
Comparison of Convolutional and Recurrent Neural Networks for the
P300 Detection
Luk
´
a
ˇ
s Va
ˇ
reka
a
NTIS - New Technologies for the Information Society, Faculty of Applied Sciences, University of West Bohemia,
Univerzitnı 8, 306 14 Plzen, Czech Republic
Keywords:
Convolutional Neural Networks, Recurrent Neural Networks, LDA, EEG, ERP, P300.
Abstract:
Single-trial classification of the P300 component is a difficult task because of the low signal to noise ratio.
However, its application to brain-computer interface development can significantly improve the usability of
these systems. This paper presents a comparison of baseline linear discriminant analysis (LDA) with convolu-
tional (CNN) and recurrent neural networks (RNN) for the P300 classification. The experiments were based on
a large multi-subject publicly available dataset of school-age children. Several hyperparameter choices were
experimentally investigated and discussed. The presented CNN slightly outperformed both RNN and baseline
LDA classifier (the accuracy of 63.2 % vs. 61.3 % and 62.8 %). The differences were most pronounced in
precision and recall. Implications of the results and proposals for future work, e.g., stacked CNN–LSTM, are
discussed.
1 INTRODUCTION
The P300 is an event-related potential (ERP) compo-
nent that can be observed in an underlying electroen-
cephalographic (EEG) signal following rare (target)
visual, auditory, or tactile stimuli in a sequence of
standard (non-target) stimuli. It can be observed as
a broad positive peak in the signal between 250 and
500 ms after the stimulus (Polich, 2007).
Detection of the P300 is a challenging task. The
amplitude of P300 is much lower than of the ongoing
EEG signal (Luck, 2005). On the other hand, appli-
cations of the P300 detection include brain-computer
interface allowing paralyzed patients to communicate
directly with brain signals, and is thus has received
much attention (McFarland and Wolpaw, 2011).
Commonly, the P300 waveform is amplified by
averaging related parts of the signal following stimuli
(epochs, trials). Since the ongoing EEG signal is ran-
dom while the P300 displays a repetitive pattern, aver-
aging can amplify the P300 and attenuate noise (Luck,
2005). However, averaging increases the time for BCI
to make a decision, thus decreasing the transfer bit-
rate. Typical steps for the P300 component detection
include preprocessing, feature extraction, and classi-
fication.
In the literature, several classification methods
a
https://orcid.org/0000-0002-5998-3676
have been discussed without any method established
as state-of-the-art. The most successful and re-
ported BCI classifiers include SWLDA, shrinkage
LDA (Blankertz et al., 2011) and Bayesian lin-
ear discriminant analysis (BLDA) (Manyakov et al.,
2011) (Lotte et al., 2018).
In recent years, research in deep learning has
rapidly developed. Its application in image process-
ing and natural language processing has led to signif-
icantly better classification rates than previous state-
of-the-art algorithms (Deng and Yu, 2014). There-
fore, there has been growing interest in applying
deep neural networks (DNN) in BCI systems. This
manuscript aims to evaluate and compare convolu-
tional neural networks (CNN) and recurrent neural
networks (RNN) for the P300 detection on a large
multi-subject publicly available dataset. This paper
extends previous work in (Va
ˇ
reka, 2020) by consider-
ing RNNs in evaluations and comparisons. In a recent
review of the field (Lotte et al., 2018), convolutional
neural networks were the most frequently used while
RNNs have not yet emerged as a frequent deep learn-
ing model in the field. To the author’s best knowl-
edge, RNNs have never been evaluated on a sizeable
multi-subject dataset.
186
Va
ˇ
reka, L.
Comparison of Convolutional and Recurrent Neural Networks for the P300 Detection.
DOI: 10.5220/0010248201860191
In Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2021) - Volume 4: BIOSIGNALS, pages 186-191
ISBN: 978-989-758-490-9
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

1.1 Hypotheses
Based on state-of-the-art and ongoing development in
deep learning, several hypotheses to investigate are
outlined:
• Convolutional neural networks have been well es-
tablished for multidimensional data such as im-
ages (Deng and Yu, 2014). For EEG, convo-
lutional filters are applied to the spatio-temporal
matrix (number of EEG channels x number of
time samples) to extract the relevant information.
Since most CNN-related experiments were per-
formed in the related work (Va
ˇ
reka, 2020), CNNs
serve mostly as the baseline neural network model
in this paper.
• Because of the temporal dynamics of EEG, recur-
rent networks may be useful in identifying regu-
lar patterns in ERPs. This hypothesis is supported
by (Sikka et al., 2020) demonstrating that RNNs
have the potential for learning the underlying tem-
poral dynamics of EEG microstates and are sen-
sitive to sequence aberrations characterized by
changes in mental processes. The P300 waveform
displays variable temporal and spatial characteris-
tics hidden by random EEG background (Polich,
2007).
• Large multi-subject dataset is used in the study to
provide a sufficient number of training examples.
It can be tested if a classifier trained on many ex-
amples can generalize to patterns from possibly
unseen participants (i.e., universal BCI). Such ef-
forts have been relatively rare in the P300-related
literature (Pinegger and M
¨
uller-Putz, 2017).
2 DATA ACQUISITION
The data used for the subsequent experiment originate
from the ’Guess the number’ (GTN) experiment. In
this experiment, the measured person is asked to pick
a number between 1 and 9. During the EEG mea-
surement phase, the person is stimulated with these
numbers (white on the black background). He/she is
silently counting the number of occurrences of the se-
lected number. The target number is supposed to trig-
ger the P300 response, in a similar way to the well-
known P300 speller (Farwell and Donchin, 1988).
After the experiment, this number is revealed and
compared with the guess of the experimenters observ-
ing averaged EEG/ERP waveforms. (Mou
ˇ
cek et al.,
2017)
250 school-age children participated in these GTN
experiments that were carried out in elementary and
Figure 1: This figure shows the ’Guess the number’ ex-
perimental design. The measured participant watches the
stimulation monitor while the experimenters control the ex-
periment and try to guess the number thought (the target
stimulus) by observing averaged waveforms.
secondary schools in the Czech Republic. EEG data
from three EEG channels (Fz, Cz, Pz) and stimuli
markers were stored. Additional metadata about the
participants were collected (gender, age, laterality, the
number thought by the participant, the experimenters’
guess, and various interesting additional information).
All related data are publicly available (Mou
ˇ
cek et al.,
2017).
3 METHODS
To include standard machine learning procedure as
the baseline, both CNN and RNN were compared
with a traditional classification pipeline based on
spatio-temporal feature extraction and LDA classifi-
cation (Blankertz et al., 2011).
3.1 Preprocessing and Feature
Extraction
The data were preprocessed as follows:
1. From each participant of the experiments, epoch
(trial) extraction was performed. The prestimu-
lus interval between -200 ms and 0 ms was used
for baseline correction, i.e., computing the aver-
age amplitude and subtracting it from the data.
1000 ms following the stimulus was considered
as the poststimulus interval. Thus given the sam-
pling frequency of 1 kHz, 11532 x 3 x 1200 (num-
ber of epochs x number of EEG channels x num-
ber of samples) data matrix was produced. Two
following two events were used for epoch extrac-
tion. One of them was the thought number (the
target class). Another one was randomly selected
Comparison of Convolutional and Recurrent Neural Networks for the P300 Detection
187

number out of the remaining stimuli between 1
and 9 (the non-target class). This guaranteed a
relatively balanced dataset. The extracted epochs
are available in (Mou
ˇ
cek et al., 2019).
2. To skip severely damaged epochs, the amplitude
threshold (Luck, 2005) was set to 100 µV . Any
epoch x[c, t] with c being the channel index and t
time was rejected if:
max
c,t
|x[c,t]| > 100 (1)
Consequently, 30.3 % of epochs were rejected.
Such a high number of rejected epochs can be ex-
plained by a high rate of eye-blinking in school-
age children and disruptive outside the laboratory
environment.
Feature Extraction. The feature extraction method
for baseline LDA was based on averaging time inter-
vals of interest and merging these averages across all
relevant EEG channels to get reduced spatio-temporal
feature vectors (Windowed means feature extraction,
WM). Traditional a priori time window for P300 BCIs
is between 300 ms and 500 ms after stimuli (Tan
and Nijholt, 2010; Vos et al., 2014). However, the
P300 in children is significantly delayed in its la-
tency to peak (Riggins and Scott, 2020). Therefore,
the time window was extended to between 300 ms
and 1000 ms for the presented experiments. It was
further divided into 20 equal-sized time intervals in
which amplitude averages were computed. Therefore,
with three EEG channels, the dimensionality of fea-
ture vectors was reduced to 60. Finally, these feature
vectors were scaled to zero mean and unit variance.
In contrast, for deep learning models, no feature
engineering was performed because of possible over-
training caused by too many trainable parameters and
low feature dimensionality. All preprocessing was
supposed to be performed using the neural network
itself.
LDA. As the baseline classifier, state-of-the-
art (Blankertz et al., 2011) LDA with eigenvalue de-
composition used as the solver, and automatic shrink-
age using the Ledoit-Wolf lemma (Ledoit and Wolf,
2004) was applied.
CNN. Convolutional neural networks were imple-
mented in Keras (Chollet et al., 2015). They were
configured to maximize classification performance
using the validation subsets. Initially, after empirical
hyperparameter tuning based on cross-validation, the
baseline parameters were selected as follows (Va
ˇ
reka,
2020):
• The first convolutional layers had six 3 x 3 fil-
ters. The filter size was set to correspond to all
three EEG channels. Both the second filter di-
mension and the number of filters were adjusted
experimentally.
• In both cases, dropout was set to 0.5.
• The convolutional layer’s output was further
downsampled by a factor of 8 using the average
pooling layer.
• ELU activation function (Clevert et al., 2016) was
used for both convolutional and dense layers as
recommended in related literature (Schirrmeister
et al., 2017).
• Batch size was set to 16.
• Cross-entropy was used as the loss function.
• Adam (Kingma and Ba, 2014) optimizer was used
for training because it is computationally efficient,
has little memory requirements, and is frequently
used in the field (Roy et al., 2019).
• The number of training epochs was set to 30.
• Early stopping with the patience parameter of 5
was used.
RNN. The following parameters were modified
when compared to CNN.
• Instead of a convolutional layer, a Long Short-
Term Memory (LSTM) layer with 25 neurons was
used as the input layer — LSTM(25). The return-
sequence parameter was set to true to output the
full sequence of hidden states.
• The fully connected layer with 50 neurons and
ELU activation followed.
• Flattening layer was used to reshape the output.
• Finally, the fully connected layer with softmax ac-
tivation was used.
Moreover, several manipulations of the original set-
tings were investigated, as listed in Table 1.
4 RESULTS
Before classification, the data were randomly split
into training (75 %) and testing (25 %) sets. Using
the training set, 30 iterations of Monte-Carlo cross-
validation (again 75:25 from the subset) were per-
formed to optimize parameters. Results using the
holdout testing set were computed in each cross-
validation iteration and averaged at the end of the
BIOSIGNALS 2021 - 14th International Conference on Bio-inspired Systems and Signal Processing
188
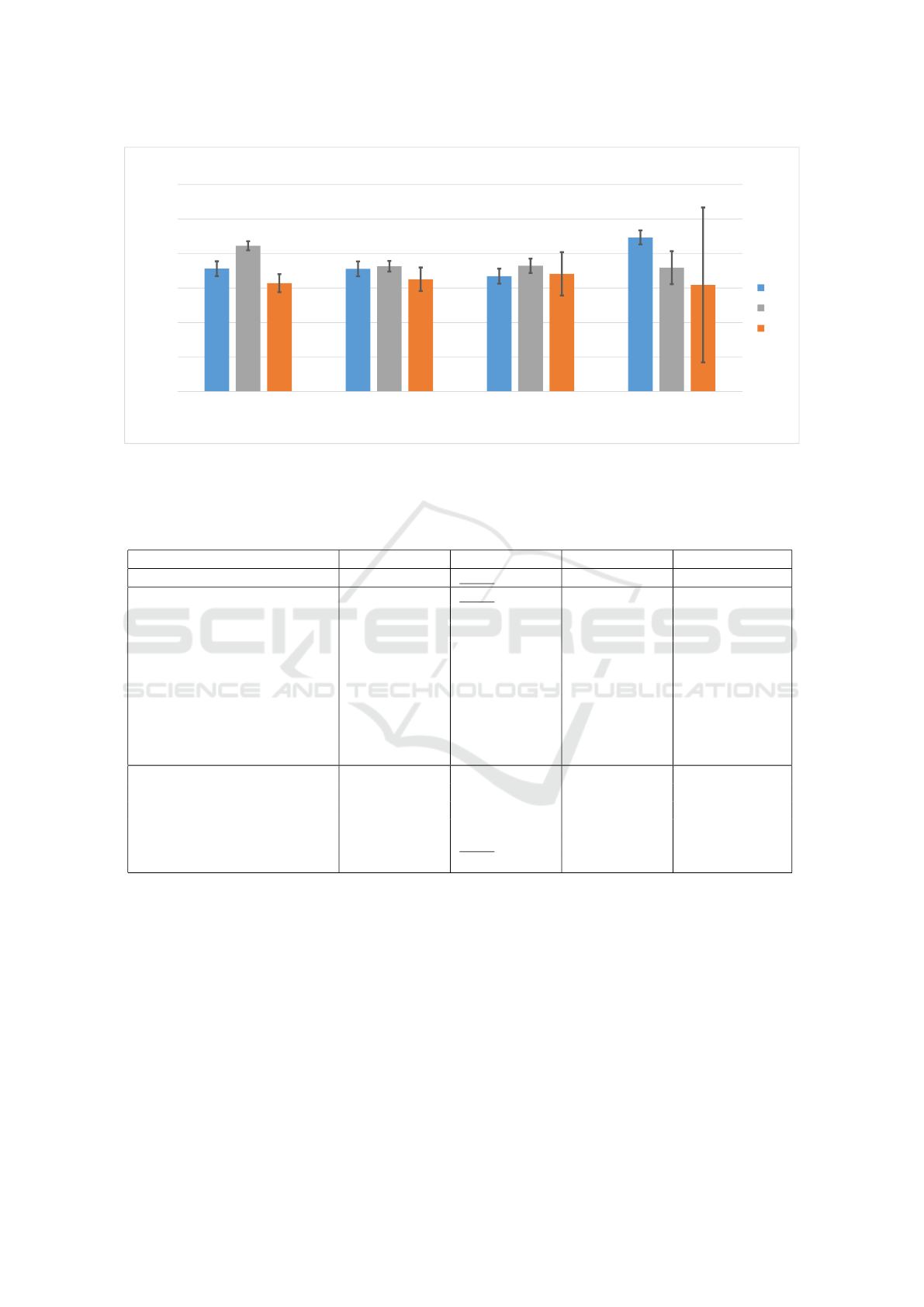
45
50
55
60
65
70
75
AUC Accuracy Precision Recall
Classification results (%)
Classification metrics
Comparison of testing single-trial classification performance
LDA
CNN
RNN
Figure 2: Classification average results in the testing phase. It can be observed that LDA and CNN slightly outperformed
RNN in accuracy. RNNs were associated with unstable recall, as seen from its high standard deviation (black line).
Table 1: Average cross-validation classification results based on the neural network parameter settings. Averages from 30
repetitions and related sample standard deviations (in brackets) are reported. The best performing models from each category
are highlighted in bold and subsequently used for testing.
Changed parameter AUC Accuracy Precision Recall
Baseline LDA (Va
ˇ
reka, 2020) 61.77 % (0.9) 61.76 % (0.91) 61.45 % (1.9) 64.64 % (1.48)
Baseline CNN (Va
ˇ
reka, 2020) 66.12 % (0.68) 62.18 % (0.94) 62.76 % (1.95) 61.34 % (2.63)
RELUs instead of ELUs 66.36 % (0.62) 61.85 % (1.15) 62.7 % (2.19) 60.1 % (3.04)
Filter size (3, 30) 65.84 % (0.49) 61.95 % (1.18) 62.7 % (2.1) 60.5 % (3.91)
12 conv. filters 66.31 % (0.51) 61.83 % (1.1) 62.3 % (2.21) 61.6 % (3.08)
No batch normalization 65.99 % (0.77) 60.55 % (1.52) 61.02 % (3.16) 61.5 % (7.21)
Dropout 0.2 67.67 % (0.65) 60.8 % (1.49) 61.33 % (2.31) 60.33 % (4.0)
No dropout 68.63 % (1.11) 59.49 % (1.2) 59.61 % (1.93) 60.7 % (4.44)
Dense (150) 66.07 % (0.8) 61.81 % (0.95) 62.33 % (1.83) 61.18 % (2.49)
Two dense l. (120-60) 65.72 % (0.77) 62.11 % (0.9) 63.14 % (2.03) 59.5 % (2.55)
Max- instead of AvgPool 64.23 % (1.15) 58.94 % (1.94) 60.22 % (4.18) 59.24 % (13.76)
Baseline RNN 65.68 % (0.85) 56.92 % (1.74) 57.61 % (2.31) 56.25 % (8.11)
LSTM(6) 65.79 % (1.04) 58.28 % (1.25) 58.33 % (2.48) 61.32 % (7)
LSTM(4) 65.41 % (1.04) 57.95 % (1.81) 58.67 % (2.56) 57 % (9.73)
LSTM(6), dropout 0.7 62.71 % (1.39) 58.99 % (1.47) 60.09 % (3.46) 58.35 % (11.02)
LSTM(6), dropout 0.8 60.63 % (1.32) 59.92 % (1.73) 61.49 % (3.81) 57.65 % (11.06)
LSTM(6), dropout 0.9 54.92 % (2.95) 56.89 % (4.86) 59.34 % (7.07) 55.8 % (25.3)
processing. No parameter decision was based on the
holdout set.
Table 1 shows results of cross-validation. The config-
uration with the highest accuracy was highlighted and
used for the testing phase. Figure 2 show the classifi-
cation results.
5 DISCUSSION
As seen from the results, CNN yielded similar per-
formance to the baseline LDA. RNN accuracy was
slightly lower. Moreover, CNN results were far more
stable, as seen from the RNN high standard deviation
of recall.
The validation set experiments revealed that a
combination of the ELU activation, batch normaliza-
tion, dropout, and average pooling was preferable for
CNN (Va
ˇ
reka, 2020). A substantial regularization
was necessary because of a large number of trainable
parameters for the RNN (120,592 for the LSTM(6)
network). In the cross-validation experiments, the
dropout of 0.8 yielded the highest classification ac-
curacy.
Comparison of Convolutional and Recurrent Neural Networks for the P300 Detection
189

Similar single-trial P300 classification perfor-
mance has been commonly reported in the litera-
ture. For example, in (Haghighatpanah et al., 2013),
65 % single-trial accuracy was achieved (using one to
three EEG channels and personalized training data).
In (Sharma, 2017), 40 % to 66 % classification accu-
racy was reported, highly dependent on the individ-
ual tested. This paper presents comparable classifica-
tion accuracy that was achieved using a multi-subject
dataset. Therefore, time-consuming training data col-
lection for each new user might be avoided, and long
training times of deep neural networks no longer pose
a problem. However, despite being successful, this
paper did not confirm their benefits over traditional
methods.
Even though LSTM did not outperform other clas-
sifiers in the presented P300 experiments, it could be-
come valuable as a layer in a more complex model.
For example, in (Ditthapron et al., 2019), an LSTM
layer has been used in a multi-task autoencoder. First,
CNN layers were used to capture spatial domain fea-
tures, and LSTM was used for temporal relationship.
The resulting latent vector was either used to recon-
struct the input or for the P300 classification. A simi-
lar approach could be applied in future work.
This study has several limitations. First, classifi-
cation results on school-age children outside the labo-
ratory environment may not be generalized to a more
typical BCI population. Moreover, despite careful
manual tuning of hyperparameters, there might be an-
other RNN architecture outperforming the presented
CNN architecture that has not been discovered.
6 CONCLUSION
The presented experiments demonstrated that suc-
cessful P300 detection is possible for a multi-subject
dataset with all presented models (LDA, CNN, RNN).
However, when directly comparing CNN and RNN,
CNN appeared superior. It yielded comparable classi-
fication accuracy, more stable results, and was easier
to configure. The presented offline experiments can
be further reproduced in an online BCI. More exper-
iments into stacking CNN and RNN layers could be
the aim of future work.
ACKNOWLEDGEMENTS
This work was supported by the University specific
research project SGS-2019-018 Processing of hetero-
geneous data and its specialized applications. Special
thanks go to Master’s students Patrik Harag and Mar-
tin Matas for their initial experiments that inspired
this work.
REFERENCES
Blankertz, B., Lemm, S., Treder, M., Haufe, S., and Muller,
K. (2011). Single-trial analysis and classification of
ERP components - a tutorial. NeuroImage, 56(2):814–
825.
Chollet, F. et al. (2015). Keras. https://keras.io.
Clevert, D.-A., Unterthiner, T., and Hochreiter, S. (2016).
Fast and accurate deep network learning by exponen-
tial linear units (ELUs). CoRR, abs/1511.07289.
Deng, L. and Yu, D. (2014). Deep learning: Methods
and applications. Foundations and Trends
R
in Sig-
nal Processing, 7(3–4):197–387.
Ditthapron, A., Banluesombatkul, N., Ketrat, S., Chuang-
suwanich, E., and Wilaiprasitporn, T. (2019). Uni-
versal joint feature extraction for p300 eeg classifi-
cation using multi-task autoencoder. IEEE Access,
7:68415–68428.
Farwell, L. A. and Donchin, E. (1988). Talking off the top of
your head: Toward a mental prosthesis utilizing event-
related brain potentials. Electroencephalography and
Clinical Neurophysiology, 70:510–523.
Haghighatpanah, N., Amirfattahi, R., Abootalebi, V., and
Nazari, B.(2013). A single channel-single trial P300
detection algorithm. In 2013 21st Iranian Conference
on Electrical Engineering (ICEE),pages 1–5.
Kingma, D. P. and Ba, J. (2014). Adam: A
method for stochastic optimization. cite
arxiv:1412.6980Comment: Published as a con-
ference paper at the 3rd International Conf. for
Learning Representations, San Diego, 2015.
Ledoit, O. and Wolf, M. (2004). Honey, i shrunk the sample
covariance matrix. The Journal of Portfolio Manage-
ment, 30(4):110–119.
Lotte, F., Bougrain, L., Cichocki, A., Clerc, M., Con-
gedo, M., Rakotomamonjy, A., and Yger, F. (2018).
A review of classification algorithms for EEG-based
brain–computer interfaces: a 10 year update. Journal
of Neural Engineering, 15(3):031005.
Luck, S. J. (2005). An introduction to the event-related po-
tential technique. MIT Press, Cambridge, MA.
Manyakov, N. V., Chumerin, N., Combaz, A., and
Van Hulle, M. M. (2011). Comparison of classi-
fication methods for P300 brain-computer interface
on disabled subjects. Intell. Neuroscience, 2011:2:1–
2:12.
McFarland, D. J. and Wolpaw, J. R. (2011). Brain-computer
interfaces for communication and control. Commun.
ACM, 54(5):60–66.
Mou
ˇ
cek, R., Va
ˇ
reka, L., Prokop, T.,
ˇ
St
ˇ
ebet
´
ak, J., and Br
˚
uha,
P. (2017). Event-related potential data from a guess
the number brain-computer interface experiment on
school children. Scientific Data, 4.
BIOSIGNALS 2021 - 14th International Conference on Bio-inspired Systems and Signal Processing
190

Mou
ˇ
cek, R., Va
ˇ
reka, L., Prokop, T.,
ˇ
St
ˇ
ebet
´
ak, J., and Br
˚
uha,
P. (2019). Replication Data for: Evaluation of con-
volutional neural networks using a large multi-subject
P300 dataset.
Pinegger, A. and M
¨
uller-Putz, G. (2017). No training, same
performance!? - a generic P300 classifier approach. In
Proceedings of the 7th International BCI Conference
Graz 2017.
Polich, J. (2007). Updating p300: an integrative the-
ory of p3a and p3b. Clinical neurophysiology,
118(10):2128–2148.
Riggins, T. and Scott, L. S. (2020). P300 develop-
ment from infancy to adolescence. Psychophysiology,
57(7):e13346.
Roy, Y., Banville, H. J., Albuquerque, I., Gramfort, A., Falk,
T. H., and Faubert, J. (2019). Deep learning-based
electroencephalography analysis: a systematic review.
CoRR, abs/1901.05498.
Schirrmeister, R. T., Springenberg, J. T., Fiederer, L. D. J.,
Glasstetter, M., Eggensperger, K., Tangermann, M.,
Hutter, F., Burgard, W., and Ball, T. (2017). Deep
learning with convolutional neural networks for brain
mapping and decoding of movement-related informa-
tion from the human EEG. CoRR, abs/1703.05051.
Sharma, N. (2017). Single-trial P300 classification using
PCA with lda, QDA and neural networks. CoRR,
abs/1712.01977.
Sikka, A., Jamalabadi, H., Krylova, M., Alizadeh, S.,
van der Meer, J. N., Danyeli, L., Deliano, M., Vicheva,
P., Hahn, T., Koenig, T., Bathula, D. R., and Wal-
ter, M. (2020). Investigating the temporal dynam-
ics of electroencephalogram (EEG) microstates using
recurrent neural networks. Human Brain Mapping,
41(9):2334–2346.
Tan, D. S. and Nijholt, A. (2010). Brain-Computer In-
terfaces: Applying Our Minds to Human-Computer
Interaction. Springer Publishing Company, Incorpo-
rated, 1st edition.
Va
ˇ
reka, L. (2020). Evaluation of convolutional neu-
ral networks using a large multi-subject P300
dataset. Biomedical Signal Processing and Control,
58:101837.
Vos, M. D., Kroesen, M., Emkes, R., and Debener, S.
(2014). P300 speller BCI with a mobile EEG sys-
tem: comparison to a traditional amplifier. Journal of
Neural Engineering, 11(3):036008.
Comparison of Convolutional and Recurrent Neural Networks for the P300 Detection
191
