effects of feature space reduction applied to
a HAR task are evaluated in the next sections us-
ing a person dependent shuffled and stratified 5-fold
cross-validation and a person independent leave-one-
person-out cross-validation. Each fold is evaluated
using a balanced accuracy (shown in Equation 1) as
the datasets are slightly imbalanced. The different
parameters are then compared using the average and
standard deviation over the folds. The baseline will
be optimized using a parameterwise grid search along
the recognizer stages using the best parameters ac-
cording to the independent evaluation for both the
next independent and dependent evaluations. Follow-
ing best practices, it starts with windowing and ends
with HMM parameters. The optimized baseline is
then compared against the best performing recognizer
with feature space reduction.
Let E donate the set of all activities a, then the
balanced or macro average accuracy is defined as de-
scribed in Equation 1.
Balanced accuracy =
1
|E|
|E|
∑
a
Accuracy
a
(1)
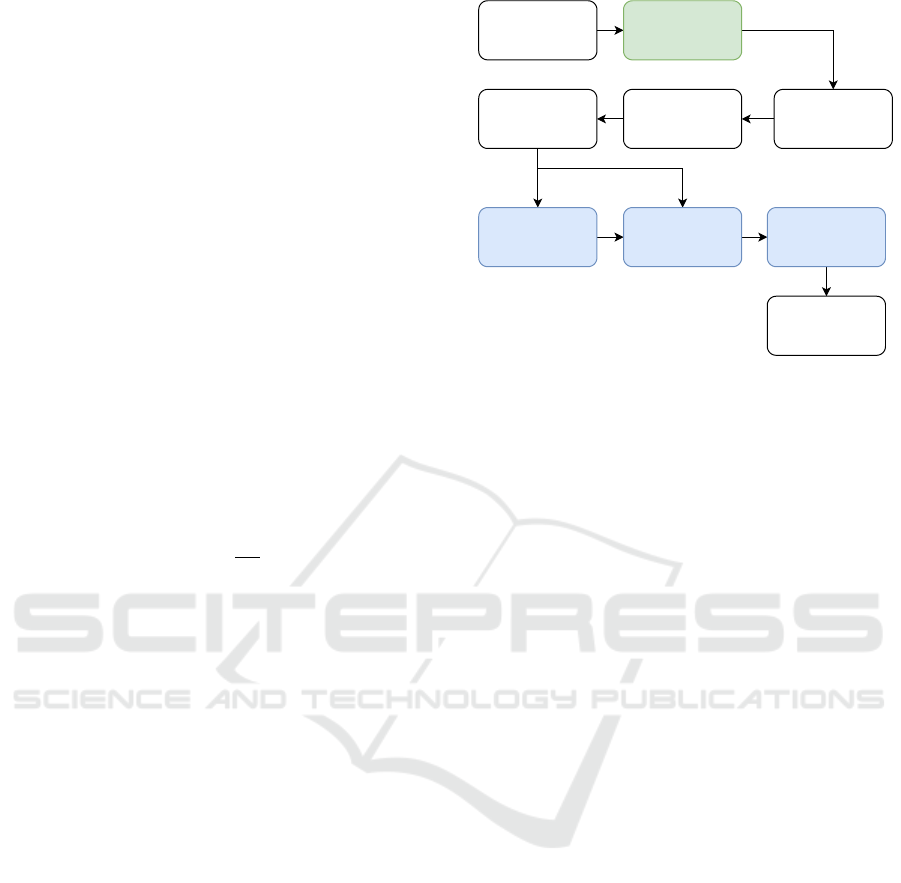
The recognizer is set up similar to our previous
work, and the different stages are displayed in Figure
1. Each data segment is windowed, and features are
calculated on each window. The normalization then
removes the mean and scales the whole segment to
a standard deviation of one. An HMM with GMMs
modeling the emission probabilities performs the final
classification. The HMM is trained with the Expec-
tation Maximisation algorithm, and the GMMs use a
merge and split algorithm. For the reduction, the three
blue steps HMM, Stacking, and LDA are enabled. For
the UniMiB, a simple mirror is implemented (dis-
played in green) to offset the smartphones’ different
rotation between pockets. If the mean of the signal is
below zero, the signal is flipped.
The blue HMM step denotes the force alignment
from the segmented sequential data to the topologies’
states. The alignment assigns each vector in the se-
quence the most likely HMM emission state. Force
alignments create a supervised vector-based problem
where each vector is assigned the activity and state
pair as a new target. Stacking refers to the process
of prepending the n previous feature vectors and ap-
pending the following n vectors to all vectors in a se-
quence, thereby increasing the time context and the
vector dimension by 2n + 1 the original dimension.
The LDA is then trained on the aligned and stacked
vectors and transforms the feature space along the
most discriminatory linear plane. During training, the
Data
Feature calculation
DecodingTraining
Normalization
HMM Stacking LDA
HMM
Axis Mirror
(UniMiB)
Windowing
Figure 1: Recognition pipeline. Green: unique steps for
UniMiB; Blue: feature space reduction steps.
HMM aligns the given segments and assigns each fea-
ture vector in the sequence its aligned activity and
state as a new target. During recognition, the activ-
ity of each segment is unknown. Therefore, the HMM
step is skipped, and the feature vectors are stacked
and transformed directly.
The recognizer was developed with SciPy (Virta-
nen et al., 2020), NumPy (Harris et al., 2020), scikit-
learn (Pedregosa et al., 2011), Matplotlib (Hunter,
2007), TSFEL (Barandas et al., 2020), and our in-
house decoder BioKIT (Telaar et al., 2014).
4 EVALUATION CSL19
First, a baseline for the CSL19 dataset is evaluated.
The HMM topology uses five states for each gait cy-
cle in an activity. Running, for example, contains
three full cycles and therefore uses fifteen states. Sit,
stand, and the transitions between them are modeled
using a single state. For the initial hyperparameters,
a Hamming window with 20ms overlap, enabled nor-
malization, and ten HMM train-iterations are chosen
based on previous work (Hartmann et al., 2020). The
window size is determined in the first experiment and
does not require an initial value. After optimiza-
tion, the best parameters are 100ms Hamming win-
dows with 50ms overlap, 10 HMM train-iterations,
and normalization enabled. The baseline achieves
a 93.7 ± 1.4% balanced accuracy in a person in-
dependent leave-one-person-out cross-validation and
97.8 ± 0.2% balanced accuracy in a stratified person
dependent 5-fold cross-validation.
The next step is to evaluate the best reduction pa-
rameters. The results of the experiment are displayed
in Figure 2. Note that the evaluation without reduc-
Feature Space Reduction for Human Activity Recognition based on Multi-channel Biosignals
217
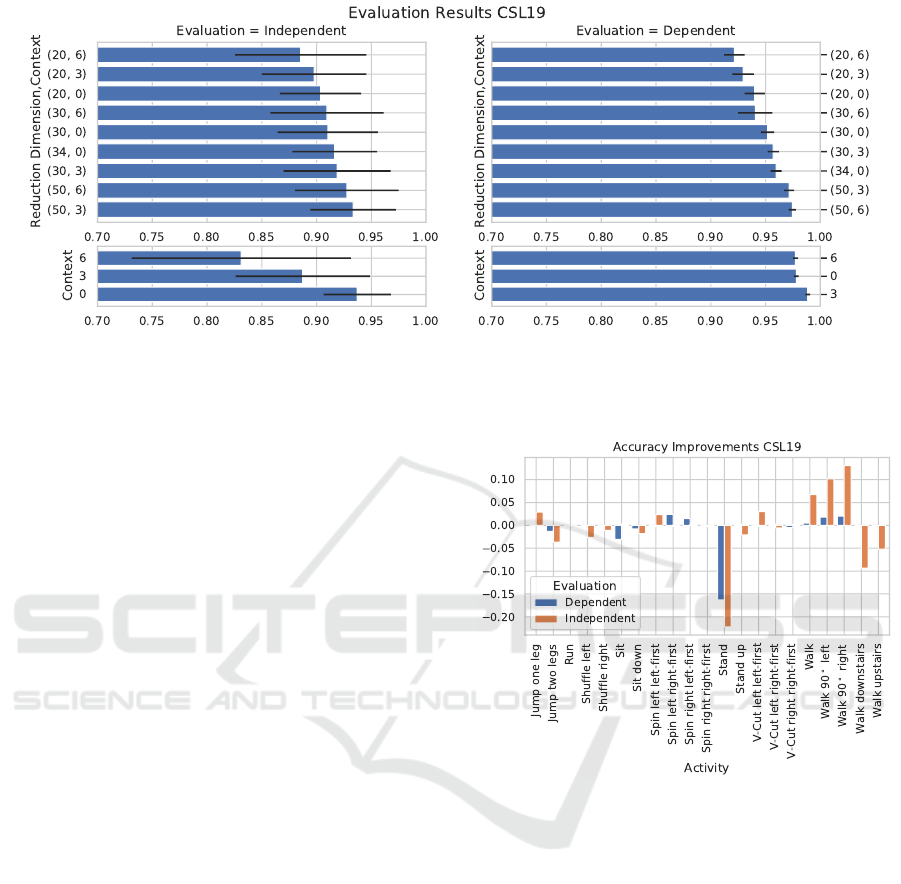
Figure 2: Evaluation results for stacking and reduction on the CSL19 dataset. Left column: person independent evaluation,
right: person dependent evaluation. Upper row: the combination of stacking and reduction (dimension target, stacking
context), lower row: stacking alone. Without stacking, there is no reduction to 50 dimensions but a transformation of the
original 34 dimensions.
tion and a stacking context of zero corresponds to the
baseline recognizer. Figure 2 shows that stacking it-
self does not improve performance, except in the de-
pendent evaluation with a context of three. Reducing
to any dimension offsets this, but not above the base-
line. Notably, the recognition accuracy increases with
higher target dimensions independent of the stacking
context. Reducing to 50 dimensions performs better
than 30 and 20, independent of the specified stacking
context.
The best performance in an independent evalu-
ation is achieved using a stacking context of three
and reducing to a 50-dimensional feature space at
93.3 ± 3.9% accuracy. Notably, this is not higher
than the 93.7 ± 1.4% achieved without reduction. A
similar observation can be made in the person depen-
dent evaluation: 97.8±0.2% without and 97.4±0.3%
with the reduction. Figure 3 shows, that while the re-
duction based recognizer can better distinguish walk
and 90
◦
curves, standing is confused for sitting more
often, resulting in the same overall performance.
Additionally, it should be noted that the LDA
is trained with 238-dimensional feature vectors (17
channels times two features times seven vectors due
to context) on 370k samples. It distinguishes between
94 classes (five phases for most of the 22 activities).
The LDA of the best performing recognizer achieves a
27% accuracy between these classes in a 10-fold per-
son dependent evaluation.
5 EVALUATION UniMiB-SHAR
The activities are segmented with a fixed-length peak
centered window rather than at the activity’s start and
Figure 3: Relative recognition accuracy improvements in
percentage points between baseline and reduction based
recognizer on the CSL19 dataset.
end. Therefore, the HMM topology is modeled with
a random state at the beginning and end of each ac-
tivity. The activities containing gait cycles are mod-
eled similarly to the CSL19 activities using five states
for each cycle. The falls are modeled with ten states,
as this performed best in a person independent cross-
validation.
The UniMiB-SHAR dataset contains fewer sam-
ples per segmented activity due to the lower sam-
pling rate. Therefore, a single grid search is feasi-
ble and executed. The best parameters are 400ms
Hamming windows with 320ms window overlap,
30 HMM train-iterations, and normalization enabled.
The baseline achieves a 59.7 ± 8.6% balanced ac-
curacy in a person independent leave-one-person-out
cross-validation.
The feature space reduction is evaluated on a grid
BIOSIGNALS 2021 - 14th International Conference on Bio-inspired Systems and Signal Processing
218
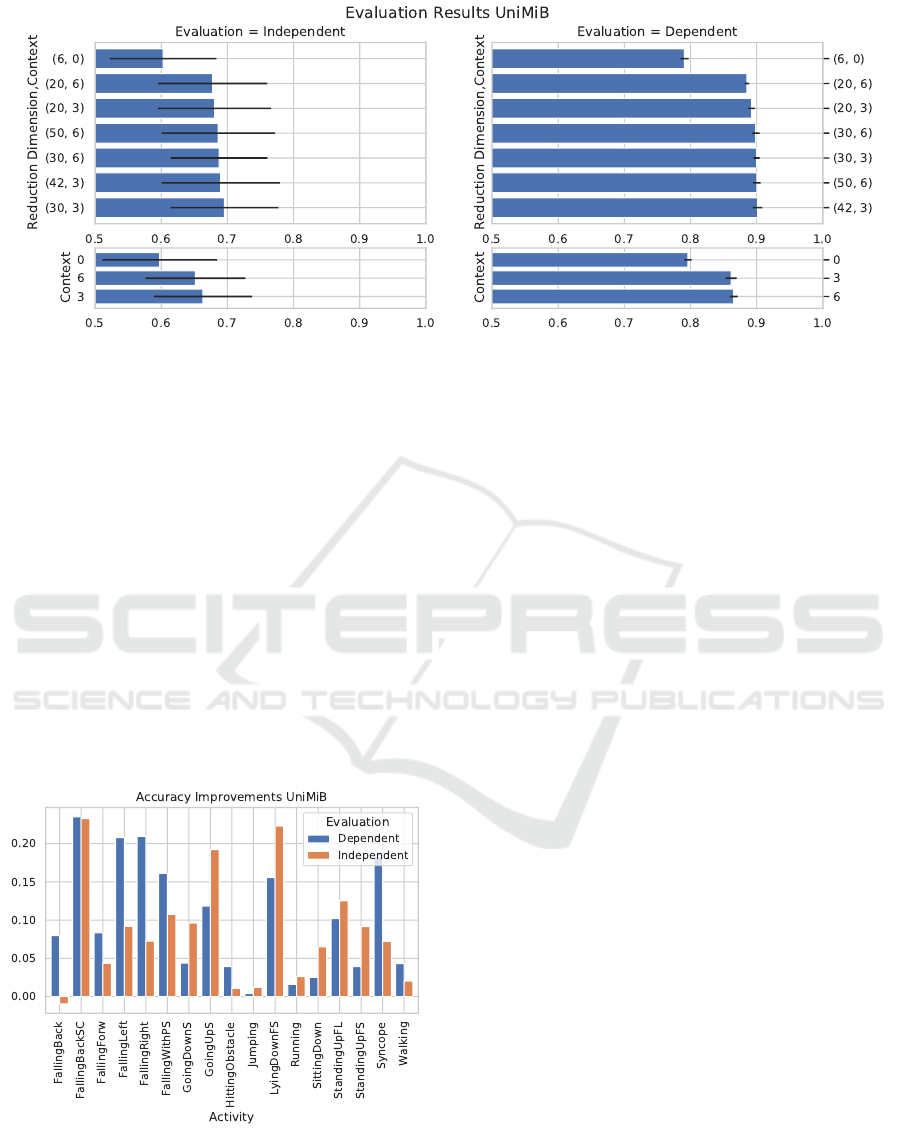
Figure 4: Evaluation results for stacking and reduction on the UniMiB dataset. Left column: person independent evaluation,
right: person dependent evaluation. Upper row: the combination of stacking and reduction (dimension target, stacking
context), lower row: stacking alone. Without stacking, there is no reduction to 20, 40, or 50 dimensions but a transformation
of the original six dimensions.
ranging from zero to six frames stacking context and
a target dimension from twenty to fifty. The results
are displayed in Figure 4. Notably, stacking alone
already increases performance, and the reduction ex-
tends this. Similar to the CSL19 dataset, the target di-
mension influences performance more than the stack-
ing context. The best performing parameter combi-
nation with a target dimension of 30 and a context of
three performs at 69.5 ± 8.1% balanced accuracy in a
leave-one-person-out cross-validation. Compared to
the 59.7±8.6% accuracy in the baseline, this is an im-
provement by ten percentage points. This difference
is also apparent in the performance improvements for
each activity, as depicted in Figure 5.
Figure 5: Relative recognition accuracy improvements in
percentage points between baseline and reduction based
recognizer on the UniMiB dataset.
For easier comparison with previous work, the ac-
curacy for the best performing recognizer with fea-
ture space reduction was calculated in addition to the
balanced accuracy. The accuracy in the independent
evaluation is 77.0%, and the difference to the bal-
anced accuracy mainly arises from the ADLs occur-
ring more often and being recognized much better
than the falls.
Similar to the CSL19 dataset, the LDA performs
poorly when classifying the different vectors. The
LDA is trained with 42-dimensional feature vectors
(three channels, two features, seven vectors due to
context) on 360k samples. It distinguishes between
148 classes (ten phases for most of the 17 activities)
and achieves a 29% accuracy between these classes in
a 10-fold person dependent evaluation.
6 EVALUATION CSL18
The CSL18 dataset has been evaluated with and with-
out reduction previously, and a performance increase
with reduction could be shown (Hartmann et al.,
2020). The experiment is repeated with a balanced
accuracy metric to keep the results comparable with
the other two datasets. Furthermore, the CSL19 rec-
ognizer’s topology is applied to the CSL18 dataset in-
stead of the fixed six-state topology used previously,
and the parameters are optimized with a person inde-
pendent evaluation rather than a dependent one. The
optimized baseline achieves 63.8±8.5% balanced ac-
curacy in an independent evaluation using the CSL19
topology, 30ms windows with 6ms overlap, and nor-
malization enabled.
Figure 6 shows the results from the stacking and
reduction experiments. In the person independent
evaluation, the best performances are achieved with-
Feature Space Reduction for Human Activity Recognition based on Multi-channel Biosignals
219
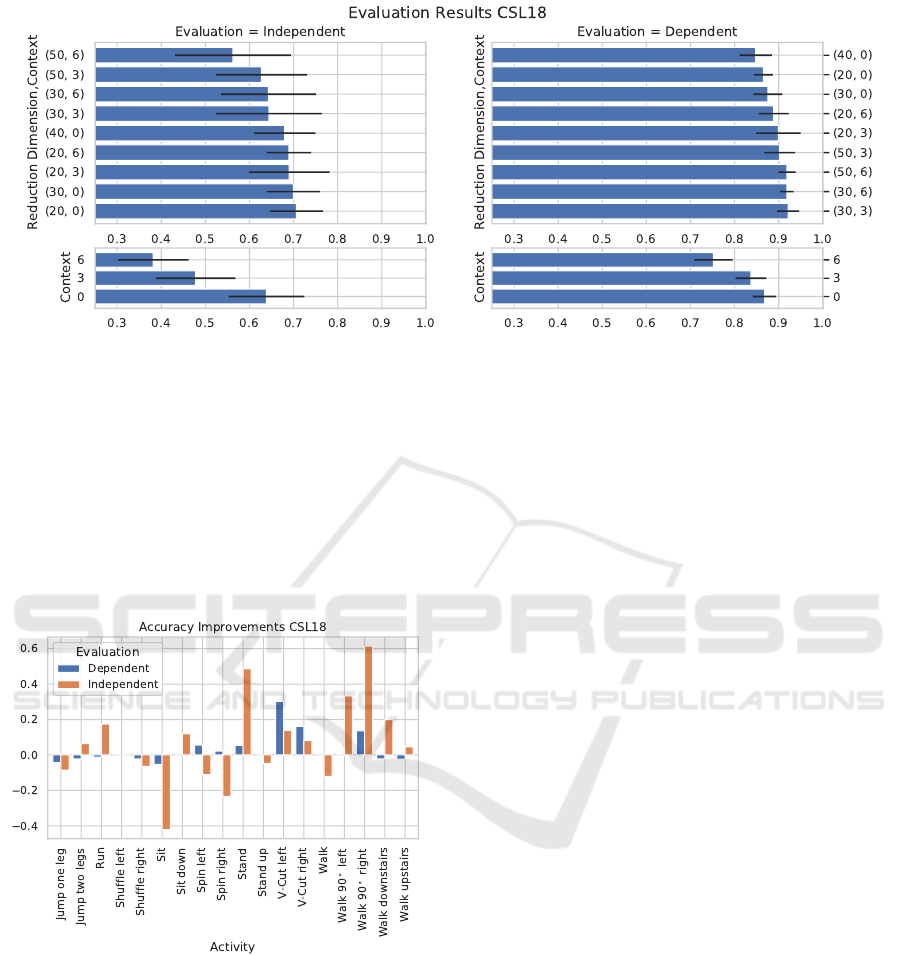
Figure 6: Evaluation results for stacking and reduction on the CSL18 dataset. Left column: person independent evaluation,
right: person dependent evaluation. Upper row: the combination of stacking and reduction (dimension target, stacking
context), lower row: stacking alone. Without stacking, there is no reduction to 50 dimensions but a transformation of the
original 40 dimensions.
out stacking at 70.6 ± 6% balanced accuracy when re-
ducing to a 20-dimensional feature space. A closer
look at the performance increases per activity shown
in Figure 7 reveals similar results to the CSL19
dataset with the walking derivatives being recognized
better, while the static activities stand and sit are con-
fused more often.
Figure 7: Relative recognition accuracy improvements in
percentage points between baseline and reduction based
recognizer on the CSL18 dataset.
7 DISCUSSION
There are several differences between the three rec-
ognizer and datasets, like the number of participants
(CSL18: 4, CSL19: 20, UniMiB: 30), the types of ac-
tivities (ADL, sport, falls), the HMM topology (gait-
phase based, fixed number of states), or the evalu-
ated window length (30, 100, and 400ms). Across
all three, the feature space reduction with an LDA
did improve recognition accuracy for several activi-
ties. The most notable exception being the two only
static activities sit and stand. The resulting overall
performance is either similar or significantly higher
compared to the baseline. Table 3 shows a summary
of the evaluation results.
The best performance on the CSL19 dataset with
93.7 ± 1.4% independent balanced accuracy fits in
with the high ninety percent accuracies reported in
other works even though distinguishing more classes
(Rebelo et al., 2013) (Demrozi et al., 2020) (Lara
et al., 2012). The 92.8% person dependent accuracy
on the CSL18 dataset is slightly lower than the 94.9%
previously reported, as the parameters were optimized
with an independent evaluation instead of the purely
person dependent optimization done previously. The
results on the UniMiB dataset are directly comparable
to current research and better or on par. The person
independent balanced accuracy of 69.5± 8.1% is sig-
nificantly higher than the 56.53% balanced accuracy
(Micucci et al., 2017), and the 77.0% independent ac-
curacy is on par with the 77.03% accuracy (Li et al.,
2018) previously reported.
The LDA does not perform as well as the HMM.
One reason might be the higher number of classes and
higher feature dimensionality compared to the HMM.
However, being trained with more than 350k samples
on both the CSL19 and UniMiB datasets, this seems
unlikely. Instead, this difference is probably caused
by the HMM modeling sequences and the LDA single
vectors, as discussed in section 1. Additionally, the
LDA is given samples that are very similar but differ-
ently labeled. Spin left with the left foot first is shifted
half a gait cycle from spin left starting with the right
BIOSIGNALS 2021 - 14th International Conference on Bio-inspired Systems and Signal Processing
220
Table 3: Baseline and reduction results for both independent and dependent evaluation on each dataset.
Person independent Person dependent
Dataset Baseline Reduction Baseline Reduction
Balanced acc. Balanced acc. Accuracy Balanced acc. Balanced acc. Accuracy
CSL18 63.8 ± 8.5% 70.6 ± 6.0% 73.2% 86.7 ± 2.6% 92.1 ± 2.4% 92.8%
CSL19 93.7 ± 1.4% 93.3 ± 3.9% 93.6% 97.8 ± 0.2% 97.4 ± 0.3% 97.4%
UniMiB 59.7 ± 8.6% 69.5 ± 8.1% 77.0% 79.6 ± 0.5% 90.1 ± 0.7% 93.6%
foot. Nevertheless, the samples are assigned different
targets.
Despite these poor LDA discrimination perfor-
mances, its transformation does contribute to better
overall recognition performance, most notably in the
UniMiB and CSL18 datasets. The reduction was ben-
eficial to the recognition of most activities, with the
notable exception of the two only static activities sit
and stand in the CSL18 and CSL19 dataset, which
are likely impacted by the normalization. The LDA’s
contribution should be investigated further in future
work, especially which activities can or cannot be im-
proved. Furthermore, the LDA should be compared
to a non-discriminatory reduction method like a PCA.
Non-discriminatory methods do not need to separate
targets and potentially handle overlapping classes bet-
ter.
These experiments show that the feature space
reduction using an LDA trained with HMM force
aligned targets can significantly improve the recogni-
tion accuracy of activities as well as overall accuracy.
8 CONCLUSION AND FUTURE
WORK
The curse of dimensionality in HAR can be addressed
in several ways. For instance, through feature se-
lection or feature space transformations into lower-
dimensional spaces. The latter was evaluated on
the three HAR datasets CSL18, CSL19, and UniMiB
using force aligned labels and an LDA combined
with stacking. Initially, baselines were developed
for each dataset. In a person independent leave-one-
person-out cross-validation, the baselines achieved
93.7 ± 1.4% on the CSL19 and 59.7 ± 8.6% on the
UniMiB, and 63.8 ± 8.5% balanced accuracy on the
CSL18 dataset. Then the best context for stack-
ing and best feature dimension target were evaluated.
The reduction did not improve performance on the
CSL19 dataset. However, reduction increased perfor-
mance by ten percentage points to 69.5± 8.1% on the
UniMiB dataset and by seven points to 70.6 ± 6% on
the CSL18 dataset.
In future work, a closer investigation of the classes
as aligned by the HMM will be made and the benefits
and downsides of the LDA-based reduction for differ-
ent activities closer investigated as well as compared
to a PCA-based reduction. Furthermore, experiments
with different feature selection methods will be con-
ducted, and their respective performance compared to
the LDA based reduction.
REFERENCES
Almaslukh, B., Artoli, A. M., and Al-Muhtadi, J.
(2018). A Robust Deep Learning Approach for
Position-Independent Smartphone-Based Human Ac-
tivity Recognition. Sensors, 18(11):3726.
Barandas, M., Folgado, D., Fernandes, L., Santos, S.,
Abreu, M., Bota, P., Liu, H., Schultz, T., and Gamboa,
H. (2020). TSFEL: Time Series Feature Extraction
Library. SoftwareX, 11:100456.
Bulling, A., Blanke, U., and Schiele, B. (2014). A tutorial
on human activity recognition using body-worn iner-
tial sensors. ACM Computing Surveys, 46(3):1–33.
Capela, N. A., Lemaire, E. D., and Baddour, N. (2015).
Feature selection for wearable smartphone-based hu-
man activity recognition with able bodied, elderly, and
stroke patients. PLoS ONE, 10(4):1–18.
Demrozi, F., Pravadelli, G., Bihorac, A., and Rashidi, P.
(2020). Human Activity Recognition using Inertial,
Physiological and Environmental Sensors: a Compre-
hensive Survey. 1(1).
Haeb-Umbach, R. and Ney, H. (1992). Linear discrimi-
nant analysis for improved large vocabulary contin-
uous speech recognition. In [Proceedings] ICASSP-
92: 1992 IEEE International Conference on Acous-
tics, Speech, and Signal Processing, volume 11, pages
13–16 vol.1. IEEE.
Harris, C. R., Millman, K. J., van der Walt, S. J., Gommers,
R., Virtanen, P., Cournapeau, D., Wieser, E., Taylor,
J., Berg, S., Smith, N. J., Kern, R., Picus, M., Hoyer,
S., van Kerkwijk, M. H., Brett, M., Haldane, A., del
R
´
ıo, J. F., Wiebe, M., Peterson, P., G
´
erard-Marchant,
P., Sheppard, K., Reddy, T., Weckesser, W., Abbasi,
H., Gohlke, C., and Oliphant, T. E. (2020). Array pro-
gramming with NumPy. Nature, 585(7825):357–362.
Hartmann, Y., Liu, H., and Schultz, T. (2020). Feature space
reduction for multimodal human activity recognition.
In BIOSIGNALS 2020 - 13th International Confer-
ence on Bio-Inspired Systems and Signal Processing,
Proceedings; Part of 13th International Joint Confer-
Feature Space Reduction for Human Activity Recognition based on Multi-channel Biosignals
221

ence on Biomedical Engineering Systems and Tech-
nologies, BIOSTEC 2020.
Hu, H. and Zahorian, S. A. (2010). Dimensionality Reduc-
tion Methods for Hmm Phonetic Recognition. 2010
IEEE International Conference on Acoustics, Speech
and Signal Processing, pages 4854–4857.
Hunter, J. D. (2007). Matplotlib: A 2D Graphics Environ-
ment. Computing in Science & Engineering, 9(3):90–
95.
Lara,
´
O. D., Labrador, M. A., Lara, O. D., and Labrador,
M. A. (2012). A Survey on Human Activity Recogni-
tion using Wearable Sensors. IEEE Communications
Surveys & Tutorials, 15(3):1192–1209.
Li, F., Shirahama, K., Nisar, M. A., K
¨
oping, L., and Grze-
gorzek, M. (2018). Comparison of feature learning
methods for human activity recognition using wear-
able sensors. Sensors (Switzerland), 18(2):1–22.
Liu, H. and Schultz, T. (2018). ASK: A Framework for Data
Acquisition and Activity Recognition. BIOSIGNALS
2018 - 11th International Conference on Bio-Inspired
Systems and Signal Processing, Proceedings; Part
of 11th International Joint Conference on Biomedi-
cal Engineering Systems and Technologies, BIOSTEC
2018, 4:262–268.
Liu, H. and Schultz, T. (2019). A Wearable Real-time Hu-
man Activity Recognition System using Biosensors
Integrated into a Knee Bandage. BIODEVICES 2019
- 12th International Conference on Biomedical Elec-
tronics and Devices, Proceedings; Part of 12th Inter-
national Joint Conference on Biomedical Engineering
Systems and Technologies, BIOSTEC 2019, pages 47–
55.
Mezghani, N., Fuentes, A., Gaudreault, N., Mitiche, A.,
Aissaoui, R., Hagmeister, N., and De Guise, J. A.
(2013). Identification of knee frontal plane kinematic
patterns in normal gait by principal component anal-
ysis. Journal of Mechanics in Medicine and Biology,
13(03):1350026.
Micucci, D., Mobilio, M., and Napoletano, P. (2017).
UniMiB SHAR: A Dataset for Human Activity
Recognition Using Acceleration Data from Smart-
phones.
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V.,
Thirion, B., Grisel, O., Blondel, M., Prettenhofer, P.,
Weiss, R., Dubourg, V., Vanderplas, J., Passos, A.,
Cournapeau, D., Brucher, M., Perrot, M., and Duch-
esnay,
´
E. (2011). Scikit-learn: Machine Learning
in Python. Journal of Machine Learning Research,
12(85):2825–2830.
Rebelo, D., Amma, C., Gamboa, H., and Schultz, T. (2013).
Human Activity Recognition for an Intelligent Knee
Orthosis. BIOSIGNALS 2013 - Proceedings of the In-
ternational Conference on Bio-Inspired Systems and
Signal Processing, pages 368–371.
Siohan, O. (1995). On the robustness of linear discrimi-
nant analysis as a preprocessing step for noisy speech
recognition. ICASSP, IEEE International Conference
on Acoustics, Speech and Signal Processing - Pro-
ceedings, 1(3):125–128.
Suto, J., Oniga, S., and Sitar, P. P. (2016). Comparison of
wrapper and filter feature selection algorithms on hu-
man activity recognition. In 2016 6th International
Conference on Computers Communications and Con-
trol (ICCCC), number Icccc, pages 124–129. IEEE.
Telaar, D., Wand, M., Gehrig, D., Putze, F., Amma, C.,
Heger, D., Vu, N. T., Erhardt, M., Schlippe, T., Janke,
M., Herff, C., and Schultz, T. (2014). BioKIT - Real-
time Decoder For Biosignal Processing. In The 15th
Annual Conference of the International Speech Com-
munication Association, Singapore, number Novem-
ber, pages 2650–2654.
Virtanen, P., Gommers, R., Oliphant, T. E., Haberland, M.,
Reddy, T., Cournapeau, D., Burovski, E., Peterson, P.,
Weckesser, W., Bright, J., van der Walt, S. J., Brett,
M., Wilson, J., Millman, K. J., Mayorov, N., Nel-
son, A. R., Jones, E., Kern, R., Larson, E., Carey,
C. J., Polat,
˙
I., Feng, Y., Moore, E. W., VanderPlas,
J., Laxalde, D., Perktold, J., Cimrman, R., Henriksen,
I., Quintero, E. A., Harris, C. R., Archibald, A. M.,
Ribeiro, A. H., Pedregosa, F., van Mulbregt, P., Vi-
jaykumar, A., Bardelli, A. P., Rothberg, A., Hilboll,
A., Kloeckner, A., Scopatz, A., Lee, A., Rokem, A.,
Woods, C. N., Fulton, C., Masson, C., H
¨
aggstr
¨
om,
C., Fitzgerald, C., Nicholson, D. A., Hagen, D. R.,
Pasechnik, D. V., Olivetti, E., Martin, E., Wieser, E.,
Silva, F., Lenders, F., Wilhelm, F., Young, G., Price,
G. A., Ingold, G. L., Allen, G. E., Lee, G. R., Au-
dren, H., Probst, I., Dietrich, J. P., Silterra, J., Webber,
J. T., Slavi
ˇ
c, J., Nothman, J., Buchner, J., Kulick, J.,
Sch
¨
onberger, J. L., de Miranda Cardoso, J. V., Reimer,
J., Harrington, J., Rodr
´
ıguez, J. L. C., Nunez-Iglesias,
J., Kuczynski, J., Tritz, K., Thoma, M., Newville, M.,
K
¨
ummerer, M., Bolingbroke, M., Tartre, M., Pak, M.,
Smith, N. J., Nowaczyk, N., Shebanov, N., Pavlyk,
O., Brodtkorb, P. A., Lee, P., McGibbon, R. T., Feld-
bauer, R., Lewis, S., Tygier, S., Sievert, S., Vigna, S.,
Peterson, S., More, S., Pudlik, T., Oshima, T., Pin-
gel, T. J., Robitaille, T. P., Spura, T., Jones, T. R.,
Cera, T., Leslie, T., Zito, T., Krauss, T., Upadhyay,
U., Halchenko, Y. O., and V
´
azquez-Baeza, Y. (2020).
SciPy 1.0: fundamental algorithms for scientific com-
puting in Python. Nature Methods, 17(3):261–272.
Weiner, J. and Schultz, T. (2018). Selecting Features for
Automatic Screening for Dementia Based on Speech.
pages 747–756.
BIOSIGNALS 2021 - 14th International Conference on Bio-inspired Systems and Signal Processing
222
