
Disambiguating Clinical Abbreviations using Pre-trained Word
Embeddings
Areej Jaber
1,2
and Paloma Mart
´
ınez
1
1
Computer Science Department, Universidad Carlos III de Madrid, Legan
´
es, Spain
2
Applied Computer Science Department, PTUK University, Tulkarm, Palestine
Keywords:
Clinical Natural Language Processing, Word Sense Disambiguation, Word Embeddings, Clinical
Abbreviations, Pre-trained Models.
Abstract:
Abbreviations are broadly used in clinical texts and most of them have more than one meaning which makes
them highly ambiguous. Determining the right sense of an abbreviation is considered a Word Sense Disam-
biguation (WSD) task in clinical natural language processing (NLP). Many approaches are applied to disam-
biguate abbreviations in clinical narrative. However, supervised machine learning approaches are studied in
this field extensively and have proven a good performance at tackling this problem. We have investigated
four strategies that integrate pre-trained word embedding as features to train two supervised machine learning
models: Support Vector Machines (SVM) and Naive Bayes (NB). Our training features include information
of the context of target abbreviation, which is applied on 500 sentences for each of the 13 abbreviations that
have been extracted from public clinical notes data sets from the University of Minnesota-affiliated (UMN)
Fairview Health Services in the Twin Cities. Our results showed that SVM performs better than NB in all four
strategies; the highest accuracy being 97.08% using a pre-trained model trained from Wikipedia, PubMed and
PMC (PubMedCentral) texts.
1 INTRODUCTION
Electronic Health Records (EHR) in hospitals and
medical centers store a high volume of patients data
in computerised systems. Part of this data is unstruc-
tured clinical narrative that is necessary to translate
into structured data to be useful in clinical decision-
making processes. Abbreviations and acronyms are
widely used in clinical notes in order to represent im-
portant clinical concepts like a disease or a procedure,
and their use continues to increase (Xu et al., 2007). It
is critical to understand these terms to extract relevant
information from clinical texts.
Abbreviations are defined as “a short form of
words or phrases” used instead of its definition and
acronyms are considered a special type of abbrevia-
tions which are created with the initial letters or sylla-
bles of other words. In this paper abbreviation term is
used for simplicity. Furthermore, abbreviations could
be global if they appear in the documents without
their expansions, whereas local ones come together
with their expansions in the document. Global abbre-
viations are highly ambiguous and are the focus of
this paper.
Abbreviations which are used in medical systems
could be standard terminology; for instance, there is
a list of commonly used medical abbreviations dictio-
nary
1
recommended by Spanish Ministry of Health.
But the main obstacle is that most used abbreviations
are specific to the medical center and sometimes also
to the doctor who writes the clinical note. Conse-
quently, it is a hard task to have an updated repository
that collects every new abbreviation with its senses.
Each abbreviation has a Short Form (SF) and a
set of Long Forms (LF) which are known as senses.
Unlike to the biomedical text, clinical abbreviations
are global abbreviations. Furthermore, there is no
standard rules for creating new abbreviations by clin-
icians and consequently the correct sense depends on
the context where the abbreviation is normally used.
Sharing EHR among hospitals and even with patients,
makes this vital because misunderstanding for these
abbreviations lead to unsatisfactory results.
(Liu et al., 2001) reported that 33.1% of abbre-
viations in the Unified Medical Language System
(UMLS), (Mcinnes et al., 2011), have more than one
sense.However, (Xu et al., 2007) showed that UMLS
only covered 35% of senses of abbreviations in hospi-
1
https://www.sedom.es
Jaber, A. and Martínez, P.
Disambiguating Clinical Abbreviations using Pre-trained Word Embeddings.
DOI: 10.5220/0010256105010508
In Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2021) - Volume 5: HEALTHINF, pages 501-508
ISBN: 978-989-758-490-9
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
501

tal admission notes at New York Presbyterian Hospi-
tal. Furthermore, 80% of the abbreviations included
in UMLS have ambiguous occurrences in Medline,
(Liu et al., 2001). (Schulz et al., 2017) found 7,439
ambiguous SNOMED-CT terms and 899 ambigu-
ous acronyms. A SF could have many LF’s not
only in medical domain but also in general domain.
For example, “ACA” could be “Acinic Cell Carci-
noma”, “Affordable Care Act” and “Anterior Cerebral
Artery”. Determining the right sense depends on the
context where “ACA” is used.
Disambiguate clinical abbreviations is considered
a WSD process, which means “computationally de-
termine which sense of a word is activated by its con-
text” (Navigli, 2009). WSD is considered one of the
most difficult tasks in NLP and much work has been
done for disambiguation of abbreviations in clinical
text (Moon et al., 2013), (Pakhomov, 2002), (Kashyap
et al., 2020) and (Lu, 2019).
Supervised Machine learning (ML) approaches
proved its performance for this task. These ap-
proaches require sense tagged corpora. WSD task
is considered as a classification problem where the
objective is to predict the correct sense of an abbre-
viations among its different senses. Apart from tra-
ditional features, in last years word embedding have
been used also as features used in classifications tasks.
A word embedding is a real-value vector that repre-
sents a single word based on the context in which it
appears (Khattak et al., 2019). These numerical word
representations could be built using different models
like (Mikolov et al., 2013), (Peters et al., 2018) and
(Devlin et al., 2019) based on different neural net-
works architectures. Fortunately, these embeddings
could be trained on large data set, saved and used in
solving other tasks; they are called pre-trained word
embeddings or pre-trained models. Word2Vec is one
of the most popular pre-trained word embeddings de-
veloped by Google that is implemented on two ways:
as a continuous bag-of-words (CBOW) and as a Skip-
Gram (SG) (Mikolov et al., 2013). The key difference
between the two approaches is if the neural network
tempts to predict a word on a given context (CBOW)
or the reverse.
In this paper two supervised machine learning
(ML) methods, SVM (Cortes and Vapnik, 1995) and
NB (Tschiatschek et al., 2014), have been trained and
tested using two different pre-trained word embed-
dings.
2 RELATED WORK
Any WSD system acts as follows: given an ambigu-
ous abbreviation, a technique which makes use of one
or more sources of knowledge associates the most ap-
propriate sense considering words in context of the
abbreviation. Knowledge sources can vary consid-
erably from corpora of texts, either unlabelled or la-
belled with word senses, to more structured resources,
such as machine-readable dictionaries, semantic net-
works, etc. Normally, WSD approaches are classified
according to the source of knowledge used to discrim-
inate among senses. Knowledge-based (KB), super-
vised, semi-supervised and unsupervised approaches
are distinguished.
KB approaches can be (1) rule based approaches
used hand coded patterns that represent regulari-
ties in texts derived by inspection of corpora and
(2) semantic-based approaches that integrated lexi-
cal knowledge bases and exploit semantic similar-
ity and graph-based approaches to disambiguate. In
similarity-based method each sense of the ambigu-
ous abbreviation is compared to those of the content
words appearing near it (context words) and the sense
with the highest similarity (for instance, using cosine
distance) is supposed to be the right one. Graph-
based methods use an entire knowledge base (such as
UMLS) during disambiguation by propagating infor-
mation to the graph.
Secondly, unsupervised machine learning algo-
rithms disambiguate by finding hidden structure
structure in unlabelled data, for instance, clustering
documents or sentences in groups each one represent-
ing a sense. Finally, supervised methods make use
of annotated corpora to derive a function that pre-
dicts the sense of an abbreviation and semi-supervised
methods use seed data in a bootstrapping process us-
ing few annotated corpora. Next, we will describe the
most significant works including in these four types of
approaches for WSD that are applied to disambiguate
clinical abbreviations.
2.1 Unsupervised Approaches
Unsupervised approaches are adequate to discover
and annotate senses not included in medical termi-
nologies. Work described in (Xu et al., 2012) used
clustering methods to build sense inventories for clin-
ical abbreviations by grouping in each cluster sen-
tences with the same sense. The objective is address-
ing the problem of infrequent senses that tend to be
included in other larger clusters; they collected infor-
mation for each ambiguous abbreviation from the sur-
rounding words in addition to section name of clini-
HEALTHINF 2021 - 14th International Conference on Health Informatics
502

cal notes being able of detecting 85% senses on av-
erage evaluating over 13 abbreviations on a corpus of
physician-typed inpatient admission notes from New
York Presbyterian Hospital.
(Wu et al., 2017) described a clinical abbrevia-
tion recognition and disambiguation (CARD) frame-
work that leverages a profile-based WSD consisting
on several steps. First, the LF (or senses) of abbrevi-
ations in the sentences of a dataset of discharge sum-
maries and admission notes from Presbyterian Hos-
pital are replaced with the corresponding SF (leav-
ing also its sense) using LRABR (UMLS abbrevia-
tion list) and other repositories of abbreviations. Sec-
ond, with these set of instances for each abbreviation
sense a feature vector for each instance in the train-
ing set is generated. Several types of features are
used: stemmed words in window size of 5 of the tar-
get abbreviation, positional information and section
headers of documents. These features are weighted
using a TFIDF schema. Definitely, each instance of
an abbreviation is considered as a document in a vec-
tor space model and each feature is a weighted term
in this document. A sense profile vector is then built
for all instances of an abbreviation sense by averaging
weight of the features across all the instances with that
sense. Finally, to disambiguate, a sense profile vector
is generated for sentences of test dataset. This vec-
tor is compared using the cosine similarity distance
to each of the sense profile vectors to get the closest
sense. This method achieved a precision of 87,5%.
2.2 Knowledge-based Approaches
Rule-based approaches are used in abbreviation
recognition by means of regular expressions that rep-
resent lexical patterns containing terms and context
represented by concepts (such as diseases, symptoms,
etc.). The winner in BARR2 shared task (S
´
anchez-
Le
´
on, 2018) implemented a set of 30 templates ex-
tracted from 500 Spanish clinical cases and 130 rules
that model SF occurring in different parts of a clini-
cal case. The system obtained 82.89 of F1 in BARR2
WSD sub-task 2 by combining templates with a n-
gram frequency based approach that compares the
content word list for each LF for a given SF to the fre-
quency profile for the clinical case text (the best scor-
ing LF is selected). This poses again the discussion
about machine learning versus rule-based approaches.
There are domains where rule-based methods work
better than machine learning ones. In specific do-
mains, global context is used to resolve ambiguity, for
instance, senses for a specific abbreviation are differ-
ent in neonatal clinical notes that in radiology reports
but this means that different set of rules are required
for different domains.
Semantic-based systems use lexical resources to
map the ambiguous abbreviation to the most feasible
definition. Consequently, KB approaches are useful
to process languages with available resources, such
as the case of English UMLS that incorporates abbre-
viations lists.
(Mcinnes et al., 2011) described a method to dis-
ambiguate biomedical abbreviations. A second-order
vector for a specific sense is created by first ob-
taining a textual description of the possible sense.
This consists of its definition, the definition of its
parent/children and narrow/broader relations and the
terms associated with the sense from UMLS. Second,
a word by word co-occurrence matrix is created where
the rows represent the content words in the descrip-
tion and the columns represent words that co-occur
with the words in the description found in MEDLINE
abstracts. Lastly, each word in the sense’s description
is replaced by its corresponding vector, as given in
the co-occurrence matrix. The average of these vec-
tors constitutes the second order co-occurrence vec-
tor used to represent the sense. The second-order co-
occurrence vector for the ambiguous term is created in
a similar fashion by using the words surrounding the
ambiguous term in the instance as its textual descrip-
tion. Then, cosine similarity was computed between
the vector representing the abbreviation and each vec-
tor representing senses. The sense vector with the
smallest angle with abbreviation vectors is chosen to
be the right sense. This method achieved an accuracy
of 89% on a dataset of 18 acronyms found in Medline
abstracts.
2.3 Supervised Approaches
Supervised approaches are ML classification mod-
els induced from semantically annotated corpora.We
have a training set that contains a number of examples
in the form of (x
1
, y
1
), ...(x
n
, y
n
) , where x
i
is a vec-
tor of features that represents the target abbreviation.
The output y
i
are the classes (sense or LF of the tar-
get abbreviation). The main drawback of supervised
methods is the large amount of manually labeled data
they require. To tackle this problem alternative semi-
supervise methods arise that use a small amount of
labeled data as seeds to automatically labeled unla-
beled data. At the end of this process, a big amount
of labeled data is available which could be used to
fully supervised approaches. Most works select a set
of ambiguous and frequently used abbreviations and
build a classifier for each abbreviation trained from
examples containing the abbreviation.
(Pakhomov, 2002) used six abbreviations and their
Disambiguating Clinical Abbreviations using Pre-trained Word Embeddings
503

LF from LRABR file (included in UMLS) to annotate
a subset of Rheumatology notes from Mayo Clinic by
replacing LF by SF. This corpus is then used to train
a Maximum Entropy (ME) classifier to disambiguate
contexts of SF (local context) and also information
about the section of the document where the abbrevia-
tion appears (global context). Indeed, a separate clas-
sifier is built for each abbreviation and are compare
with the results of only one classifier for all abbrevia-
tions (with similar results around 89% Accuracy).
(Wu et al., 2015b) derived their WSD features
from neural word embedding trained on public clin-
ical notes MIMIC II (Amir, Weber, Beard, Bomyea,
2011). Two feature vectors are created for this study.
One of them is generated by taking the MAX score
of each embedding dimension over all the surround-
ing words (MAX SBE), the second is generating by
summing up the embedding row vectors of the sur-
rounding words from each side (LR SBE). (Wu et al.,
2015b) combined these features with traditional set
of WSD features and trained a SVM to disambiguate
a set of abbreviations from UMN (Moon et al., 2012)
and VUH (Wu et al., 2013). The best accuracy for this
model achieved 95.97% on UMN dataset, and 93.01%
on VUH dataset, when (MAX SBE) with standard
WSD features are merged.
Recent research investigated deep features in this
domain, (Joopudi et al., 2018) applied convolu-
tional neural network (CNN) on different data sets
from PubMed and and clinical notes; the model im-
proved the accuracy in the range of 1 to 4 percent-
age points. (Lu, 2019) trained ElMo (Peters et al.,
2018) on the MIMIC-III corpus and implemented
a neural topic-attention model to disambiguate clin-
ical abbreviations. Their model results achieved
74.76%. (Kashyap et al., 2020) applied logistic re-
gression, BERT (Devlin et al., 2019) and BioBERT
(Lee et al., 2020) to predict senses for ambiguous ab-
breviation from PubMed Central where 150 papers
per unique abbreviation-sense pair were used. Then
the model was used to predict the senses from 1000
MIMIC-III clinical notes for each abbreviation. The
model achieved best overall accuracy 76.92% using
BioBERT. Furthermore, based on this study, 57.29%
of abbreviations could be overlapped in both clinical
and biomedical domains.
2.4 Semi-supervised Approaches
Fully supervised ML bottleneck is that a large size
of manual annotated dataset is required. Since it
is a time-consuming and expensive task, many re-
searchers tried to overcome this problem by applying
semi-supervised approaches which differ from fully
supervised approaches in how training data is col-
lected (Pakhomov, 2002), (Finley et al., 2016) and
(Wu et al., 2017). Supervised approaches require
manually annotated training and testing data sets. In
semi-supervised approaches training data is automat-
ically generated but testing data needs to be manually
annotated.
(Finley et al., 2016) applied this approach by auto
generating the training data using reverse substitution
methods on large clinical data repository in Fairview
Health Services system. Then, the model was tested
on manually annotated dataset .(Finley et al., 2016)
applied NB, Multinomial logistic regression, SVM
and cosine similarity with bag of words and hyper-
dimensional indexing for representing the features.
The system get 94.2% and 96.2% for NB and SVM,
respectively.
(Wu et al., 2017) applied semi-supervised clus-
tering with profile-based WSD to develop an open
source framework for recognizing and disambiguat-
ing clinical abbreviations. They implemented fea-
ture vectors to represent different senses in vector
space model. The system is evaluated in Vanderbilt
University Medical Center (VUMC) corpus and the
ShARe/CLEF 2013 challenge corpus, and achieved a
F score of 76% , 29% respectively.
3 METHOD
An overview of our method is shown in Figure 1.
Three phases are distinguished; first, dataset gener-
ation where different pre-processing tasks are per-
formed to prepare and clean data, second training a
machine learning model for classification (SVM and
NB methods) using different types of features and fi-
nally, testing the model over the test dataset. The
following subsections will detail the various steps in-
volved in disambiguation the clinical abbreviations
using different strategies integrating pre-trained mod-
els over two supervised machine learning algorithms:
SVM and NB classifiers.
3.1 Data Set
In this study a publicly annotated clinical notes
dataset from the University of Minnesota-affiliated
(UMN) Fairview Health Services in the Twin Cities
was used (Moon et al., 2012). It was collected from
admission notes, inpatient consult notes, operative
notes, and discharge summaries with window size 12
for each size. The whole dataset contains 75 abbrevia-
tions of the most frequent acronyms and abbreviations
HEALTHINF 2021 - 14th International Conference on Health Informatics
504
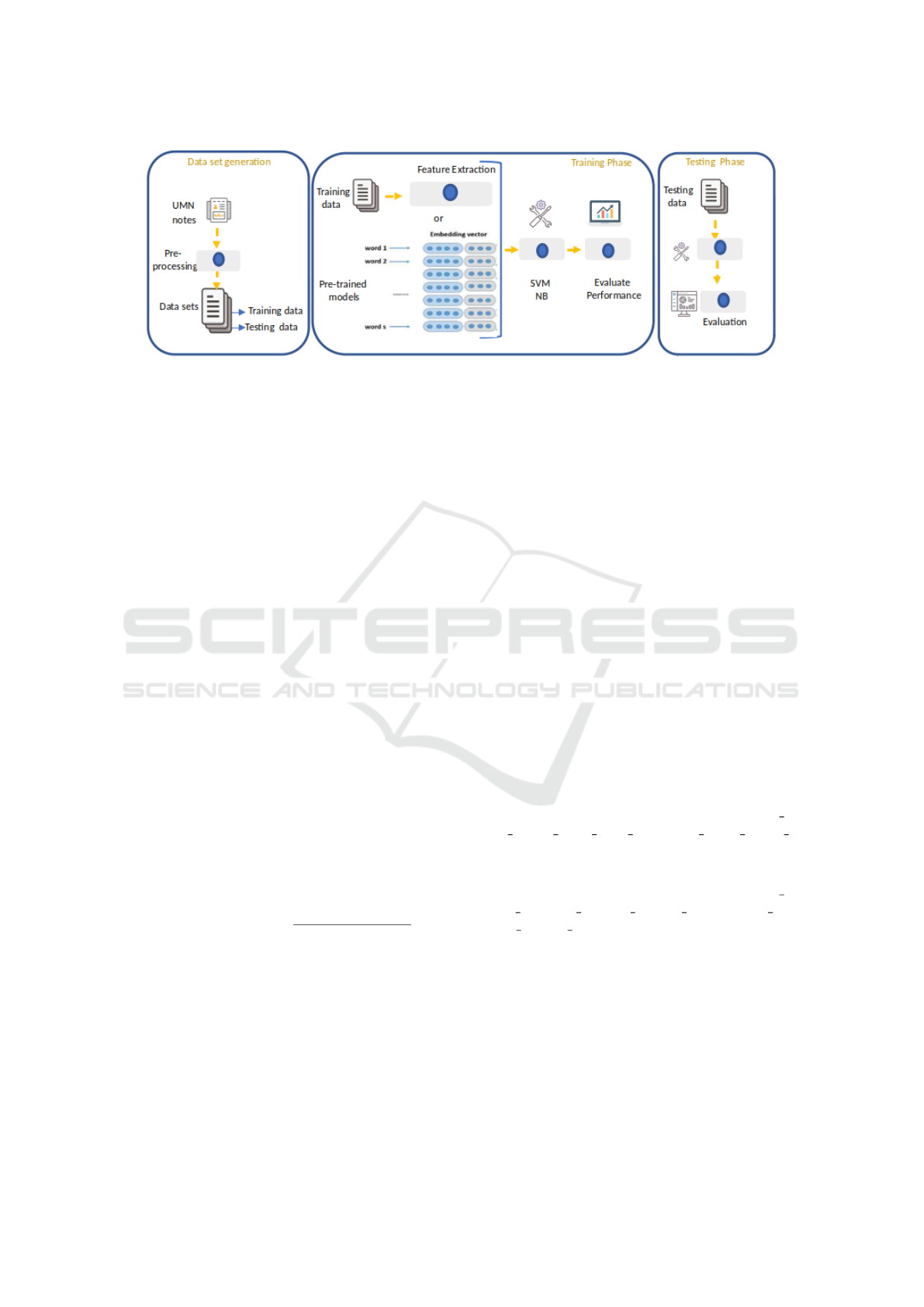
Figure 1: Overview of our approach to disambiguate clinical abbreviations. Training and testing phases are repeated for each
abbreviations in the dataset.
with 351 senses in total with an average of 4.7 senses
per abbreviation.
Table 1 summarizes the data for 13 abbreviations
chosen for this study with a total of 6588 instances;
88 instances are excluded due to annotation errors.
The abbreviations are shown in the first column; sec-
ond and third columns show the number of sentences
and tokens per abbreviation in the dataset. Fourth
column lists the senses found in this dataset. Then,
the fifth and sixth columns show the number of oc-
currences per sense and the frequency percentage for
each sense. The dataset contains 33 different senses
with an average of 2.5 senses per abbreviation.
3.2 Model
The objective of this study is to compare the effi-
ciency of two ML algorithms with different types
of features, particularly using pre-trained word em-
bedding. NB classifier is a probabilistic approach
to estimate probabilistic parameters which has a
long history of success in WSD. This approach is
based on Bayes theorem to compute the conditional
probability for each sense of an abbreviation for
which a set of features is defined (x
1
, x
2
, .., x
m
). Let
(P(sense)andP(x
i
|sense) are the probabilistic param-
eters of the model and they can be estimated from the
training set using relative frequency counts (equation
1).
argmaxP(sense|x
1
, ..., x
m
) = argmax
P(x
1
, ..., x
m
|sense)P(sense)
P(x
1
, ..., x
m
)
= argmaxP(sense)
m
∏
i=1
P(x
i
|sense)
(1)
On the other hand, SVM separates positive samples
from negative ones based on the idea of linear hyper-
plane from labeled data set differentiating between
samples into true or false categories. SVM is adapted
to multi-class classification for word sense disam-
biguation. It is then converted into binary classifi-
cation problem of the type sense Si versus all other
senses.
A set of WSD features used as a baseline are de-
scribed below. Then, different strategies of combin-
ing features were tested using pre-trained word em-
beddings of window size 5 for each side. For both
algorithms, a separate model was trained for each of
the 13 abbreviations.
3.3 WSD Features
Several features were used to disambiguate clinical
abbreviations considering both left and right contexts
of the target abbreviation. In this study, we imple-
mented traditional features that have been successful
in WSD (Wu et al., 2015a) with window size 5 for
each side. These features are described using next
sentence as an example: ” ...Last time she was dis-
charged AMA and since she ...”.
1. Word Features- stemmed words within a window
size 5 for each side of the target abbreviation.
{last, time, she, wa, discharg, and, sinc, she }.
2. Word features with direction- The relative direc-
tion (left or right side) of stemmed words. {l last,
l time, l she, l wa, l discharg, r and, r sinc, r she
}.
3. Position features- The distance between the fea-
ture word and the target abbreviation. {l5 last,
l4 time, l3 she, l2 wa, l1 discharg, r1 and ,
r2 sinc, r3 she }.
4. Word formation features from the abbreviation it-
self including special characters, capital letters
and numbers.
3.4 Word Embedding Features
Two pre-trained models which were trained with
Word2vec using skip-gram with a window size 5 to
create 200-dimensional vectors (Pyysalo et al., 2013)
are used in this study, see table 2.
Disambiguating Clinical Abbreviations using Pre-trained Word Embeddings
505
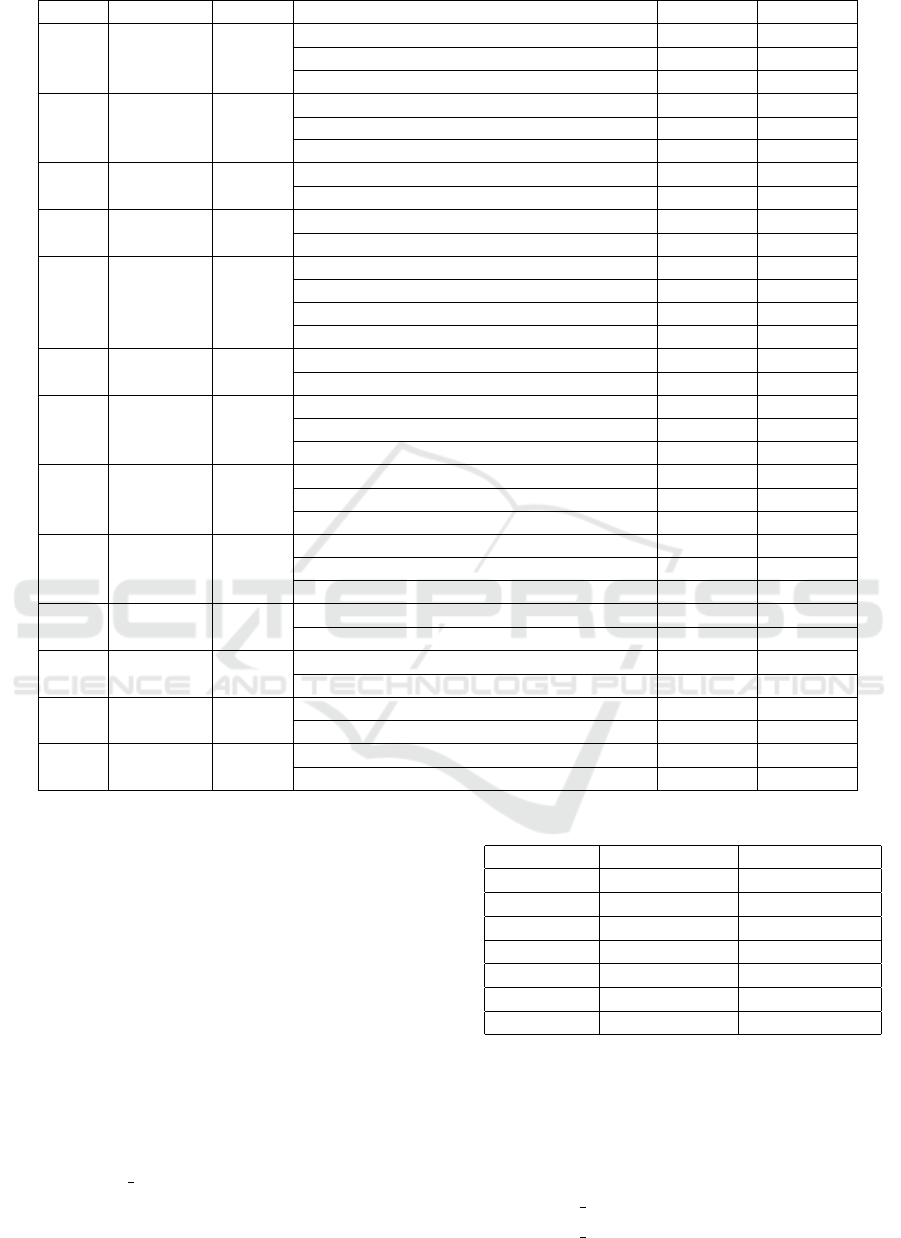
Table 1: List of the 13 abbreviations that are used in this study with their senses from UMN dataset.
Abb Sentences Tokens Senses Sense No Sense(%)
AMA 2881 37887
against medical advice 444 88.8
advanced maternal age 31 6.2
antimitochondrial antibody 25 5.0
ASA 6117 37047
acetylsalicylic acid 404 80.41
American Society of Anesthesiologists 93 18.98
aminosalicylic acid 3 .61
BAL 3267 38483
bronchoalveolar lavage 457 91.4
blood alcohol level 43 8.6
BK 3721 37687
BK (virus) 343 68.35
below knee 157 31.65
C3 3270 39901
cervical (level) 3 249 49.8
(complement) component 3 243 48.6
propionylcarnitine 6 1.2
(stage) C3 2 0.4
CVA 5212 36616
cerebrovascular accident 278 55.6
costovertebral angle 222 44.4
CVP 3919 37573
central venous pressure 436 87.2
cyclophosphamide, vincristine, prednisone 62 12.4
cardiovascular pulmonary 2 0.4
CVS 2224 36722
chorionic villus sampling 457 91.4
cardiovascular system 41 8.2
customer, value, service 2 0.4
ER 3199 37013
emergency room 448 89.52
extended release 34 6.85
estrogen receptor 18 3.63
FISH 3129 39248
fluorescent in situ hybridization 449 89.8
GENERAL ENGLISH TERM 51 10.2
NAD 6417 41364
no acute distress 377 75.30
nothing abnormal detected 123 24.70
OTC 6173 37356
over the counter 469 93.8
ornithine transcarbamoylase 31 6.2
SBP 3867 38000
spontaneous bacterial peritonitis 417 83.4
systolic blood pressure 83 16.6
The first model was trained on a collection of
unlabelled data extracted from PMC articles. The
second was trained on a combination of unlabelled
data extracted from approximately four million En-
glish Wikipedia articles, PubMed abstracts (nearly 23
million abstracts) in addition to PMC. We investigate
four methods for integrating word embedding to dis-
ambiguate clinical abbreviations and how the differ-
ent training parameters can affect the model. These
methods are discussed below.
The summation of the embedding row vector of
surrounding words for the abbreviation with window
size 5 from each size is calculated as shown in equa-
tion 2.
SUM W E(w) =
j+5
∑
i= j−5
Emb(S(i)) (2)
Where w is the target word to disambiguate, j is the
Table 2: Pre-trained models Details.
Details Model 1 Model 2
Language English English
Resource PMC PMC, PubMed
Documents 672,589 22,792,858
Sentences 105,194,341 229,810,015
Tokens 2,591,137,744 5,487,486,225
Vector size 200 200
Algorithms Skip-gram Skip-gram
index of w, S is the sentence containing w and S(i)
is the word indexed by position i in sentence S. Sec-
ond and third strategies are computing by taking the
maximum and the minimum value for each embed-
ding dimension for the surrounding words. As shown
in equations 3, 4 respectively.
MAX W E(w)
j
= MAX{Emb
j
S(i)
} (3)
MIN W E(w)
j
= MIN{Emb
j
S(i)
} (4)
HEALTHINF 2021 - 14th International Conference on Health Informatics
506
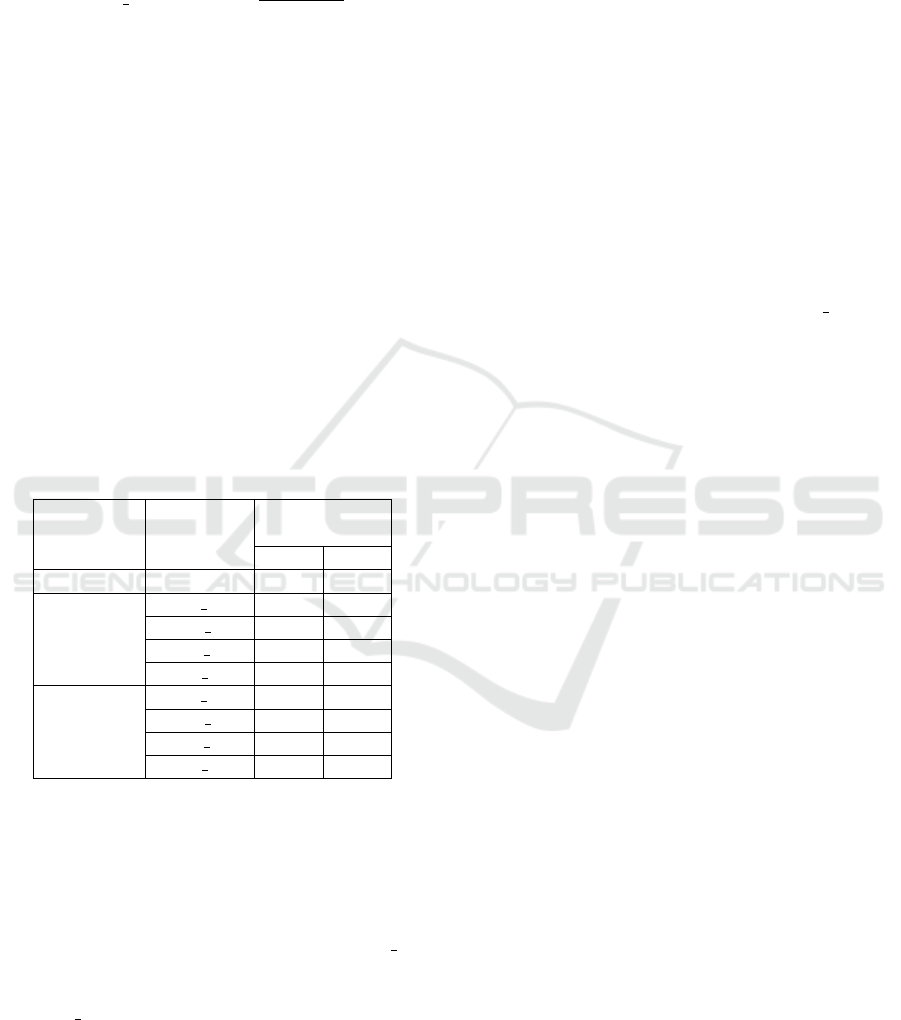
The last strategy is generated by computing the aver-
age for the word embedding vectors surrounding the
abbreviations, as is shown in equation 5
AV G W E(w) =
j+5
∑
i= j−5
Emb(S(i))
2W
(5)
4 RESULTS & DISCUSSION
For each abbreviation in the dataset, SVM and NB
were trained using conventional features as a baseline.
Then, in order to realize the effect of each trained rep-
resentation feature, the model was retrained using the
four strategies for word embedding. Each dataset was
randomly split in the 80/20 fashion (training/testing).
We then reported the accuracy across the 13 abbrevi-
ations which are used in this study.
Table 3 shows the macro accuracy for each abbre-
viation in both SVM and NB algorithms. The SVM
classifier achieved 94.3% in average and NB achieved
91.82% in the baseline.
Table 3: Average accuracy of the WSD systems using pre-
trained word embedding on 13 abbreviations selected from
UMN dataset.
Pre-trained
Resource
Features
Average
Accuracy(%)
SVM NB
Standard Baseline 94.30 91.82
PMC
MIN WE 96.61 92.91
MAX WE 96.15 93.00
SUM WE 96.47 90.59
AVG WE 95.99 84.59
Wikipedia
PubMed
PMC
MIN WE 97.07 92.91
MAX WE 97.08 93.34
SUM WE 96.69 90.82
AVG WE 96.30 86.60
Remarkably, SVM clinical abbreviation disam-
biguation system performs better than NB in all ex-
periments. All the four types of embedding features
improved the average accuracy for both pre-trained
models that are used in this study with improvement
ranging from 2 to 3 percentage. The best performance
for PMC pre-trained word embedding is MIN WE
with 96.61%, while the highest accuracy is gotten
from Wikipedia, PubMed and PMC pre-trained model
on MAX WE features with 97.08%.
Our work differs from (Wu et al., 2015b) because
they trained their own word embedding model from
MIMIC II and we used pre-trained ones generated
from a combination of biomedical and general re-
sources. Furthermore, they got the best result from
using both traditional and word embedding features.
Our best result is got using the pre-trained word em-
bedding features.
5 CONCLUSIONS
In this paper we have investigated the effect of word
embedding features for disambiguate clinical abbre-
viations. Four strategies to integrate word embedding
features from two pre-trained models were tested us-
ing SVM and NB classifiers on 13 clinical abbrevia-
tions that were manually annotated with their expan-
sions. The results showed that SVM outperforms NB
in all four strategies and the best system performance
was achieved when using a pre-trained model that is
generated from PubMed, PMC biomedical literature
and Wikipedia dump data sets with MAX WE fea-
ture.
Some issues still remain uncovered processing ab-
breviations in medical text. The problem of low re-
sourced languages (both annotated corpora and se-
mantic resources like dictionaries or ontologies), de-
mands new approaches to extract new abbreviations
with their definitions from clinical narrative to pop-
ulate medical databases. Clinical narrative has pecu-
liarities (misspellings, incomplete sentences, abuse of
negation, high ambiguity, etc.) that require methods
different than those used in biomedical texts such as
Medline scientific literature.
ACKNOWLEDGEMENTS
Thanks to Palestine Technical University-Kadoorie
and DeepEMR project (TIN2017-87548-C2-1-R) for
partially funding this work.
REFERENCES
Amir, Weber, Beard, Bomyea, T. (2011). Multiparameter
Intelligent Monitoring in Intensive Care II. Crit Care
Med, 23(1):1–7.
Cortes, C. and Vapnik, V. (1995). Support-vector networks.
Machine learning, 20(3):273–297.
Devlin, J., Chang, M. W., Lee, K., and Toutanova, K.
(2019). BERT: Pre-training of deep bidirectional
transformers for language understanding. NAACL
HLT 2019 - 2019 Conference of the North American
Chapter of the Association for Computational Lin-
guistics: Human Language Technologies - Proceed-
ings of the Conference, 1(Mlm):4171–4186.
Disambiguating Clinical Abbreviations using Pre-trained Word Embeddings
507

Finley, G. P., Pakhomov, S. V. S., McEwan, R., and Melton,
G. B. (2016). Towards Comprehensive Clinical Ab-
breviation Disambiguation Using Machine-Labeled
Training Data. AMIA ... Annual Symposium proceed-
ings. AMIA Symposium, 2016:560–569.
Joopudi, V., Dandala, B., and Devarakonda, M. (2018).
A convolutional route to abbreviation disambiguation
in clinical text. Journal of Biomedical Informatics,
86(June):71–78.
Kashyap, A., Burris, H., Callison-Burch, C., and Boland,
M. R. (2020). The CLASSE GATOR (CLini-
cal Acronym SenSE disambiGuATOR): A Method
for predicting acronym sense from neonatal clinical
notes. International Journal of Medical Informatics,
137(October 2019):104101.
Khattak, F. K., Jeblee, S., Pou-Prom, C., Abdalla, M.,
Meaney, C., and Rudzicz, F. (2019). A survey of word
embeddings for clinical text. Journal of Biomedical
Informatics: X, 4(October):100057.
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., and
Kang, J. (2020). BioBERT: A pre-trained biomedi-
cal language representation model for biomedical text
mining. Bioinformatics, 36(4):1234–1240.
Liu, H., Lussier, Y. A., and Friedman, C. (2001). A study
of abbreviations in the UMLS. Proceedings. AMIA
Symposium, pages 393–7.
Lu, X. (2019). Deep Contextualized Biomedical Abbrevia-
tion Expansion.
Mcinnes, B. T., Pedersen, T., Liu, Y., Pakhomov, S. V., and
Melton, G. B. (2011). Using second-order vectors in
a knowledge-based method for acronym disambigua-
tion. CoNLL 2011 - Fifteenth Conference on Compu-
tational Natural Language Learning, Proceedings of
the Conference, (June):145–153.
Mikolov, T., Sutskever, I., Chen, K., Corrado, G., and Dean,
J. (2013). Distributed representations ofwords and
phrases and their compositionality. Advances in Neu-
ral Information Processing Systems, pages 1–9.
Moon, S., Berster, B.-T., Xu, H., and Cohen, T. (2013).
Word Sense Disambiguation of clinical abbrevia-
tions with hyperdimensional computing. AMIA ...
Annual Symposium proceedings. AMIA Symposium,
2013:1007–16.
Moon, S., Pakhomov, S., and Melton, G. B. (2012). Au-
tomated disambiguation of acronyms and abbrevia-
tions in clinical texts: window and training size con-
siderations. AMIA ... Annual Symposium proceedings
/ AMIA Symposium. AMIA Symposium, 2012:1310–
1319.
Navigli, R. (2009). Word sense disambiguation: A survey.
ACM Computing Surveys, 41(2).
Pakhomov, S. (2002). Semi-supervised Maximum Entropy
based approach to acronym and abbreviation normal-
ization in medical texts. (July):160.
Peters, M., Neumann, M., Iyyer, M., Gardner, M., Clark,
C., Lee, K., and Zettlemoyer, L. (2018). Deep Con-
textualized Word Representations. pages 2227–2237.
Pyysalo, S., Ginter, F., Moen, H., Salakoski, T., and Anani-
adou, S. (2013). Distributional Semantics Resources
for Biomedical Text Processing. Aistats, 5:39–44.
S
´
anchez-Le
´
on, F. (2018). ARBOREx: Abbreviation resolu-
tion based on regular expressions for BARR2? CEUR
Workshop Proceedings, 2150:303–315.
Schulz, S., Mart
´
ınez-Costa, C., and Mi
˜
narro-Gim
´
enez, J. A.
(2017). Lexical ambiguity in SNOMED CT. CEUR
Workshop Proceedings, 2050.
Tschiatschek, S., Paul, K., and Pernkopf, F. (2014). Integer
bayesian network classifiers. pages 209–224.
Wu, Y., Denny, J. C., Rosenbloom, S. T., Miller, R. A.,
Giuse, D. A., Song, M., and Xu, H. (2013). A pro-
totype application for real-time recognition and dis-
ambiguation of clinical abbreviations. International
Conference on Information and Knowledge Manage-
ment, Proceedings, pages 7–8.
Wu, Y., Denny, J. C., Rosenbloom, S. T., Miller, R. A.,
Giuse, D. A., Song, M., and Xu, H. (2015a). A Prelim-
inary Study of Clinical Abbreviation Disambiguation
in Real Time. Applied Clinical Informatics, 6(2):364–
374.
Wu, Y., Denny, J. C., Trent Rosenbloom, S., Miller, R. A.,
Giuse, D. A., Wang, L., Blanquicett, C., Soysal, E.,
Xu, J., and Xu, H. (2017). A long journey to short
abbreviations: Developing an open-source framework
for clinical abbreviation recognition and disambigua-
tion (CARD). Journal of the American Medical Infor-
matics Association, 24(e1):e79–e86.
Wu, Y., Xu, J., Zhang, Y., and Xu, H. (2015b). Clini-
cal Abbreviation Disambiguation Using Neural Word
Embeddings. (BioNLP):171–176.
Xu, H., Stetson, P. D., and Friedman, C. (2007). A study of
abbreviations in clinical notes. AMIA ... Annual Sym-
posium proceedings. AMIA Symposium, pages 821–5.
Xu, H., Wu, Y., Elhadad, N., Stetson, P. D., and Friedman,
C. (2012). A new clustering method for detecting rare
senses of abbreviations in clinical notes. Journal of
Biomedical Informatics, 45(6):1075–1083.
HEALTHINF 2021 - 14th International Conference on Health Informatics
508
