
Using Geometric Graph Matching in Image Registration
Giomar O. Sequeiros Olivera
1 a
, Aura Conci
2 b
and Leandro A. F. Fernandes
2 c
1
Anhanguera Educacional, Niter
´
oi, RJ, Brazil
2
Institute of Computing, Fluminense Federal University, Niter
´
oi, RJ, Brazil
Keywords:
Image Registration, Graph Matching, Edit Distance, Infrared Images.
Abstract:
Image registration is a fundamental task in many medical applications, allowing interpreting and analyzing
images acquired using different technologies, from different viewpoints, or at different times. The image
registration task is particularly challenging when the images have little high-frequency information and when
average brightness changes over time, as is the case with infrared breast exams acquired using a dynamic
protocol. This paper presents a new method for registering these images, where each one is represented in a
compact form by a geometric graph, and the registration is done by comparing graphs. The application of the
proposed technique consists of five stages: (i) pre-process the infrared breast image; (ii) extract the internal
linear structures that characterize arteries, vascular structures, and other hot regions; (iii) create a geometric
graph to represent such structures; (iv) perform structure registration by comparing graphs; and (v) estimate
the transformation function. The Dice coefficient, Jaccard index, and total overlap agreement measure are
considered to evaluate the results’ quality. The output obtained on a public database of infrared breast images
is compared against SURF interest points for image registration and a state of the art approach for infrared
breast image registration from the literature. The analyzes show that the proposed method outperforms others.
1 INTRODUCTION
Medical image processing has become a fundamen-
tal tool in healthcare, being increasingly used in tasks
such as diagnostic, treatment planning, surgeries, and
disease follow-up (Rahman, 2018). In medical image-
based applications, it is often interesting to analyze
more than one set of data simultaneously since im-
ages obtained with different acquisition technologies
may reveal complementary information about struc-
tures of interest (Balakrishnan et al., 2018). More-
over, same patient images taken at different times can
help to monitor abnormalities or assist their treatment.
In both cases, image registration (IR) is an essential
task in the processing of medical images, being cru-
cial in all applications that need to combine, compare,
or merge visual information (Brock et al., 2017; Conci
et al., 2015; Gonz
´
alez et al., 2018).
The IR process consists of estimating a func-
tion that allows mapping one of the images to the
other (Zitov
´
a and Flusser, 2003). For this purpose,
a wide variety of IR techniques can be seen in the
a
https://orcid.org/0000-0002-7172-6525
b
https://orcid.org/0000-0003-0782-2501
c
https://orcid.org/0000-0001-8491-793X
literature, that in general are divided into intensity-
based and feature-based methods (Zitov
´
a and Flusser,
2003). For the second approaches, it is necessary
to manually, semi-automatically, or automatically ex-
tract feature points from the images (Ma et al., 2016).
The manual and semi-automatic selection of feature
points are usually time-consuming and often imprac-
tical. Consequently, it is important to develop tech-
niques that allow the automatic identification of these
points. Moreover, those techniques must be stable to
ensure consistency and reproduction of the results.
Unfortunately, the most used approaches for auto-
matic identification of feature points in natural images
are not adequate for IR of low contrast images such as
infrared images (a.k.a. thermograms or thermal im-
ages) (Falco et al., 2020). Similarly, some approaches
use anatomical structures present in the medical im-
ages to perform the registration (Deng et al., 2010).
When applied to infrared breast images, the prob-
lem in this type of IR approach is to represent the
anatomical structures properly. In this sense, geomet-
ric graphs emerge as a powerful tool to represent ob-
jects and their spatial relationships (Garcia-Guevara
et al., 2018). Graph matching has been applied in sev-
eral domains to solve problems such as data retrieval,
Olivera, G., Conci, A. and Fernandes, L.
Using Geometric Graph Matching in Image Registration.
DOI: 10.5220/0010239200870098
In Proceedings of the 16th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2021) - Volume 4: VISAPP, pages
87-98
ISBN: 978-989-758-488-6
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
87

graph classification, pattern discovery, and structural
data characterization (Pinheiro et al., 2017) but are
still little explored for IR (Tong et al., 2017).
In this paper, we present a method for extract-
ing the location of internal linear structures from in-
frared breast images and represent them as geometric
graphs. The internal linear structures may be arteries,
vascular structures, or hot regions. We also present
an algorithm to match the graph representations of
the structures extracted from two images. The pro-
posed matching procedure is an adaptation of the edit
distance (Armiti and Gertz, 2014) that, in addition to
considering the structural relationships of a vertex and
its neighbors, uses local image descriptors around the
edge that connects two vertices to improve the match-
ing. We have applied this proposition for registering
breast examinations and have observed significant ad-
vantages when compared it to previous techniques.
The remaining of this paper is organized as fol-
lows: Section 2 presents a literature review of related
works. The proposed approach is detailed in Sec-
tion 3. Section 4 presents the experiments and their
results. Finally, Section 5 concludes the paper and
points to directions for future exploration.
2 RELATED WORK
There are few approaches in the literature for breast
infrared IR. To the best of our knowledge, none of
them uses graph-based techniques to perform the task.
Thus, we claim that this is one of the original contri-
butions of our work. Next, we discuss some works
that extend traditional IR methods to thermograms.
Agostini et al. (2009) recorded about 500 frames
of thermal images. Prior acquisition, they glued black
and white markers of 5mm diameter on the patient’s
skin. The white markers are used for estimating
the transformations between image pairs, while the
black markers are used to measure the quality of the
method. The alignment of the set of frames is per-
formed by taking the first image of the sequence as the
reference, being the others transformed to it. Since
the white markers guide registration, the method be-
gins with the automatic identification of this kind of
marker. After that, each marker is manually labeled
and used to solve the linear transformation that bet-
ter explains their location in the image pair. The effi-
cacy of this method is measured by the signal-to-noise
ratio (SNR). In this case, the signal analyzed along
the series is formed by the temperatures in the posi-
tions of the black markers in the first frame, together
with the temperature of the same positions in the other
images. The noise calculated by the measurement is
characterized by the change throughout the series of
the temperature values in the observed locations.
Lee et al. (2010) also proposed the IR of infrared
images by using markers previously placed on the
patient. These markers are automatically identified
by the Harrys’ corners detection method (Harris and
Stephens, 1988). After that, the association between
markers in both images is manually set by the user.
Through these association, a transformation by Thin
Plate Spline function (Holden, 2008) is calculated and
used to align the sensitive to the reference image.
In the end, the transformation function is refined by
the simplex method. In subsequent work, Lee et al.
(2012) used the Harris corner coefficient to detect fea-
ture points from heat patterns on the thermal images.
Registration is made by taking the first image as a ref-
erence and estimating transformations that explain the
location of the feature points on other image of the set.
To evaluate the proposed approach, numbered mark-
ers were placed on a patient, and a sequence of ten
images of that patient was acquired. During the ac-
quisition, the patients are instructed to perform small
movements, simulating the displacements that occur
in examinations that take large time intervals.
Silva et al. (2016) used an IR process as part of
a methodology that aims to analyze thermograms us-
ing time series. In their methodology, thermograms
are sequentially acquired, forming a set of twenty im-
ages per patient. During the five minutes of acqui-
sitions, the patient performs small involuntary move-
ments for breathing. These movements lead to differ-
ences from one thermogram to another. In Silva et al.
(2016) work, the first thermogram of the sequence
is considered the reference image, and the others are
deemed sensitive images. Thus, for the examination
of a patient, the registration process is executed nine-
teen times since the sensitive thermograms of the se-
quence must be registered to the first one. Silva et al.
(2016) technique uses Mutual Information (MI) as a
global measure of similarity of pixel intensities to es-
timate translation, rotation, and scale transformations
between the images.
Falco et al. (2020) used the same dataset of Silva
et al. (2016) for IR. All the serial thermograms of
the same patient are analyzed, creating a relationship
among them. The reference image for registration is
chosen as the one that is more similar to all the others.
Then, all the other thermograms of a patient are regis-
tered systematically, creating a new and more similar
set of thermograms. Initially, the images to be regis-
tered are classified as a reference or sensitive image.
After that, all images are processed to estimate the sil-
houette of the patient’s body. Through the silhouette,
feature points are identified. These points are used to
VISAPP 2021 - 16th International Conference on Computer Vision Theory and Applications
88

estimate the transformation that better explains the lo-
cation of the feature points from one image to another.
With these transformations, a new set of serial images
are created, one for each type of transformation com-
puted. Finally, the generated images are compared
with the reference image, and the set of registered im-
ages that presented the best result is chosen as the out-
put of the registration method.
It is important to note that the techniques men-
tioned above either rely on markers to perform the
registration or local information that may change
from one image to another, such as the infrared in-
tensity or the patient’s silhouette. Our approach, on
the other hand, performs the registration based on the
topological relationship between the main sources of
heat identified in the image, which are related to the
arteries, vascular structures, and other hot regions.
3 THE PROPOSED APPROACH
Our method for infrared breast IR consists of five
steps: (i) pre-process a given infrared breast image;
(ii) extract the internal linear structures; (iii) repre-
sent the internal structures using a geometric graph;
(iv) perform graph matching to register the graph of
the current image to the graph of another thermogram;
and (v) estimate the transformation for image registra-
tion. The following subsections describe these steps.
3.1 Pre-processing
Infrared images are formed by a temperature matrix
representing the thermal pixel values. By using a
min-max mapping, temperatures values may be rep-
resented in the [0, 255] range as a conventional 8-bit
grayscale image. Such an image can also be treated
as a heightmap, i.e., the gray intensity value assigned
at pixel location (x, y) also represents the distance of
displacement or elevation of a surface, with 0 (black)
representing minimum height and 255 (white) repre-
senting maximum height. Figures 1 (a) and (b) exem-
plify, respectively, an infrared breast image and the
region enclosed by the rectangle in (a) represented as
a relief. When the whole image is considered, the
highest portions of the terrain are the warmest regions
of the patient’s body, with part of the ridges indicat-
ing the location of thicker blood vessels. Global mini-
mum regions correspond to the background, i.e., areas
outside the patient’s body.
The purpose of the pre-processing step is to pre-
pare the image to facilitate the extraction of internal
linear structures in the next step. According to our
experience, global minimum and more accentuated
(a)
Local Minimum
Regions
(b)
Removal of
Local Minimum Regions
(c)
Figure 1: Three-Dimensional representation of the yellow
portion of a thermogram (a) as a height map before (b) and
after (c) applying the H-minima transform. Notice that the
relief around local minimum in (b) have changed in (c).
local minima affect the quality of the segmentation
algorithm that is used in the next step to extract in-
ternal linear structures. To solve this problem, we
apply the H-minima transform (Ismail et al., 2016)
as a pre-processing to suppresses all minima in the
grayscale image whose intensity is less than h. Fig-
ures 1 (b) and (c) illustrate the relief induced by the
thermogram before and after the application of the H-
minima transform. Through empirical experimenta-
tion, we observed that the value h = 8 meets the needs
of the proposed technique.
3.2 Extraction of Linear Structures
The input of this step is the image resulting from the
pre-processing stage. The result is a binary image
where 1-pixels represent the location of the internal
linear structures, which correspond to ridges of the
relief induced by the thermogram.
In this work, we use the watershed algorithm by
flooding (Kornilov and Safonov, 2018) to achieve the
objective of the internal linear structures’ extraction
step. This algorithm simulates relief flooding from
water sources located at local minimum. When the
Using Geometric Graph Matching in Image Registration
89
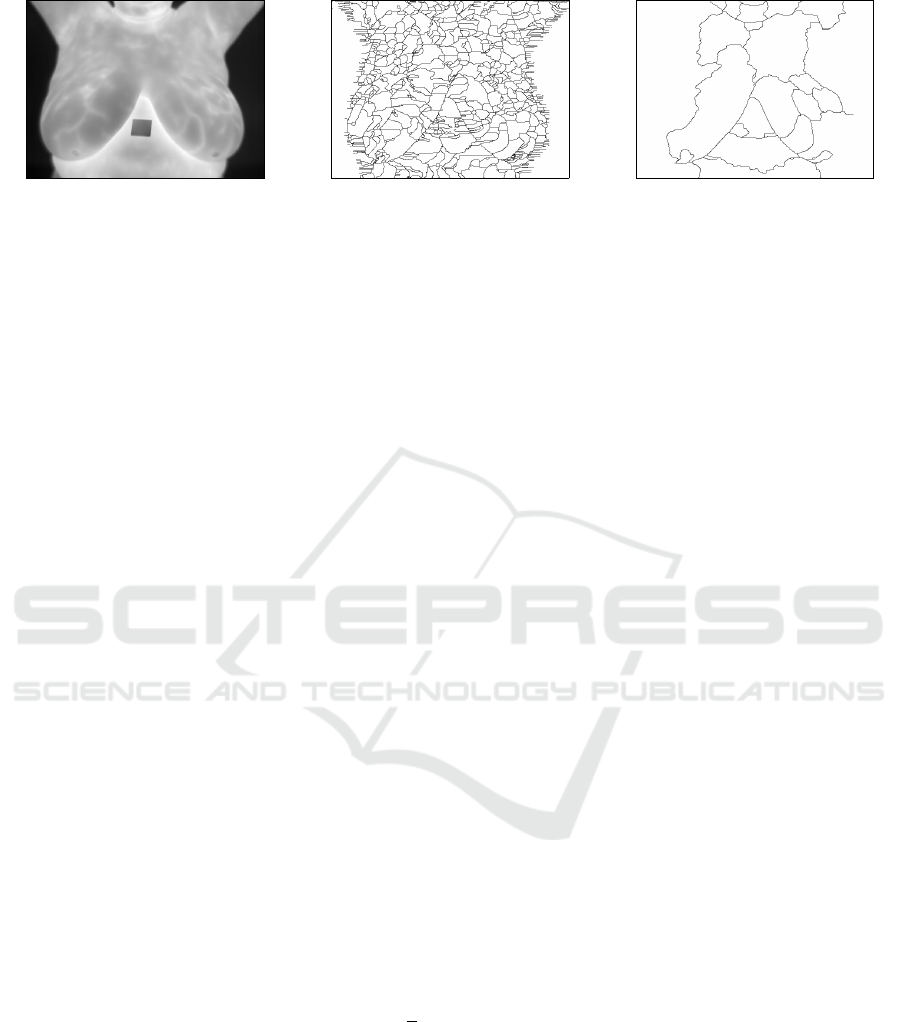
(a) (b) (c)
Figure 2: From left to right: (a) the original thermogram; (b) internal structures for (a); and (c) internal structures for the
image resulting from the H-minima transform of (a).
water rises, retention basins are created. At the end
of the flooding process, neighboring basins define the
watershed lines. Figure 2 (b) shows the watershed
of the non pre-processed gray-level image in Fig-
ure 2 (a). As it is possible to observe, many lines
were defined as the internal linear structures because
many retention basins were created. Figure 2 (c)
shows the result of the watershed segmentation ap-
plied to the version of Figure 2 (a) pre-processed by
the H-minima transform. This result presents a much
cleaner set of linear features.
The binary image produced by the watershed al-
gorithm is subsequently processed by the thinning
procedure described by Zhang and Suen (1984). The
objective is to obtain a binary image with structures
having the thickness of one pixel.
3.3 Linear Structure Representation
Once the internal structure is extracted, it must be
turned into a geometric graph. For this, we start with
an 8-connected neighborhood representation of the
binary image obtained in the previous step.
Let G = (V, E) be an undirected graph, where V
is the set of vertices and E the set of edges. The ver-
tices v ∈V correspond to the 1-pixels of the given bi-
nary image, and the 8-connected neighborhood of 1-
pixels defines the edges e = (v
i
, v
j
) ∈ E for any pair
of neighbor vertices v
i
, v
j
∈V . The graph is geomet-
ric because vertices carry the (x, y) pixels’ location,
and edges’ weight is given by the Euclidean distance
between the pixels of the vertices that define them.
Thus, the weight of an edge can be equal to 1 or
√
2.
One problem that arises in representing the inter-
nal linear structures using G is the creation of too
many vertices, which can compromise the perfor-
mance of matching algorithms (Zheng et al., 2013).
To mitigate this issue, we create the geometric graph
G
0
= (V
0
, E
0
) from the graph G = (V, E). Here,
V
0
⊆V is the new set of vertices formed by endpoints,
corners, and junction points of the linear structures
in the given binary image. The new set of edges
E
0
⊆V
0
×V
0
allows connecting vertices in V
0
consid-
ering the shortest paths in V . Additionally, the new
graph is enhanced with the inclusion of features at the
vertices and edges. Below we describe the processes
for obtaining the new sets of vertices and edges, and
how to extract the features to be assigned to elements
of the new graph.
Determination of the New Set of Vertices. The
candidate vertices for graph G
0
= (V
0
, E
0
) must sat-
isfy one of the three rules below:
1. v ∈V is an endpoint, i.e., degree(v) = 1;
2. v ∈V is a corner, i.e., degree(v) = 2 and the ver-
tex v and its direct neighbors are not collinear;
3. v ∈V is a junction point, i.e., degree(v) ≥ 3;
were degree(v) is the number of vertices directly con-
nected to v. To avoid creating many close vertices in
G
0
, we only include in V
0
the candidate vertices that
do not have another candidate vertex within a radius
bounded by the threshold d. In our experiments, we
observed that the value d = 20 meets the needs of the
proposed technique for the image database we have
using. This value must be adjusted for other databases
regarding the resolution of the input thermograms and
the patient’s distance to the camera.
Determination of the New Set of Edges. The set
of edges E becomes obsolete when the new collec-
tion V
0
of vertices is defined as a subset of V . There-
fore, we need to build E
0
to reconnect the vertices that
were included in the graph G
0
. We use the graph G in
this process to not lose the overall structure of the lin-
ear features depicted in the input binary image. More
specifically, we create edges e
0
= (v
0
i
, v
0
j
) ∈ E
0
using
the Dijkstra algorithm to determine the shortest path
between v
0
i
and v
0
j
in the original graph G. An edge is
created whenever there is no vertex in V
0
positioned
between vertices v
0
i
and v
0
j
in such path.
VISAPP 2021 - 16th International Conference on Computer Vision Theory and Applications
90

Figure 3: Example of visual feature extraction.
Feature Extraction. The purpose of this step is to
create spatial features vectors and visual feature vec-
tors for the elements in the geometric graph G
0
.
Spatial features are associated with vertices. For
a given vertex v
0
∈V
0
, its spatial features are encoded
by a cyclic string of tuples, F = [ f
1
, f
2
, ··· , f
n
], where
n = degree(v
0
) and f
i
is a tuple (|e
0
i
|, < e
0
i
e
0
i−1
). Here,
|e
0
i
| represents the length of the edge e
i
= (v
0
, v
0
i
),
where v
0
i
is the neighbor vertex reached from v
through e
i
, and < e
0
i
e
0
i−1
denotes the angle between
the edges e
0
i
and e
0
i−1
in counterclockwise order.
Visual feature vectors are created to encode visual
information around the graph edges. For this purpose,
we use Histogram of Oriented Gradients (HOG) fea-
tures (Dalal and Triggs, 2005), generally applied in
pattern recognition and image processing to detect or
recognize objects. In the present case, the objects
represent variations of intensities nearby the thicker
blood vessels in the original infrared breast image.
We compute one HOG feature from the 8 ×8 grid
placed at the center point of each edge of the graph G
0
.
The cells in this grid are grouped into 2 ×2 blocks,
and the orientation of the gradients of each pixel of
the image is transformed in such a way that they are
mapped to the [−180
◦
, 180
◦
) range, with a 40
◦
in-
terval, i.e., the orientations assume values in the dis-
crete set {−180
◦
, −140
◦
, −100
◦
, −60
◦
, −20
◦
,20
◦
,
60
◦
,100
◦
,140
◦
}. After this process, the magnitude of
each pixel is used as a weighting factor for calculat-
ing the average orientation of each cell. Finally, each
block is represented by a histogram of average orien-
tations. Histograms of 9 bins are created, and since
we have 4 blocks, the whole feature vector will have
size 36. Figure 3 illustrates the computation of the
HOG features.
3.4 Graph Matching
This step aims to perform the comparison of graphs
G
0
and F
0
representing, respectively, the reference and
the sensitive images, to obtain the best correspon-
dence between their vertices. From this correspon-
dence, it will be possible to estimate the transforma-
tion that will allow registering the thermograms.
Algorithm 1 estimates the resulting matching ma-
trix. We use the Hungarian method (Riesen et al.,
2018) to find the best matching between the vertices
of G
0
and F
0
from the matrix M, whose entry M
i, j
represents the cost of transforming the information
related to the i-th vertex v
0
∈ G
0
into the information
assigned to the j-th vertex u
0
∈ F
0
(see Algorithm 2).
The vertex transformation cost is computed by
Algorithm 3 as the vertex edit distance (Armiti and
Gertz, 2014). Editing distance is a measure of similar-
ity and represents a powerful approach within error-
tolerant methods for correspondence between graphs.
This distance involves basic operations such as re-
moving, adding, or replacing vertices and edges. This
distance is calculated using dynamic programming
with complexity O(nm
2
), where n = degree(v
0
) and
m = degree(u
0
) (Armiti and Gertz, 2014). We imple-
ment the substitution, insertion, and removal opera-
tions applied in the edit distance algorithms follow-
ing Armiti and Gertz (2014).
The following are the edit operations on the edges:
substitution, insertion, and removal. Given two ver-
tices v and u, let be the edge e
i
the neighbor of v and
the edge e
j
the neighbor of u. The substitution cost
between two edges is defined as:
γ(e
i
→ e
j
) = d
L
(e
i
, e
j
) + d
S
(e
i
, e
j
), (1)
where d
L
(e
i
, e
j
) returns the Euclidean distance be-
Algorithm 1: Geometric graph matching.
1 SetAlFnt
Input: Graphs G
0
= (V
0
, E
0
) and F
0
= (U
0
, D
0
)
Output: The matching matrix
2 foreach v
0
∈V
0
do
3 i ← the index of v
0
;
4 foreach u
0
∈U
0
do
5 j ← the index of u
0
;
6 M
i, j
← computeMinimalDistance(v
0
, u
0
);
7 return HungarianMethod(M);
Using Geometric Graph Matching in Image Registration
91

tween the visual features of the edges e
i
and e
j
that
were extracted by using HOG features. Similarly,
d
S
(e
i
, e
j
) returns the spatial distance based on angles
and length of the edges:
d
S
(e
i
, e
j
) =
(
c(e
i
, e
j
) , for |θ
e
i
−θ
e
j
| ≤ π,
c(e
i
, e
j
) + 2 max{l
e
i
, l
e
j
} , otherwise,
(2)
where
c(e
i
, e
j
) =
q
l
2
e
i
+ l
2
e
j
−2l
e
i
l
e
j
cos(|θ
e
i
−θ
e
j
|), (3)
and θ
e
k
is the angle between an edge and the previous
one and l
e
k
is the length of the edge.
The cost of substitution is defined as the distance
required so that the neighboring vertex of the edge
e
i
is aligned with the neighboring vertex of the edge
e
j
, which can be seen as the polar distance between
them. The cost for insertion and removal operations
are defined by:
γ(λ → e
i
) = γ(e
i
→ λ) =
(
c(e
i
) + d
L
(e
i
) , for θ
e
i
≤ π,
c(e
i
) + 2l
e
i
+ d
L
(e
i
) , otherwise,
(4)
where
c(e
i
) =
q
l
e
i
+ l
e
i−1
−2l
e
i
l
e
i−1
cos(|θ
e
i
|). (5)
3.5 Image Registration
In this work, we assume that the transformation used
to register the sensitive image to the reference im-
age is a planar homography. The result of the pre-
vious step is the best match between the vertices of
two geometric graphs representing thermograms. In
some cases, incorrect matches (outliers) may occur,
introducing errors in the homography that would be
estimated if all corresponding vertices were consid-
ered. Consequently, the image registration step uses
the RANSAC algorithm (Fischler and Bolles, 1981)
to remove outliers and estimate the best homogra-
phy between the actual corresponding vertices (in-
liers). The resulting transformation function serves
to change the sensitive image, making it more similar
to the reference one. For this, it is necessary to use in-
terpolation techniques that map the continuous values
of the transformation function into discrete values of
the image representation domain (Pan et al., 2012).
4 EXPERIMENTS AND RESULTS
Following Falco et al. (2020) and Silva et al. (2015), a
sample from the DMR-IR database (Silva et al., 2016)
Algorithm 2: Compute minimal distance.
1 SetAlFnt
Input: Vertices v
0
and u
0
Output: minimal distance
2 word1 ← spatial and visual features from v
0
;
3 word2 ← spatial and visual features from u
0
;
4 n ← degree(v
0
);
5 m ← degree(u
0
);
6 if n <m then
7 swap(word 1,word2);
8 minimalDistance ← ∞;
9 for i ← 1 to m do
10 word2 ←rotate(word 2);
11 distance ← editDistance(word1,word2);
12 if distance >minimalDistance then
13 minimalDistance ← distance;
14 return minimalDistance;
Algorithm 3: Edit distance.
1 SetAlFnt
Input: Spatial and visual features
word1andword2
Output: The edit distance stored in D
n+1,m+1
2 n ← length(word1);
3 m ← length(word2);
4 for i ← 1 to n do
5 D
i,1
← i;
6 for j ← 1 to m do
7 D
1, j
← j;
8 for i ← 1 to n do
9 c1 ← the i-th entry of word1;
10 for j ← 1 to m do
11 c2 ← the j-th entry of word2;
12 if c1 = c2 then
13 D
i+1, j+1
← D
i, j
;
14 else
15 D
i+1, j+1
← min{
D
i, j
+ SubstitutionCost(c1, c2),
D
i, j+1
+InsertRemoveCost(c1, c2),
D
i+1, j
+InsertRemoveCost(c1, c2)};
16 return D
n+1,m+1
;
was used to carry out the experiments and to validate
the proposed approach. From these, 11 healthy pa-
tients and 12 with disease diagnoses were selected
due their diversity of body shapes. Each one has a set
of 20 frontal images captured every 15 seconds during
5 minutes according to a protocol proposed by Silva
et al. (2016). This time is long enough for the patient
to perform involuntary movements in such a way that
becomes necessary the IR before analysis.
We have considered the first thermogram of each
patient as the reference image, and the remaining 19
thermograms have been considered the sensitive im-
ages that will be registered to the first one. Due to
VISAPP 2021 - 16th International Conference on Computer Vision Theory and Applications
92
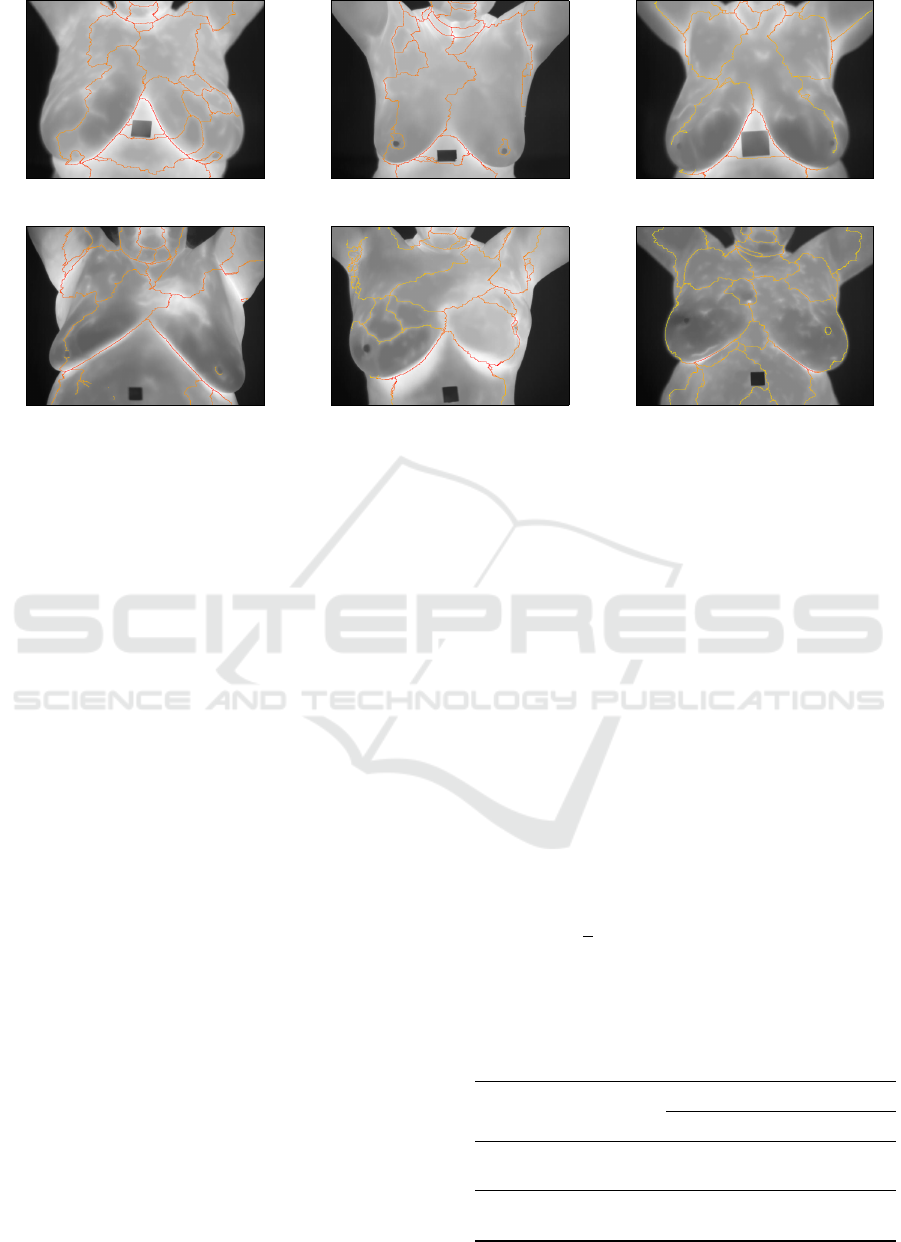
(a) (b) (c)
(d) (e) (f)
Figure 4: Examples of structures extracted from thermograms of healthy patients (a)-(c), and from thermograms of patients
diagnosed with a disease in the breast (d)-(f).
the non-deterministic nature of the RANSAC algo-
rithm, we have performed the registration of each
pair of images ten times. We have considered the
average transformation as the expected transforma-
tion. By doing so, we avoid small variations from
one registration to another in the same pair of im-
ages. Thus, having 23 patients with 20 images each,
19 ×10 = 190 RANSAC evaluations were performed
per patient, leading to a total of 4,370 registrations.
For simplicity, through this section, the patients
are identified by the numbering P1, P2, . . . , P23.
Patients P1 to P11 correspond to those with healthy
breasts, while patients P12 to P23 present some breast
disease diagnoses. Figure 4 shows the linear struc-
tures (orange lines) extracted from some of such im-
ages, of which Figures 4 (a)-(c) are from healthy pa-
tients, and Figures 4 (d)-(f) are from breast disease
patients. It is important to mention that each im-
age shown in Figure 4 represents the first in a se-
ries of 20 images acquired by using a dynamic pro-
tocol proposed by Silva et al. (2016). The diagnoses
were obtained through other exams (mammography
or biopsy) and the visual differences are not necessar-
ily evident enough to distinguish between healthy and
diseased patients by analysing just one thermogram.
An example of visual differences between healthy and
diseased patients is the presence of asymmetries heat
distribution in the left and right breast. Figure 5 shows
the geometric graphs defined from the linear struc-
tures presented in Figure 4.
Next, we discuss the complexity of the geometric
graphs constructed using the proposed approach and
two other suggested solutions, the performance of the
proposed technique in the registration of images of
each patient, and its performance comparison to other
IR techniques.
Number of Vertices. Table 1 compares the aver-
age number and the standard deviation (SD) of the
number of vertices in three types of geometric graphs
computed per patient’s condition on images of the
dataset used in our experiments. Recall that we are
considering 23 patients, with 20 images each, mak-
ing 460 geometric graphs, where 220 are related to
healthy patients, and 240 are related to patients with
some breast disease.
In Table 1, the graph type N-8 considers the bi-
nary image representing the thermogram’s watershed
structure as a graph where each 1-pixel is a vertex.
The 8-connectivity between pixels defines edges of
length 1 or
√
2. In graph type Harris, the Harris cor-
ner detector (Harris and Stephens, 1988) was applied
to the watershed image to identify the graph’s ver-
Table 1: Average and standard deviation (SD) of the number
of vertices per patient’s condition.
Condition Measure
Graph Type
N-8 Harris Ours
Healthy
Average 3319.55 422.73 173.27
SD 477.67 58.22 25.02
With
Disease
Average 4039.25 523.00 209.75
SD 825.86 104.71 43.45
Using Geometric Graph Matching in Image Registration
93
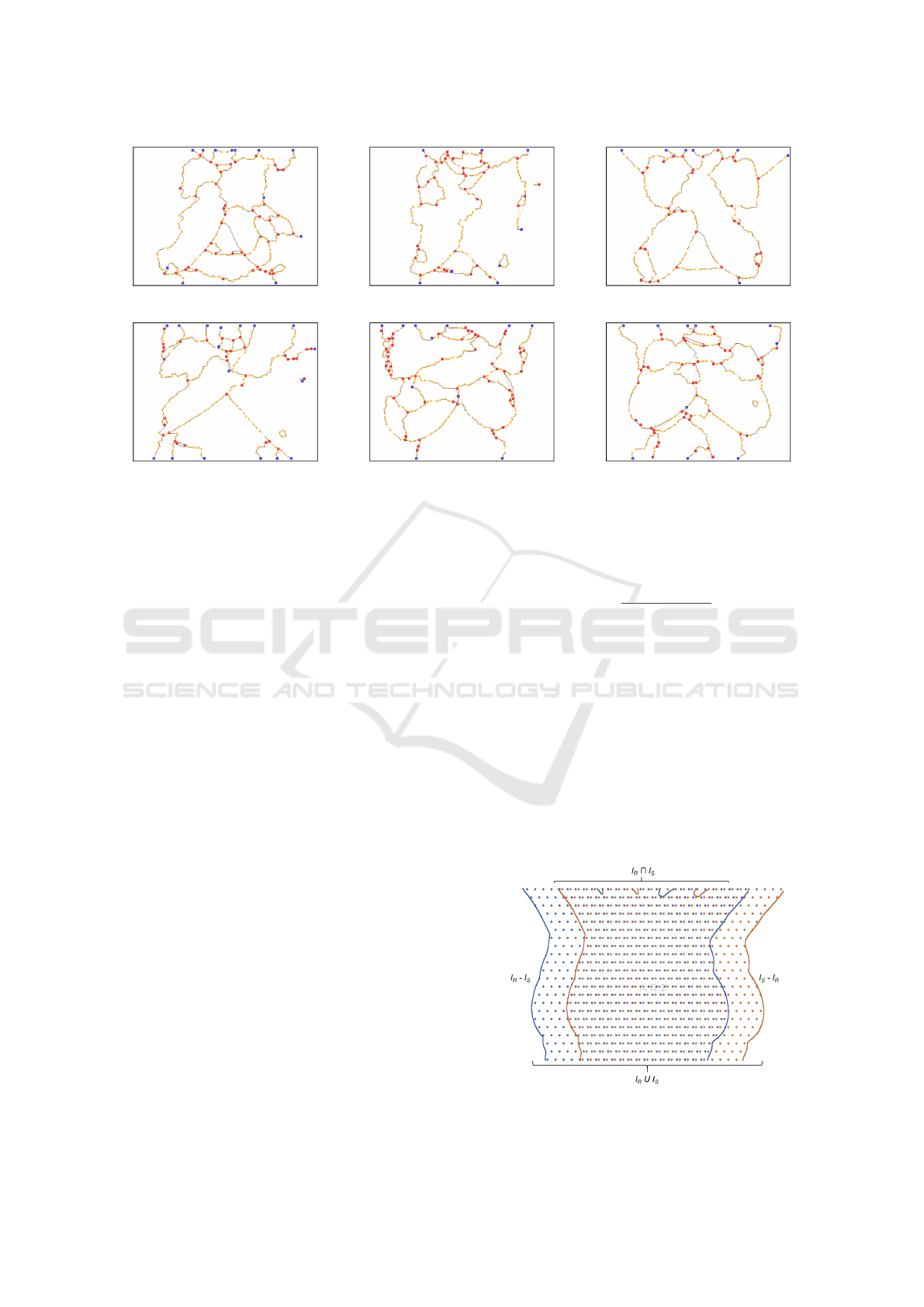
(a) (b) (c)
(d) (e) (f)
Figure 5: Geometric graphs computed from the thermograms presented in Figure 4. Blue, orange and red vertices correspond
to vertices created from, respectively, endpoints, corners, and junction points.
tices as the detected points of interest. Notice that
the average number and the SD of vertices produced
by the proposed approach is much smaller than the
values resulting from graphs of type N-8 and Harris.
It is necessary to have about 173 vertices on average
to represent the internal structures of the thermogram
with healthy diagnosis and 210 vertices for patients
with some disease. The importance of having a small
number of vertices to represent the structures prop-
erly resides in the fact that graph matching algorithms
are NP-complete (Riesen et al., 2018). Thus, be able
to describe the structures present in the thermograms
with less information is a desirable property of the
proposed approach. Such a property allows obtaining
a considerable computational performance gain in the
execution of graph matching algorithms.
Performance of the Registration. Table 2 presents
the performance of the proposed graph-based IR
approach per patient. The analysis considers the
mean Dice coefficient (Dice, 1945), mean Jaccard in-
dex (Jaccard, 1912), and mean Total Overlap Agree-
ment (TOA) measure (Klein et al., 2009) achieved
before and after registration. Table 2 also presents
the number of times in which the evaluation measure
gets improved after IR. These values have been high-
lighted for convenience. Similarly, Table 3 shows the
standard deviation per patient before and after regis-
tration. In this case, the increments of standard devi-
ation after registration are highlighted, and they indi-
cate greater dispersion in relation to the mean.
The Dice coefficient is defined as:
Dice =
2N (I
R
∩I
S
)
N (I
R
) + N (I
S
)
, (6)
where I
R
and I
S
are the binary representation of, re-
spectively, the reference and sensitive images. These
binary images are considered as sets whose elements
are the pixels that form the patient’s body. Here, N (S )
is the number of elements in the set S. Thus, it gives
the area of the objects formed by 1-pixels in the given
image, that can be seen in Figure 6. To that extent,
Dice coefficient equal to 1 means the total overlap, the
greater similarity between the images, while 0 means
no overlap.
The Jaccard index represents the percentage of
Figure 6: Comparison between reference image I
R
and sen-
sitive image I
S
.
VISAPP 2021 - 16th International Conference on Computer Vision Theory and Applications
94

Table 2: Mean performance results per patient before and after using the proposed IR technique.
Healthy
Patient
Dice Jaccard TOA
Before After Before After Before After
P1 0.97275 0.98511 0.98617 0.99249 0.98528 0.98879
P2 0.88114 0.92625 0.93563 0.96160 0.94110 0.94836
P3 0.92052 0.95638 0.95836 0.97766 0.96167 0.98204
P4 0.91520 0.91190 0.95534 0.95379 0.96774 0.93614
P5 0.97410 0.98584 0.98684 0.99285 0.98311 0.99066
P6 0.96601 0.97545 0.98269 0.98755 0.98795 0.98357
P7 0.90388 0.96251 0.94897 0.98082 0.94532 0.97394
P8 0.94357 0.97285 0.97094 0.98592 0.96495 0.97987
P9 0.94280 0.97477 0.97034 0.98720 0.96922 0.98075
P10 0.95459 0.97377 0.97674 0.98670 0.96572 0.98209
P11 0.97641 0.96841 0.98805 0.98393 0.98979 0.97793
Improved/Subjects 9/11 9/11 8/11
Patient
with Disease
Dice Jaccard TOA
Before After Before After Before After
P12 0.97569 0.97634 0.98768 0.98770 0.98209 0.98256
P13 0.91645 0.94692 0.95610 0.97266 0.95920 0.95609
P14 0.93368 0.95948 0.96537 0.97926 0.96919 0.98325
P15 0.97468 0.96117 0.98717 0.98008 0.98828 0.97179
P16 0.97230 0.95491 0.98593 0.97682 0.98993 0.97251
P17 0.97370 0.97787 0.98665 0.98879 0.97607 0.98210
P18 0.96957 0.96970 0.98454 0.98456 0.98576 0.98474
P19 0.93046 0.92758 0.96385 0.96236 0.96490 0.94261
P20 0.97691 0.96566 0.98831 0.98251 0.99157 0.97271
P21 0.94886 0.95382 0.97348 0.97582 0.97446 0.96801
P22 0.96275 0.96974 0.98098 0.98461 0.97918 0.97741
P23 0.97168 0.97826 0.98561 0.98900 0.98915 0.98338
Improved/Subjects 8/12 8/12 3/12
Total Improved/Subjects 17/23 17/23 11/23
overlap of two sets in relation to their union:
Jaccard =
N (I
R
∩I
S
)
N (I
R
∪I
S
)
. (7)
Like the Dice coefficient, Jaccard index equal to 1
means greater similarity between the images, while
0 indicates no similarities.
The TOA measure for a given registration is:
TOA =
N (I
R
∩I
S
)
N (I
S
)
, (8)
whose value also range from 0 to 1.
We have used similarity measures based on bi-
nary images following works in the literature. This
allows our technique to be compared with other ap-
proaches. Besides, the intensity information on the
patient’s thermographs varies over time because of the
dynamic protocol (Silva et al., 2016). This variation
can introduce errors in the analysis when the intensi-
ties are compared directly after registration.
From Table 2, it is possible to observe that, ac-
cording to the Dice coefficient and Jaccard index,
the proposed IR approach improved the registration
of images of 9 out 11 healthy patients and 8 out
12 patients with a disease. For the Dice coefficient,
the evaluation measure values increased up to 0.0586
units in success cases (subject P7) and decreased up
to 0.0174 units in the other cases (subject P16). For
the Jaccard index and TOA measure, the most signif-
icant performance improvement was of 0.0166 (sub-
ject P13) and 0.0286 units (subject P7), respectively,
while the greatest deterioration in performances were
of, respectively, 0.0091 (subject P16) and 0.0316
units (subject P4). These results show that the bene-
fits of the proposed technique outweigh the problems
it may introduce.
From a more detailed look at the registration re-
sults, it was observed that, in a general way, the pro-
posed method performed well, achieving IR improve-
ments in 77% of the pairs of images of healthy pa-
Using Geometric Graph Matching in Image Registration
95
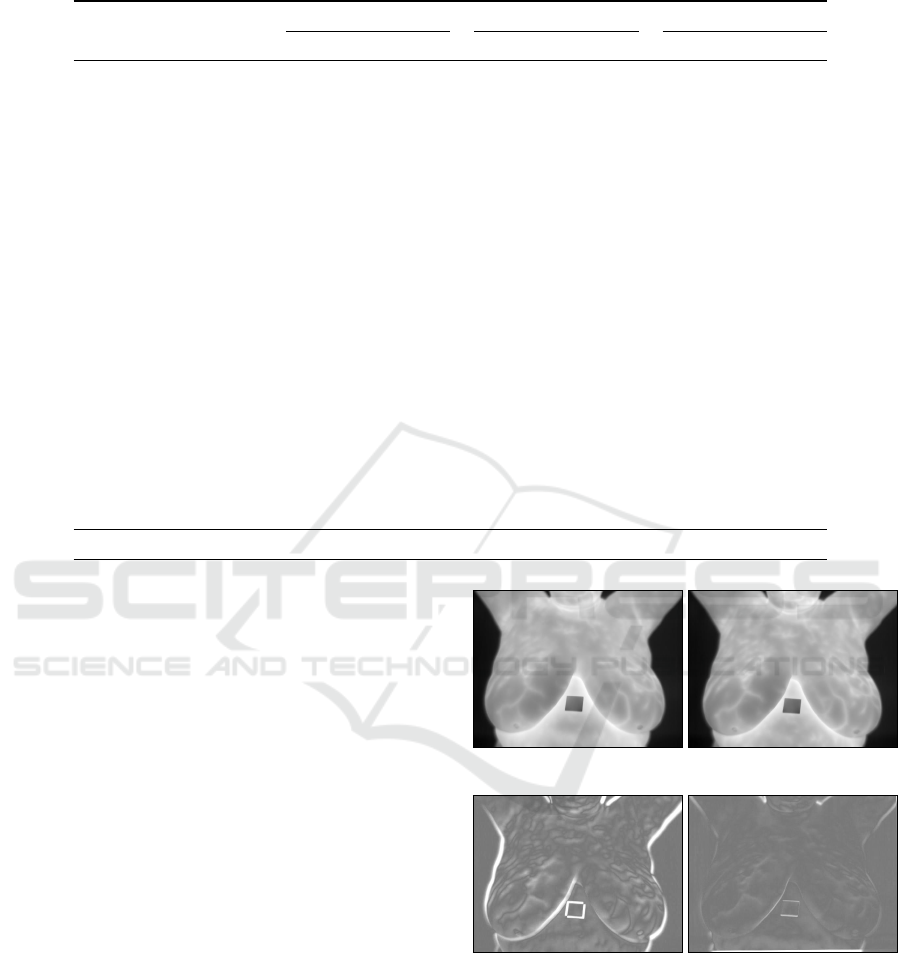
Table 3: Standard deviation performance results per patient before and after using the proposed IR technique.
Patient
Dice Jaccard TOA
Before After Before After Before After
P1 0.00654 0.00581 0.00335 0.00295 0.00356 0.00457
P2 0.06300 0.02134 0.03744 0.01144 0.04407 0.01878
P3 0.03094 0.01257 0.01684 0.00660 0.01693 0.00767
P4 0.03740 0.02177 0.02067 0.01183 0.02031 0.01820
P5 0.01336 0.00924 0.00687 0.00472 0.00900 0.00800
P6 0.00877 0.01035 0.00452 0.00531 0.00704 0.00891
P7 0.04465 0.01687 0.02453 0.00880 0.02387 0.01480
P8 0.01067 0.03501 0.00563 0.01864 0.00800 0.02189
P9 0.02912 0.01002 0.01548 0.00515 0.01553 0.00883
P10 0.01141 0.00614 0.05970 0.00315 0.00693 0.00473
P11 0.00673 0.00861 0.03440 0.00443 0.00400 0.00686
P12 0.00797 0.03590 0.00410 0.01940 0.00593 0.03573
P13 0.03378 0.01736 0.01834 0.00909 0.01549 0.01522
P14 0.03536 0.01510 0.01943 0.00795 0.02024 0.00935
P15 0.00676 0.02167 0.00346 0.01128 0.00233 0.01748
P16 0.00859 0.02164 0.00441 0.01148 0.00308 0.01165
P17 0.00904 0.00939 0.00463 0.00485 0.00889 0.00797
P18 0.00826 0.01486 0.00426 0.00776 0.00411 0.00570
P19 0.02218 0.01626 0.01194 0.00874 0.01467 0.01506
P20 0.00600 0.00853 0.00308 0.00442 0.00344 0.00757
P21 0.03275 0.04485 0.17470 0.02512 0.01858 0.02642
P22 0.01191 0.01046 0.00615 0.00537 0.00748 0.00869
P23 0.01000 0.00592 0.00517 0.00302 0.00436 0.00614
Total Improved/Subjects 14/23 16/23 10/23
tients. In the case of patients with diseases, there were
improvement in 56% of the image pairs.
Figures 7 (a) and (b) exemplify, respectively, the
reference image and 19
th
sensitive image for patient
P1. Figure 7 (c) shows the pixelwise difference be-
tween (a) and (b) without performing IR, while Fig-
ure 7 (d) shows the difference after performing the
proposed IR technique. In both images (c) and (d),
darker and lighter regions correspond to more signif-
icant (signed) differences. Results were mapped to
the [0, 255] range to improve visualization. A notice-
able superposition improvement can be observed after
IR, where most of the image (d) takes a medium gray
tone, which represents a difference close to zero.
Processing Time. The testbed implementation of
our approach was not tailored for performance. Even
so, it is capable of performing image registration in
less than one second on a PC with Intel Core i5 6500
CPU and 8GB of RAM.
Comparison to Other Approaches. Figure 8
shows a summary of the performance achieved by
the proposed method, the method described by Falco
et al. (2020), and the traditional use of SURF to ex-
ecute IR. As one can see, SURF performed worse,
not being competitive with either of the other two
(a) (b)
(c) (d)
Figure 7: Example of a thermogram taken as the reference
image (a) and the 19
th
sensitive image (b) of a given pa-
tient. Images (c) and (d) show the normalized pixel by pixel
difference between images (a) and (b) before and after ap-
plying the proposed IR technique, respectively.
techniques. It is because SURF and related feature
extraction techniques, e.g., SIFT and ORB, require
high-frequency information to characterize textured
regions properly. By using the Dice coefficient as the
VISAPP 2021 - 16th International Conference on Computer Vision Theory and Applications
96
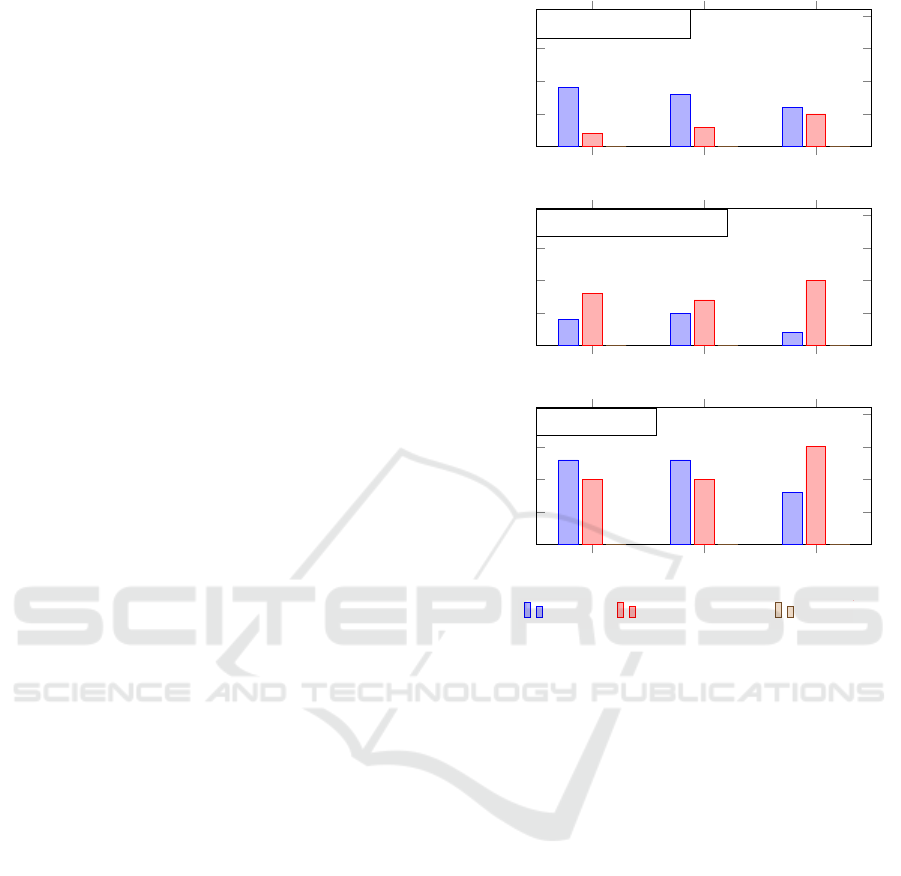
evaluation measure, our method performs better regis-
tering of healthy patients in 9 out of 11 cases, against
2 out of 11 cases where Falco’s et al. approach per-
formed better (Figure 8, a). For the Jaccard index, the
mean performance of our approach was higher for 8
against 3 subjects. The TOA measure shows a score
of 6 against 5 cases of better performance of the pro-
posed technique over Falco’s et al. approach. When
patients with a disease are considered, results show
that the Falco et al. method is slightly superior (Fig-
ure 8, b). But considering all subjects (Figure 8, c),
our approach is superior in two of the three evalua-
tion measures assumed.
5 CONCLUSIONS
This paper proposed an automatic method for infrared
breast IR by using a geometric graph matching ap-
proach. The graph that was created has a reduced
number of vertices that make it computationally ef-
ficient. The execution of our registration method per-
formed well, especially in healthy patients. In these
patients, the temperature change during the dynamic
image acquisition protocol seems to be more stable.
On the other hand, in patients with breast disease,
more significant changes were observed in the appar-
ent internal linear structures, leading to substantial
changes in the graphs’ structure and, consequently,
affecting the matching process. Nevertheless, the re-
sults presented in this paper are interesting since they
indicate that the first technique to use graphs for in-
frared breasts IR is promising.
Several works may emerge from the proposed ap-
proach. For instance, it is possible to modify the edit
distance model to assign different weights to the in-
sert, replace, and remove operations to allow different
priorities to be set to the operations.
Another possible direction of future work is using
other types of visual features to characterize the local
information around a vertex or an edge, including the
use of features extracted by artificial neural networks.
Finally, our approach could be adapted to differ-
ent types of medical images with linear and vascular
structures such as fundus images widely used to diag-
nose ocular diseases or diseases that have global ef-
fects on the vascular system (Bhatkalkar et al., 2020).
ACKNOWLEDGEMENTS
The Brazilian research agency CAPES sponsored
Giomar O. S. Olivera. Aura Conci is par-
tially supported by MACC-INCT, CNPq (grants
Dice Jaccard TOA
9
8
6
2
3
5
0 0 0
Number o Wins
(a) Healthy Patients
Dice Jaccard TOA
4
5
2
8
7
10
0 0 0
Number o Wins
(b) Patients with Disease
Dice Jaccard TOA
13 13
8
10 10
15
0 0 0
Number o Wins
(c) All Patients
Proposed Falco et al. (2020) SURF
Figure 8: Summary of the comparison of results for healthy
patients, patients with breast disease, and the total number
of patients using the proposed approach, the IR technique
described by Falco et al. (2020), and SURF.
402988/2016-7 and 305416/2018-9), and FAPERJ
(project SIADE-2 and 210.019/2020). Leandro A.
F. Fernandes is partially supported by the Brazilian
research agencies CNPq (grant 424507/2018-8) and
FAPERJ (grant E-26/202.718/2018).
REFERENCES
Agostini, V., Knaflitz, M., and Molinari, F. (2009). Motion
artifact reduction in breast dynamic infrared imaging.
IEEE Trans. Biomedical Engineering, 56(3):903–906.
Armiti, A. and Gertz, M. (2014). Vertex similarity - a basic
framework for matching geometric graphs. In Pro-
ceedings of the LWA, pages 111–122.
Balakrishnan, G., Zhao, A., Sabuncu, M. R., Guttag, J., and
Dalca, A. V. (2018). An unsupervised learning model
for deformable medical image registration. In Pro-
ceedings of the IEEE Conference on Computer Vision
and Pattern Recognition, pages 9252–9260.
Bhatkalkar, B., Joshi, A., Prabhu, S., and Bhandary, S.
(2020). Automated fundus image quality assessment
Using Geometric Graph Matching in Image Registration
97

and segmentation of optic disc using convolutional
neural networks. International Journal of Electrical
& Computer Engineering, 10:2088–8708.
Brock, K. K., Mutic, S., McNutt, T. R., and H. Li and, M.
L. K. (2017). Use of image registration and fusion
algorithms and techniques in radiotherapy: report of
the AAPM Radiation Therapy Committee Task Group
No. 132. Medical Physics, 44(7):43–76.
Conci, A., Galv
˜
ao, S. S., Sequeiros, G. O., Saade, D. C., and
MacHenry, T. A. (2015). A new measure for compar-
ing biomedical regions of interest in segmentation of
digital images. Discrete Appl. Math., 197:103–113.
Dalal, N. and Triggs, B. (2005). Histograms of oriented
gradients for human detection. In Proceedings of
the IEEE Conference on Computer Vision and Pattern
Recognition, pages 886–893.
Deng, K., Tian, J., Zheng, J., Zhang, X., Dai, X., and Xu,
M. (2010). Retinal fundus image registration via vas-
cular structure graph matching. International Journal
of Biomedical Imaging, 2010:906067.
Dice, L. R. (1945). Measures of the amount of ecologic
association between species. Ecology, 26(3):297–302.
Falco, A. D., Galv
˜
ao, S., and Conci, A. (2020). A non lin-
ear registration without the use of the brightness con-
stancy hypothesis. In Proc. Intern. Conference Sys-
tems, Signals and Image Processing, pages 27–32.
Fischler, M. A. and Bolles, R. C. (1981). Random sample
consensus: a paradigm for model fitting with appli-
cations to image analysis and automated cartography.
Communications of the ACM, 24(6):381–395.
Garcia-Guevara, J., Peterlik, I., Berger, M. O., and Cotin, S.
(2018). Biomechanics-based graph matching for aug-
mented CT-CBCT. International Journal of Computer
Assisted Radiology and Surgery, 13(6):805–813.
Gonz
´
alez, J. R., Pupo, Y., Hernandez, M., Conci, A.,
Machenry, T., and Fiirst, W. (2018). On image reg-
istration for study of thyroid disorders by infrared ex-
ams. In Proc. Intern. Conf. Image Proc., Computer
Vision, & Pattern Recognition, pages 151–158.
Harris, C. and Stephens, M. (1988). A combined corner
and edge detector. In Proceedings of the Alvey Vision
Conference, pages 147–151.
Holden, M. (2008). A review of geometric transformations
for nonrigid body registration. IEEE Transactions on
Medical Imaging, 27(1):111–28.
Ismail, N. H. F., Zaini, T. R. M., Jaafar, M., and Pin,
N. C. (2016). H-minima transform for segmentation
of structured surface. In Proceedings of the MATEC
Web of Conferences, page 25.
Jaccard, P. (1912). The distribution of the flora in the alpine
zone. New Phytologist, 11(2):37–50.
Klein, A., Andersson, J., Ardekani, B. A., Ashburner, J.,
Avants, B., Chiang, M. C., Christensen, G. E., Collins,
D. L., Gee, J., Hellier, P., Song, J. H., Jenkinson,
M., Lepage, C., Rueckert, D., Thompson, P., Ver-
cauteren, T., Woods, R. P., Mann, J. J., and Parsey,
R. V. (2009). Evaluation of 14 nonlinear deformation
algorithms applied to human brain MRI registration.
NeuroImage, 49(3):786–802.
Kornilov, A. and Safonov, I. (2018). An overview of wa-
tershed algorithm implementations in open source li-
braries. Journal of Imaging, 4(10):123.
Lee, C., Chang, Z., Lee, W., Lee, S., Chen, C., Chang,
Y., and Huang, C. (2012). Longitudinal registration
for breast IR image without markers. In Proceed-
ings of the International Conference on Quantitative
InfraRed Thermography.
Lee, C., Chuang, C. C., Chang, Z. W., Lee, W. J., Lee,
C. Y., C, S., Lee, Huang, C. S., Chang, Y. C., and
Chen, C. M. (2010). Quantitative dual-spectrum in-
frared approach for breast cancer detection. In Pro-
ceedings of the International Conference on Quanti-
tative InfraRed Thermography.
Ma, J., Zhao, J., and Yuille, A. L. (2016). Non-rigid point
set registration by preserving global and local struc-
tures. IEEE Trans. Image Processing, 25(1):53–64.
Pan, M. S., Yang, X. L., and Tang, J. T. (2012). Research
on interpolation methods in medical image process-
ing. Journal of Medical Systems, 36(2):777–807.
Pinheiro, M. A., Kybic, J., and Fua, P. (2017). Geometric
graph matching using Monte Carlo tree search. IEEE
Transactions on Pattern Analysis and Machine Intel-
ligence, 39(11):2171–2185.
Rahman, M. M. (2018). Literature-based biomedical image
retrieval with multimodal query expansion and data
fusion based on relevance feedback (RF). In Proc. In-
tern. Conference Image Processing, Computer Vision,
& Pattern Recognition, pages 103–107.
Riesen, K., Fischer, A., and Bunke, H. (2018). On the
impact of using utilities rather than costs for graph
matching. Neural Processing Letters, 48(2):691–707.
Silva, L. F., Santos, A. A., Bravo, R. S., Silva, A. C.,
Muchaluat-Saade, D. C., and Conci, A. (2016). Hy-
brid analysis for indicating patients with breast cancer
using temperature time series. Computer Methods and
Programs in Biomedicine, 130:142–153.
Silva, L. F., Sequeiros, G., Santos, M. L., Fontes, C. A. P.,
Muchaluat-Saade, D., and Conci, A. (2015). Ther-
mal signal analysis for breast cancer risk verifica-
tion. Studies in Health Technology and Informatics,
216:746–50.
Tong, Y., Udupa, J. K., Ciesielski, K. C., Wu, C., Mc-
Donough, J. M., Mong, D. A., and Campbell Jr, R. M.
(2017). Retrospective 4D MR image construction
from free-breathing slice acquisitions: a novel graph-
based approach. Medical Image Analysis, 35:345–
359.
Zhang, T. Y. and Suen, C. Y. (1984). A fast parallel algo-
rithm for thinning digital patterns. Communications
of the ACM, 27(3):236–239.
Zheng, W., Zou, L., Lian, X., Wang, D., and Zhao, D.
(2013). Graph similarity search with edit distance
constraint in large graph databases. In Proceedings
of the ACM International Conference on Information
& Knowledge Management, pages 1595–1600.
Zitov
´
a, B. and Flusser, J. (2003). Image registration
methods: a survey. Image and Vision Computing,
21(11):977–1000.
VISAPP 2021 - 16th International Conference on Computer Vision Theory and Applications
98
