
An Extension of Chronicles Temporal Model with Taxonomies:
Application to Epidemiological Studies
Johanne Bakalara
1,2
, Thomas Guyet
2,3
, Olivier Dameron
2
, Andr
´
e Happe
4
and Emmanuel Oger
1
1
Univ. Rennes, EA-7449 REPERES, France
2
Univ. Rennes, Inria, IRISA-UMR6074, France
3
Institut Agro, Rennes, France
4
CHRU Brest, France
Keywords:
Temporal Query, Medico-administrative Databases, Sequences of Events, Chronicles, Semantic Web.
Abstract:
Medico-administrative databases contain information about patients’ medical events, i.e. their care trajecto-
ries. Semantic Web technologies are used by epidemiologists to query these databases in order to identify
patients whose care trajectories conform to some criteria. In this article we are interested in care trajecto-
ries involving temporal constraints. In such cases, Semantic Web tools lack computational efficiency while
temporal pattern matching algorithms are efficient but lack of expressiveness. We propose to use a temporal
pattern called chronicles to represent temporal constraints on care trajectories. We also propose an hybrid
approach, combining the expressiveness of SPARQL and the efficiency of chronicle recognition to query care
trajectories. We evaluate our approach on synthetic data and real large data. The results show that the hybrid
approach is more efficient than pure SPARQL, and validate the interest of our tool to detect patients having
venous thromboembolism disease in the French medico-administrative database.
1 INTRODUCTION
Pharmaco-epidemiology (PE) studies the conditions
and consequences of health products, i.e. drugs or
medical devices usage at the population scale in real
situations using methodologies developed in general
epidemiology.
Modern PE relies on administrative databases to
perform such studies on care trajectories, i.e. on
patient-centered sequences of drugs deliveries, med-
ical procedures and hospitalizations. The use of
medico-administrative databases (MADB) is useful in
PE studies, since data are readily available and cover
a large population.
The problem with MADB is the semantic gap be-
tween raw data and the epidemiological question. On
the one side, epidemiologists are looking for medi-
cal events. For instance, they would like to iden-
tify patients suffering from venous thromboembolism
(VTE). On the other side, raw data are related to reim-
bursements of medical acts or drug deliveries. There
is no exploitable diagnosis available in administrative
databases and no clinical results related to medical
acts or exams.
The challenge for epidemiologists is to define phe-
notypes of medical events (Hong et al., 2019), i.e. a
combination of information available in the database
that reveals an occurrence of a medical event. For
instance, a patient having a lower limbs doppler ul-
trasonography exam and few days after a delivery of
anticoagulant drugs for 3 to 6 or 12 months is prob-
ably suffering from VTE. As MADB record medical
exam and drugs deliveries, the above description may
be used as a proxy of VTE.
The Semantic Web offers a relevant framework for
representing complex data patterns and linking them
with domain knowledge. Semantic Web data lan-
guage (e.g. RDF) is suitable to represent structured
data of MADB (Rivault et al., 2019). Moreover, link-
ing raw data to standard medical taxonomies is in-
teresting to enrich the description of cares with for-
malized expert knowledge (for instance, ICD-10
1
for
diagnosis or ATC
2
for drugs). Once care trajectories
have been represented in standard Semantic Web for-
mat, SPARQL query engines can be used to enumer-
ate all situations that match a query. A query can be
seen as a phenotype. However, if the expressiveness
of SPARQL is interesting to specify complex care tra-
jectories as a phenotype, the drawback is their com-
putation time. In the following, we assume the reader
1
ICD-10: Inter. Classification of Diseases, 10th Revision.
http://bioportal.bioontology.org/ontologies/ICD10
2
ATC: Anatomical Therapeutic Chemical. https://bioportal.
bioontology.org/ontologies/ATC
Bakalara, J., Guyet, T., Dameron, O., Happe, A. and Oger, E.
An Extension of Chronicles Temporal Model with Taxonomies: Application to Epidemiological Studies.
DOI: 10.5220/0010236601330142
In Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2021) - Volume 5: HEALTHINF, pages 133-142
ISBN: 978-989-758-490-9
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
133

to be familiar with RDF and SPARQL, but a thorough
introduction to semantic web can be found in (Hitzler
et al., 2009). The example of VTE illustrates that such
query may be a complex arrangement of cares in a pa-
tient care trajectory. These arrangement involve tem-
poral relations between events (quantitative delays).
Thus, we are interested in specifying complex tempo-
ral patterns that may occur in care trajectories. Taking
into account numerical filters in metric temporal con-
straints is not efficient in SPARQL queries, and we
can not expect to achieve reasonable computational
efficiency on large MADB.
This article addresses the problem of enumerating
the occurrences of a complex temporal pattern in a
dataset of care trajectories.
We focus on a class of temporal patterns called
chronicles and propose a template of SPARQL query
that is both expressive and efficient. A chronicle is
an expressive temporal pattern. It is defined as a
set of events linked with temporal constraints. De-
spite its lack of taxonomy handling, this temporal
model is suitable to represent complex temporal care
pathways. One of its interests is its efficiently to
be recognized in a sequence of events (Dousson and
Le Maigat, 2007).
Our contribution is threefold: (i) we show how
chronicles can be encoded as SPARQL queries to enu-
merate all their occurrences in a sequence of events
represented in RDF; (ii) we propose HYCOR, an hy-
brid method combining the expressiveness of Seman-
tic Web and the efficiency of a pattern occurrence
enumeration algorithm; (iii) we evaluate HYCOR on
a real case study of enumerating VTE events in the
French MADB.
2 RELATED WORK
In this section, we review some approaches to enu-
merate occurrences of temporal patterns in sequences
of events and their connection to Semantic Web.
Temporal databases and querying tools (Snod-
grass and llsoo Ahn, 1986) address a part of the prob-
lem by extending the notion of database to times-
tamped data. They cover data representation prob-
lems but also specific querying language problems.
This family encompasses the temporal extension of
relational databases (e.g. TSQL) but also Seman-
tic Web approaches which combine query language
(SPARQL) extended to temporal data with Allen’s re-
lations (Wang et al., 2010). These approaches de-
fines relative temporal constraints between intervals
which are not relevant for the MADB query problems
(Pacaci et al., 2018).
Rivault et al. (Rivault et al., 2019) used RDF to
represent care trajectories and shown that querying
care trajectories can be achieved with the SPARQL
query language. Semantic Web is a relevant approach
for our problem: it does not explicitly address the
problem of timed queries, but it is relevant to deal
with data representation and taxonomies querying.
RDF also enables ontology management with OWL
based on the Description Logic (DL) (Baader et al.,
2003) allowing ontology-mediated query answering
(OMQA) (Bienvenu, 2016). For instance, O’Connor
et al. (O’Connor et al., 2009) developed a tool based
on OWL for research data management with a tem-
poral reasoning in a clinical trial system. This aspect
could be added to the presented method.
Some approaches proposed to extend RDF/SPARQL
with temporal queries in a generic way. For instance,
Zhang et al. (Zhang et al., 2019) propose SPARQL[t]
and EP-SPARQL (Anicic et al., 2011a) which is a
SPARQL extension of event processing. Finally, ON-
TOP is an ontology-based data access framework that
has been extended for temporal data (Kalayci et al.,
2019). However, these generic tools turn out to lack
practical efficiency. This calls for investigating more
algorithmic solutions.
Several temporal models have been highlighted
in literature, one of the most promising is the Com-
plex Event Processing (CEP) (Giatrakos et al., 2017)
which aims at processing a stream of event logs with
patterns. CEP processes these logs to detect or to lo-
cate complex events (or patterns) defined by the user.
These models emphasis on the effectiveness of pro-
cessing and the expressivity of patterns. Some expres-
sive formalisms, e.g. ETALIS (Anicic et al., 2011b) or
logic-based event recognition (Giatrakos et al., 2017)
propose very expressive representations of complex
events, including reasoning techniques (encompass-
ing ontologies).
While the complex event descriptions of ETALIS
are based on Allen’s logic, temporal constraint net-
works (Cabalar et al., 2000) and Chronicles (Dous-
son and Le Maigat, 2007) propose complex event de-
scriptions with more permissive temporal constraints.
These temporal models are also interesting for their
graphical representation, but are restricted to patterns
which do not involve taxonomies. It has been mainly
used to discover patterns in biomedical data (Daux-
ais et al., 2017; Sahugu
`
ede et al., 2018) or in logs of
industrial processes (Sellami et al., 2018).
HEALTHINF 2021 - 14th International Conference on Health Informatics
134
3 SEQUENCES AND
TAXONOMIES
In this work, we adopt a longitudinal view of a
MADB. Each patient is represented by a sequence of
timestamped cares, so called events. We first intro-
duce the definition of event and taxonomy of event
labels. Then, we introduce the notion of temporal se-
quence of events, or sequence for short.
Formally, an event is a pair (e,t) where e is an
event label and t ∈ N is a timestamp (in days). In the
following, (E,≤
E
) denotes the totally ordered set of
events. Usually, labels of medical events are related
to taxonomies such as ATC
2
for drugs or ICD-10
1
for
diseases.
Definition 1 (Event Taxonomy). An event taxonomy
is an ordered set of equivalence relations (R
i
)
i∈[n]
,
where n is the number of levels of the taxonomy, such
that:
∀(i, j), i < j, ∀(e,e
0
) ∈ E, eR
j
e
0
=⇒ eR
i
e
0
. (1)
We denote by c
j
i
the i-th equivalent class at level
j induced by the taxonomy. By definition, we have
that e ∈ E ⇔ ∃!i, c
0
i
= e. C denotes the set of all
equivalent classes.
An event label e ∈ E is a c ∈ C, denoted e c, iff
e is in the equivalent class of c. By extension, c ∈ C
is a c
0
∈ C, denoted c
0
c iff e c
0
⇒ e c for all
e ∈ E. And then (E,≤
E
, ) denotes a set of ordered
event labels equipped by a taxonomy relation.
In the Table 1, events are represented by ATC
codes. Each ATC code is a class in the ATC taxon-
omy. For example, the ATC code A01AA01 is a sub-
class of A (A01AA01 A).
Let us now introduce the formal definition of a
temporal sequence of events.
Definition 2 (Sequence). A sequence s is a finite list
of events h(e
1
,t
1
), (e
2
,t
2
), ··· ,(e
n
,t
n
)i where e
i
is an
event label which is a taxonomy class. Events in a
sequence are ordered by their timestamps and then
their label: i ≤ j ⇔ t
i
< t
j
∨(t
i
= t
j
∧e
i
≤
E
e
j
), ∀i, j ∈
{1,..., n}.
A dataset of sequences is a finite unordered set
of sequences, S = {s
1
,..., s
m
}. Tab. 1 illustrates six
sequences where each event is a drug delivery where
event labels are issued from the ATC taxonomy.
4 CHRONICLE OCCURRENCES
ENUMERATION
In this section, we propose an extended definition
of chronicles (Dauxais et al., 2017; Dousson and
A01,1
B01A,2
[-1,3]
C,3
[-3,5]
[-2,2]
C,4
[1,3]
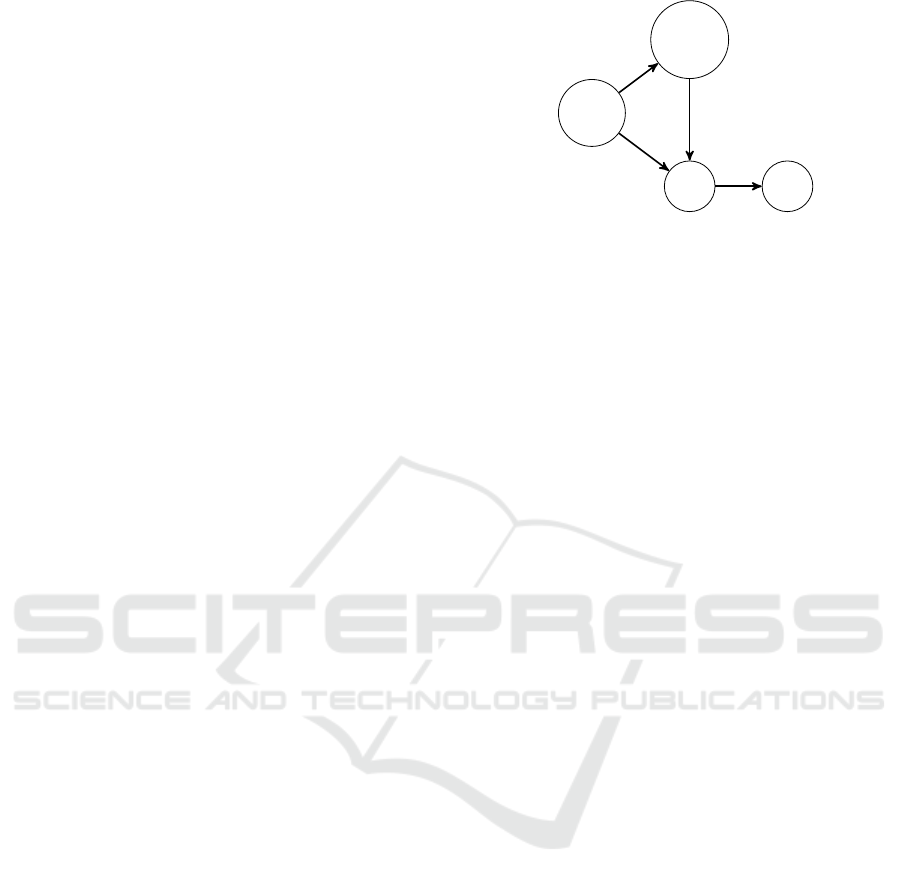
Figure 1: Chronicle example with 4 events (vertices) and 4
temporal constraints (edges with temporal intervals). Vertex
labels give the event label (ATC codes).
Le Maigat, 2007; Sahugu
`
ede et al., 2018) with events
belonging to taxonomy classes. Then, we define
formally a chronicle occurrence in a sequence and
the enumeration of all chronicle occurrences in a se-
quence.
A chronicle is a set of events and a set of tempo-
ral constraints between pairs of events (Dousson and
Le Maigat, 2007). In our applied context, chronicle
enables to represent a phenotype. The enumeration
of chronicles occurrences aims at localizing where
this medical pattern occurs in a patient care trajec-
tory. This paper proposes a chronicle extension where
event may have label belonging to the equivalence
class of an event label.
Definition 3 (Chronicle). A chronicle C is a pair
(E, T ) where
• E is an ordered set of events
{(c
1
,1),·· · ,(c
m
,m)} , where for all
i ∈ {1,...,m}, c
i
∈ E is an event label. i
designates the index of the i-th event index.
• T is a set of temporal constraints, i.e. expressions
of the form (c
j
, j)[t
−
,t
+
](c
k
,k) such that
– (c
j
, j),(c
k
,k) ∈ E,
– t
−
,t
+
∈ R ∪ {+∞,−∞} and
– For all (c
j
, j),(c
k
,k) ∈ E, j < k,
c
j
c
k
=⇒ ∃(c
j
, j)[t
−
,t
+
](c
k
,k) ∈ T
s.t. [t
−
,t
+
] ⊆ [1, +∞[
(2)
The chronicle size is m (number of events).
A temporal constraint (c
j
, j)[t
−
,t
+
](c
k
,k) en-
forces an event (c
k
,k) to occur with a temporal delay
in between t
−
and t
+
from an occurrence of (c
j
, j).
Note that several events can have the same label. A
chronicle event may also have its label belonging to
the equivalent class of another event label. In these
cases, Eq. 2 enforces event occurrences to be ordered
by their index.
The Fig. 1 illustrates graphically the following 4-
sized chronicle C = (E,T ):
An Extension of Chronicles Temporal Model with Taxonomies: Application to Epidemiological Studies
135

Table 1: Example of a dataset of six sequences (longitudinal view on six patients). Each sequence is made of drug deliveries
events (couples of label and timestamp). Labels are ATC codes, i.e. the code of a delivered drug in the ATC toxonomy.
id Sequence
s
1
(A01AA01, 1), (B01AA01, 3), (A01AB14, 4), (C01AA01, 5), (C02AC01, 6), (D01AA01, 7)
s
2
(B01AA01, 2), (D01AA01, 4), (A01AA01, 5), (C01AA01, 7)
s
3
(A03AA01, 1), (B01AA01, 4), (C01AA01, 5), (B01AA01, 6), (C01AA01, 8), (D01AA01, 9)
s
4
(B01AA01, 4), (A01AB14, 6), (N01AA01, 8), (C01AA01, 9)
s
5
(B01AA01, 1), (A01AA01, 3), (C01AA01, 4)
s
6
(C01AA01, 4), (B01AA01, 5), (A01AA01, 6), (C01AA01, 7), (D01AA01, 10)
• E = {(A01, 1), (B01A,2),(C,3),(C,4)}
• T = {(A01,1)[−1,3](B01A, 2) ,
(A01,1)[−3,5](C, 3) ,
(B01A,2)[−2,2](C, 3) , (C,3)[1,3](C,4)}
where event labels belong to the ATC taxonomy. No-
tice that temporal constraints may have negative val-
ues. The temporal constraint (A01,1)[−3,5](C,3)
states that an event with label in the equivalence class
of A01 must occur from 3 days before occurrence of
a C to 5 days after this occurrence. Thus, the chron-
icle Fig. 1 means: “An event A01 is followed by an
event B01A within a delay of [−1,3] units of time
(ut). The later is followed by an event C within a de-
lay of [−2,2] ut. In addition the delay between this
event C and the event labeled A01 must be in [−3, 5]
ut. Finally, C event is followed by an another event C
within a delay of [1,3] ut”.
In the following, we introduce the definition of a
chronicle occurrence in a sequence. Then, one can be
interested in two different tasks: enumerating all oc-
currences of a chronicle in a sequence (chronicle enu-
meration), or deciding whether a chronicle occurs at
least once in the sequence (chronicle recognition). In
the following, we focus on the chronicle enumeration
task.
Definition 4 (Chronicle occurrence). Let
s = h(e
1
,t
1
),(e
2
,t
2
),... , (e
n
,t
n
)i
be a sequence of length n and
C = (E = {(c
1
,1),.. . , (c
m
,m)},T )
be a chronicle of size m over a set of labels
(E,≤
E
, ).
An occurrence of C in s is a subsequence of s of
length m, denoted ˜s = h(e
ε
1
,t
ε
1
), . ..,(c
ε
m
,t
ε
m
)i, where
(ε
i
)
i=1..m
are indices of an event in s and s.t.
1. e
ε
i
c
i
2. t
ε
j
−t
ε
i
∈ [t
−
,t
+
]
whenever (c
i
,i)[t
−
,t
+
](c
j
, j) ∈ T .
(ε
i
)
i=1..m
describes ˜s a subsequence of s. The first
condition ensures that the i-th event label of ˜s is a sub-
class of c
i
. The second condition ensures that tempo-
ral constraints are satisfied. Note that Eq. 2 enforces
to have a strict order between events c
k
, c
j
whenever
c
k
c
j
. Thus, all ε
i
, i ∈ [1, n] are distinct.
The chronicle of Fig. 1 occurs in sequences s
1
and s
6
of the dataset in Table 1. For instance,
{(A01AA01,1), (B01AA01,3), (C01AA01,5),
(C02AC01,6)} is an occurrence of C in s
1
. This
occurrence is the subsequence of s
1
with indices
h1,2,4,5i. The chronicle does not occur in s
2
nei-
ther in s
4
because of unsatisfied temporal constraints.
It does not occur in s
5
as there is only one event with
a type of class C in the sequence and the chronicle
requires two different events. It does not occur in s
3
because there is not an event in the subgroup of A01.
5 SEMANTIC WEB FOR
CHRONICLE RECOGNITION
Semantic Web is a framework designed to represent,
share and manipulate structured data. The keystones
of Semantic Web are (i) formal data representations,
such as the RDF language, and (ii) query languages,
such as SPARQL. Semantic Web is particularly suit-
able to represent taxonomies.
Semantic Web is suitable to represent sequences
and to encode chronicle enumeration with SPARQL.
So, we propose to represent sequences in RDF and
to encode a chronicle enumeration in SPARQL. We
propose two approaches for chronicle enumeration:
the first approach fully uses Semantic Web technolo-
gies; the second approach is an hybrid tool combining
SPARQL query and a dedicated algorithm.
5.1 Sequence Representation in RDF
Sequences are represented in a RDF-Graph (Fig. 2).
We remind that our concrete objective is to query a
dataset of sequences, where each patient care trajec-
tory is represented as a sequence.
HEALTHINF 2021 - 14th International Conference on Health Informatics
136

seq:seq5 seq:hasEvent seq:seq5evt0 .
seq:seq5evt0 seq:evtLabel atc:B01AA01 ;
seq:evtDate '1'ˆˆxsd:integer .
seq:seq5 seq:hasEvent seq:seq5evt1 .
seq:seq5evt1 seq:evtLabel atc:A01AA01 ;
seq:evtDate '3'ˆˆxsd:integer .
seq:seq5 seq:hasEvent seq:seq5evt2 .
seq:seq5evt2 seq:evtLabel atc:C01AA01 ;
seq:evtDate '4'ˆˆxsd:integer .
Figure 2: Example of sequence representation in RDF
graph (see sequence s
5
in Table 1).
SELECT DISTINCT * WHERE{
?sequence patdb:hasEvent ?evt1 .
?evt1 seq:evtDate ?date1 .
?evt1 seq:evtlabel ?atc1 .
?atc1 rdfs:subClassOf* atc:A01 .
?sequence patdb:hasEvent ?evt2 .
?evt2 seq:evtDate ?date2 .
?evt2 seq:evtlabel ?atc2 .
?atc2 rdfs:subClassOf* atc:B01A .
?sequence patdb:hasEvent ?evt3 .
?evt3 seq:evtDate ?date3 .
?evt3 seq:evtlabel ?atc3 .
?atc3 rdfs:subClassOf* atc:C .
?sequence patdb:hasEvent ?evt4 .
?evt4 seq:evtDate ?date4 .
?evt4 pseq:evtlabel ?atc4 .
?atc4 rdfs:subClassOf* atc:C .
FILTER ( ?date2 - ?date1 >= -1)
FILTER ( ?date2 - ?date1 <= 3)
FILTER ( ?date3 - ?date1 >= -3)
FILTER ( ?date3 - ?date1 <= 5)
FILTER ( ?date3 - ?date2 >= -2)
FILTER ( ?date3 - ?date2 <= 2)
FILTER ( ?date4 - ?date3 >= 1)
FILTER ( ?date4 - ?date3 <= 3)
}
Figure 3: Example of a SPARQL query for chronicle enu-
meration.
In RDF, each event (e
j
,t
j
) in a sequence s
i
is en-
coded by three tuples:
• seq:sequencei seq:hasEvent seq:sequenceievtj de-
notes existence of an event e
j
in sequence s
i
• seq:sequenceievtj seq:eventLabel atc:lk denotes
event e
j
has the label l
k
. Note atc referees to the
ATC taxonomy
2
where l
k
is a leaf-class
• seq:sequenceievtj seq:eventDate '1'ˆˆ xsd:integer
denotes event e
j
has a date t
j
equal to 1.
Fig. 1 illustrates the representation in RDF by provid-
ing the representation of sequence s
5
(see Table 1).
5.2 SPARQL for Chronicle Occurrences
Enumeration
This section presents the chronicle recognition task
with SPARQL. SPARQL queries RDF sequences
where all sequences are in the same RDF named
Graph.
Figure 3 gives the SPARQL query for the chroni-
cle in Fig. 1. The query has three types of variables:
• ?sequence denotes an identifier of a sequence
• ?evtj corresponds the j-th element of the chronicle
set.
• ?datej is the date of the j-th element of an occur-
rence (t
ε
j
with notation of Definition 4).
The taxonomy of event labels are handled by
rdfs:subClassOf* pattern operator. This operator is
equivalent to the operator defined in Def. 1. Tem-
poral constraints are expressed in FILTER clauses. For
instance, the temporal constraint (C, 3)[1, 3](C, 4) is
translated in a couple of constraints between ?date4
and ?date3.
SPARQL is expressive enough for enumerating
chronicle occurrences. However the enumeration of
chronicle occurrences is a very computational task.
A SPARQL query can not compete with dedicated
enumeration algorithms as its solver strategy is not
optimised for this task (see experiments in Sect. 6).
Therefore, we propose an hybrid approach to benefit
from the best of both fields: efficiency of dedicated
approaches and expressiveness of Semantic Web.
5.3 HYCOR for Chronicle Recognition
HYCOR (Hybrid-Chronicle Occurrences Recogni-
tion) combines SPARQL and a specific algorithm
to enumerate efficiently occurrences of a chronicle.
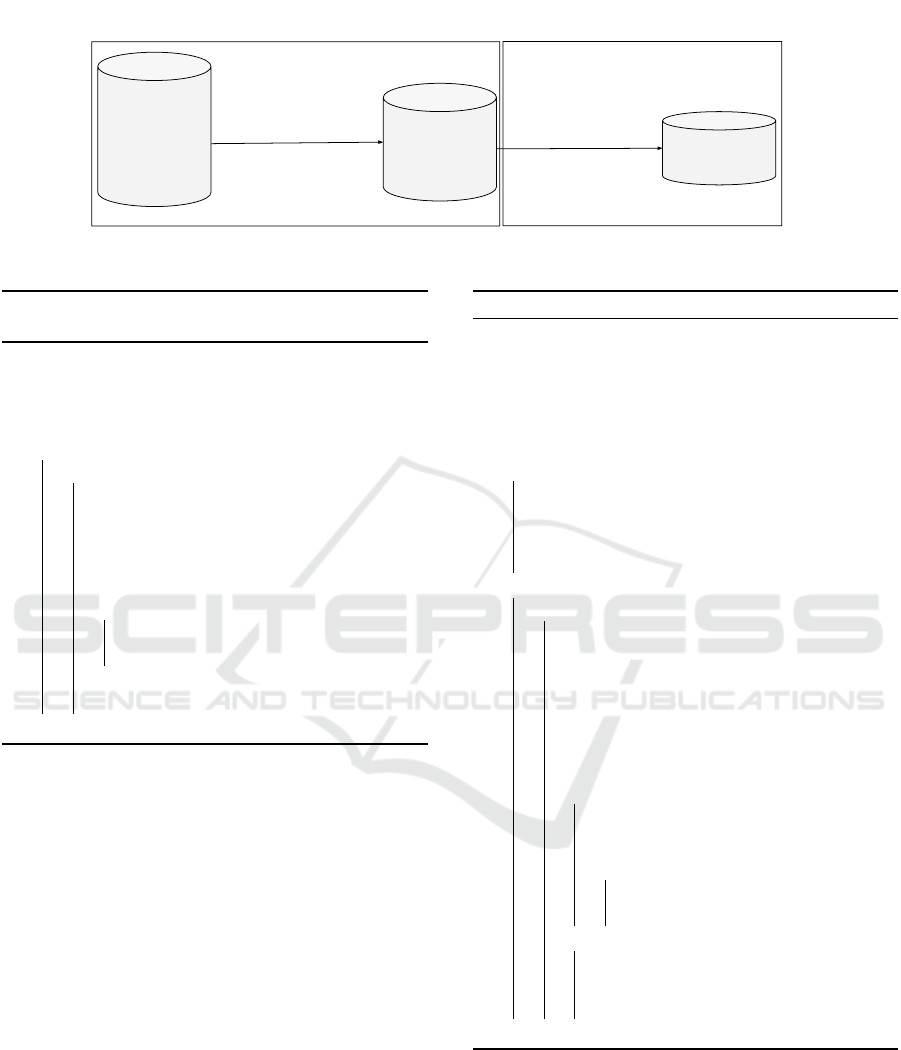
Fig. 5 illustrates the HYCOR process.
First (left box of Fig. 5), a SPARQL query yields
flattened sequences . A flattened sequence contains
only the sequence events that belong to the equivalent
class of at least one event label of E, i.e. the chronicle
events (see Def. 3). Such a query for chronicle of
Fig. 1 is on 4:
SELECT DISTINCT ?seq ?date ?label where{
values ?label { atc:A01 atc:B01A atc:C }
GRAPH patdb:onto { ?l rdfs:subClassOf* ?label }.
GRAPH ?seq{
?event patdb:drugDelivered ?l.
?event patdb:deliveryDate ?date.}}
Figure 4: Illustration of SPARQL query to map sequences
on the set of events of chronicle in Fig. 1.
Second (right box of Fig. 5), HYCOR applies Al-
gorithm 1 to enumerate chronicle occurrences in the
flattened sequences.
The algorithm’s principle is to refine progressively
intervals in which a chronicle event (c
i
,i) ∈ E may
An Extension of Chronicles Temporal Model with Taxonomies: Application to Epidemiological Studies
137
Sequences
with only
events of the
chronicle C
sequences
verifying C
SPARQL mapping
events of C
taking in account
taxonomies
Recognition Algorithm
verifying time
constraints events of C
Sequences
Figure 5: Schema of the HYCOR process to enumerate occurrences of a chronicle C .
Algorithm 1: Occurrences of a chronicle C in
a sequence s.
Input: C = (E = {(c
1
,1),.. . , (c
m
,m)},T ),
s = h(e
1
,t
1
) .. . (e
n
,t
n
)i
Output: occs: a set of occurrences of C in s
1 occs ←
/
0 // Set of occurrences
2 foreach (e,t) ∈ s do
3 if e = c
1
then
// create a set of admissible
positions π of size m
4 π ← {[t,t], [−∞, ∞], ...,[−∞,∞]};
// propagate chronicle
constraints
5 foreach (c
1
,1)[t
−
,t
+
](c
p
, p) ∈ T do
6 π
p
=
[max(t
1
,t +t
−
),min(t + t
+
,t
n
)];
7 occs ← occs∪
RECENUMERATE(π,1,C ,s);
8 return occs
occur in s. These intervals are called admissible po-
sitions. The algorithm goes through the set of events
(e
i
,t
i
) ∈ s and propagates the temporal constraints of
the chronicle to narrow position intervals until inter-
vals are only single position. Thus admissible posi-
tion designates a subsequence of s, i.e., an occurrence
of the chronicle. Algorithm 1 makes recursive calls
to Algorithm 2. The later assumes that the k − 1 first
events have been located in s. This means that the
k first intervals of the admissible intervals π are sin-
gleton intervals. The recursive call looks for event c
k
in the admissible positions of s for k-th event (lines
5-6). If found, it is a candidate for further refine-
ments and temporal constraints of the chronicle are
propagated. The constraint (c
k
,k)[t
−
,t
+
](c
p
, p) is a
constraint from (c
k
,k) event to the event c
p
at posi-
tion p. It is used to possibly narrow the admissible
positions of event p (line 11). In case the new posi-
tions are inconsistent (line 12) then this candidate oc-
currence can not satisfy the temporal constraints and
is discarded (satis f iable is set to f alse). If all con-
Algorithm 2: RECENUMERATE(π,k, C ,s).
Input: π: admissible positions, k: recursion
level,
C = (E = {(c
1
,1),.. . , (c
m
,m)},T )),
s = h(e
1
,t
1
) .. . (e
n
,t
n
)i
Output: occs: a set of occurrences of C in s
1 occs ←
/
0 // Set of occurrences
2 if k = m + 1 then
// An occurrence has been found
3 occ ← {(e
k
i
,t
k
i
) ∈ s | c
i
= e
k
i
,π
i
= t
k
i
,i =
1..m};
4 return {occ}
5 foreach (e,t) ∈ s s.t. t ∈ π
k
do
6 if e = c
k
then
// create a copy of
admissible positions π
7
˜
π ← π;
8
˜
π
k
← [t,t];
// propagate chronicle
constraints
9 satis f iable ← true;
10 foreach (c
k
,k)[t
−
,t
+
](c, p) ∈ T do
11
˜
π
p
← [max(
˜
π
−
p
,t +
t
−
),min(
˜
π
+
p
,t +t
+
)];
12 if
˜
π
−
p
>
˜
π
+
p
then
13 satis f iable ← f alse;
14 break;
15 if satis f iable then
// Recursive call
16 occs ← occs∪
RECENUMERATE(
˜
π,k + 1,C ,s);
17 return occs
straints are satisfied, the recursive call attempts to re-
fine further these positions (line 16). Note that only
forward constraints are propagated. Indeed, backward
constraints (i.e. constraint to event at position lower
than k in the set) have already been taken into account
in parent calls.
Let us illustrate the algorithm on a sim-
HEALTHINF 2021 - 14th International Conference on Health Informatics
138

ple example. Let s = h(B,2) (C,3) (A,5)
(B,6) (C,7) (C, 9) (C, 10)i and C = ({(A,1),(B,2),
(C,3)}, {(A,1)[−2,2](B, 2), (A, 1)[−3, 5](C,3),
(B,2)[−1,3](C, 3)}).
1. Processing of event A
• generates a single tuple of admissible positions
π = ([5, 5],[−∞,∞],[−∞,∞])
• constraints propagation:
– (A,1)[−2,2](B, 2): π = ([5, 5], [3, 7], [−∞,∞])
– (A,1)[−3,5](C, 3): π = ([5, 5], [3, 7], [2,10])
2. Processing of event B
• narrows positions with occurrences: (B,2) is
invalid (2 /∈ [3,7]), but (B, 6) satisfies the ad-
missible positions [3, 7] so the admissible posi-
tions can be updated (π = ([5,5], [6,6], [2, 10]))
• constraints propagation:
– (B,2)[−1,3](C, 3): π = ([5,5],[6,6],[2,10] ∩
[5,9]) = ([5,5],[6,6],[5,9])
3. Processing of event C
• narrows intervals with occurrences: (C,3) and
(C,10) are invalid, but (C, 7) and (C,9) are
valid, then the both subsequences where the
chronicle occurs are obtained by updating the
admissible positions (([5,5],[6,6], [7,7]) and
([5,5],[6, 6], [9, 9])).
6 EXPERIMENTS
In this section, we compare execution times of
SPARQL and HYCOR on synthetic datasets. All ex-
periments have been executed with a TDB-graph for-
mat for RDF and Jena-Fuseki as SPARQL engine.
The HYCOR algorithm is implemented in Python.
The computer has 16Go RAM and an SSD.
6.1 Synthetic Datasets Generation and
Plan of Experiments
Several synthetic datasets have been generated. Each
dataset contains a set of sequences where event labels
are randomly chosen at the lowest level of ATC tax-
onomy. The ATC taxonomy contains 1900 classes.
In addition, occurrences of ten 15-sized chronicles
3
are embedded in the dataset. For each chronicle, a
constraint is generated for each pair of events without
inconsistency between the temporal constraints.
3
A n-sized-chronicle denotes a chronicle of size n.
The synthetic dataset generation process ensures
that each chronicle occurs in about 20% of the se-
quences. Chronicles contain event labels from sev-
eral levels of ATC following this probability:
1
15
level
1 (ex: N),
2
15
level 2 (ex: N02),
3
15
level 3 (ex: N02B),
3
15
level 4 (ex: N02BE),
6
15
level 5 (ex: N02BE01).
We introduce the notation D
ns,ne
to denote a syn-
thetic dataset with ns sequences and ne care events
per sequence (all sequences have the same num-
ber of events). For the following experiments, 25
synthetic datasets have been generated where ns ∈
{1000, 5 000, 10 000, 15 000,20 000} and ne ∈ {100,
200,300,400,500}. Each dataset is encoded in RDF.
The ATC taxonomy is attached to the dataset.
6.2 Experiments and Results
The following experiments evaluate the impact of two
main parameters on execution times of SPARQL and
HYCOR: the size of the dataset (number of sequences
and number of events per sequence) and the chronicle
size.
Fig. 6 compares the execution times of SPARQL
and HYCOR with respect to the length of the se-
quences. It shows that HYCOR is at least one or-
der of magnitude more efficient than pure SPARQL.
6 shows that SPARQL does not scale up for datasets
containing more than 50 000 sequences. The HYCOR
SPARQL query language does not have the same lim-
itation. Indeed, the pure SPARQL query uses filters
to deal with temporal constraints on each admissible
event while SPARQL HYCOR query only uses val-
ues to find admissible event. So, the use of filters on
a large scale of admissible events seems to be ineffi-
cient in SPARQL for this kind of use.
We also evaluate the part of HYCOR execution
times spent by the SPARQL mapping and the chroni-
cle enumeration algorithm. On average, the SPARQL
query execution represents 85%±3.47 of the total ex-
ecution time.
Experiments now focus on the HYCOR evalua-
tion. Fig. 7 illustrates the impact of the number of
events per sequence on the execution times. We ob-
serve that time increases linearly with the number of
events and with the number of sequences. Outliers
and variance of the computing time can be explained
by the variability of the number of events occurrences
in the sequence that influence the time execution of
the Algorithm 2. The more an event occurs in a se-
quence, the more candidate occurrences, therefore the
longer the time spent in the algorithm. Nonetheless,
we can notice in the hardest condition: D
20000,500
,
enumeration of a chronicle with 15 events takes in av-
erage less than a minute.
An Extension of Chronicles Temporal Model with Taxonomies: Application to Epidemiological Studies
139
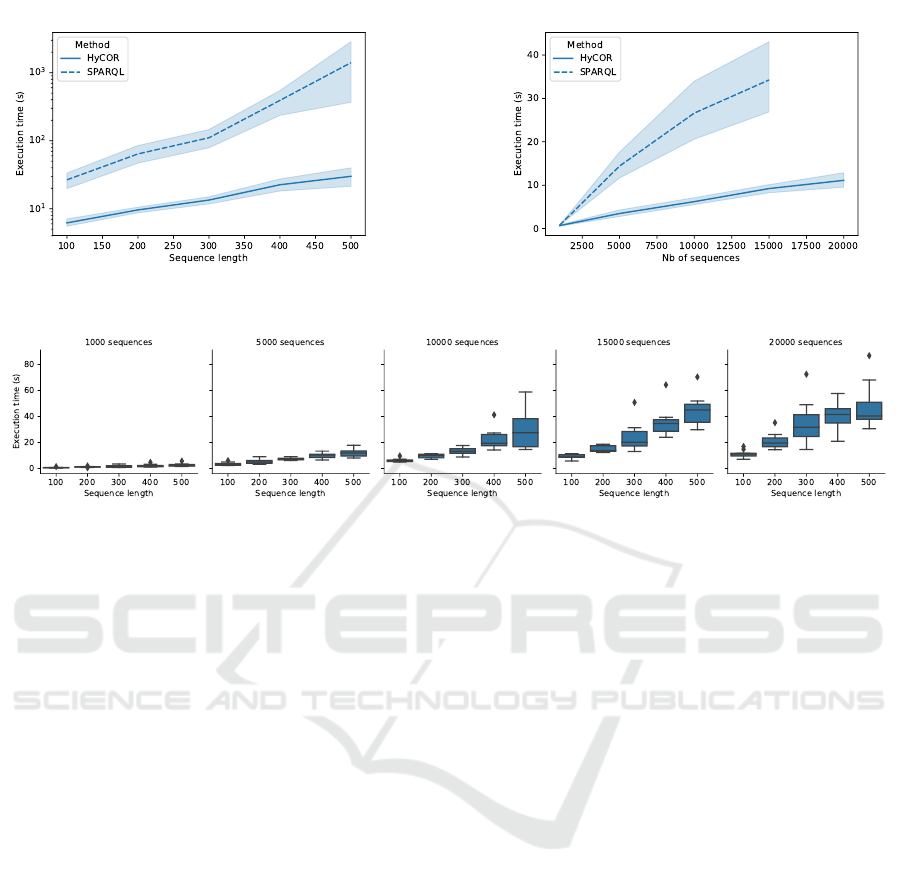
Figure 6: Execution times (in seconds) of SPARQL and HYCOR wrt sequences length on 10 000 sequences (on the left) and
wrt number of sequences (with length 100).
Figure 7: Execution times of chronicle occurrences enumeration wrt sequences length.
Fig. 8 presents execution time of HYCOR wrt
chronicle size. HYCOR is run on a unique dataset
(ns = 10000 and ne = 100) and seven sets of 10
chronicles with sizes 2 to 14. We observe execution
time linearly increases with the chronicle size.
HYCOR outperforms pure-SPARQL for the
chronicle enumeration task. Its execution time in-
creases with the chronicle sizes and the dataset size,
but it still offers impressive results for large datasets.
7 USE CASE ON THE SNDS TO
FIND PATIENTS WITH
THROMBOEMBOLISM
Our use case proposes to find patients diagnosed
with venous thromboembolism (VTE) in the SNDS.
The SNDS is the french national health insurance
database, which covers most of the french population
(above 65 million inhabitants). The advantage of this
database is to gather information about most the reim-
bursed medical events, from drug deliveries to nurse
home cares, specialist consultations, etc. The range
of medical events that are recorded in the database
makes it suitable to conduct a wide variety of health
studies (Tuppin et al., 2017). However, SNDS has
been designed for administrative purposes (care reim-
bursements) and does not contain detailed medical in-
formation such as medical reports, laboratory results
or diagnosis.
This use case uses a geographical-based SNDS
subset (the north western French Brittany population)
which contains 377 359 individuals. For the use case
application, rdf:type have been added in the RDF
event triples to speed up access to event labels. We
used five different types: DrugDeliveries (e.g., drug
deliveries from pharmacy), Cares (e.g., nurse assis-
tance, domestic assistance), Medical acts (e.g., radi-
ology, surgery), Biologies (e.g., blood withdrawal),
Hospitalisations.
VTE is identified by epidemiologists in SNDS
when a patient matches the following description:
In clinical practice facing a suspicion of VTE
physicians first prescribe anticoagulant and
then confirm or not the diagnosis through spe-
cific medical acts: e.g. Doppler ultrasonogra-
phy or CT scan. Patients with suspected PE
are often hospitalized whereas patients with
suspected DVT are managed on an ambula-
tory basis. If the suspicion is confirmed, an-
ticoagulant deliveries continues for 3 to 12
months (once per month) or sometimes longer
duration. Hence, the diagnosis (through med-
ical act) is preceded or followed by anticoagu-
lant initiation within a time window of at most
0 to 7 days, keeping in mind that PE suspicion
leads to hospitalisation during which medical
acts to confirm the diagnosis are performed
and then anticoagulant delivery is observed
HEALTHINF 2021 - 14th International Conference on Health Informatics
140
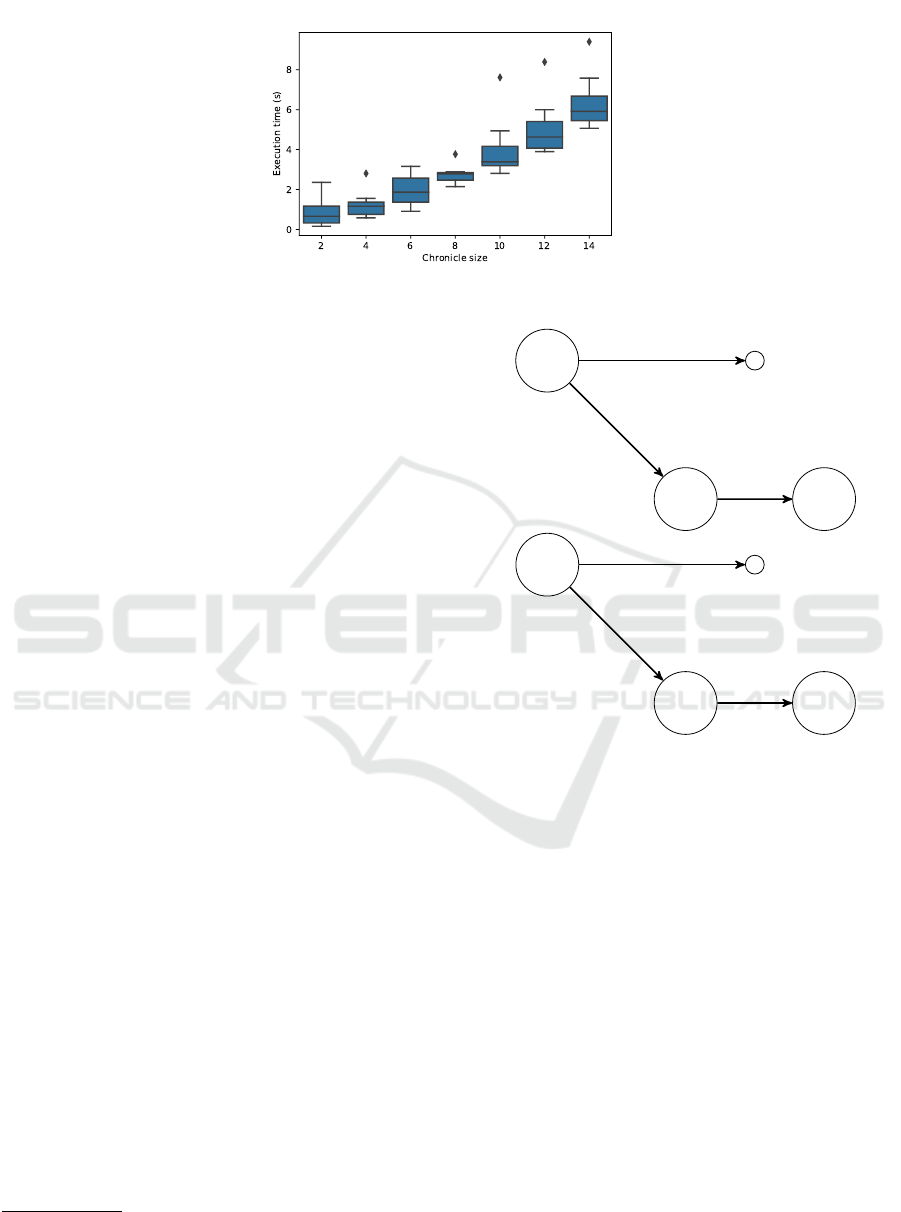
Figure 8: Execution time (in seconds) wrt chronicle size (ns = 10 000 and ne = 100).
only after the patient comes back home.
Fig. 9 illustrates two chronicles defining the phe-
notype of “patients with VTE”. Each chronicle speci-
fies temporal constraints (within 7 days and one anti-
coagulant delivery per month) but also takes into ac-
count some details on the anti-coagulant class and on
the medical acts on ambulatory or in hospital. An-
ticoagulant are identified with the ATC code B01.
We also added to the knowledge base a class named
ccam:thrombose which is defined as an union
of 36 different codes of medical acts issued of the
CCAM taxonomy
4
. Furthermore, some VTE are
identified in hospital through ICD-10 codes
1
, they are
gathered in a class called cim:diagthrombose.
For this experiment, we load RDF graphs, we
query them with both chronicles, one by one, and
evaluate computing time. Note that we do not have
the ground truth. Thus, we are not interested in the
accuracy of the chronicles to identify true VTE. In
this experiments, we compare the computational effi-
ciency to enumerate the same set of VTE occurrences.
The first result is that the corresponding pure-
SPARQL query does not finish the enumeration
within a day of computation. This is due to the scale
up limitation shown in Sect. 5.2.
HYCOR finds 2 686 patients having VTE in
105.06s. The first chronicle finds 2568 patients in
56.21s (of which 52.86s in algorithm execution), the
second chronicle finds 118 patients in 48.85s (of
which 48.46s in algorithm execution). Use case time
execution is even faster than expected by experiments
on synthetic data. The real patient sequences length
is about 100 events in average. So if we refer to
Fig. 6, on the right, the expected execution time is
about 11min41s. We explain this difference by the
lower size of the chronicles in our use case query.
4
CCAM : common classification of medical acts used by
the french social security
B01A
[-2,2]
B01A
[1,31]
B01A
[1,31]
ccam:thrombose
B01A
[-2,2]
B01A
[1,31]
B01A
[1,31]
cim:thrombose
Figure 9: Chronicles for representing VTE phenotype.
8 CONCLUSION
In this article, we extended the model of chroni-
cle with taxonomies to enumerate complex tempo-
ral patterns in sequences. This problem is moti-
vated by the need for phenotyping patients in medico-
administrative databases. We proposed HYCOR, an
hybrid approach that combines the expressiveness of
SPARQL and the efficiency of a dedicated algorithm.
The results show that HYCOR is one order of mag-
nitude faster than pure SPARQL queries on both syn-
thetic and real dataset. As a perspective, chronicles
should be extended with negation to denote the ab-
sence of event. Furthermore, it could be interesting to
compare the efficiency of different SPARQL engines.
An Extension of Chronicles Temporal Model with Taxonomies: Application to Epidemiological Studies
141

REFERENCES
Anicic, D., Fodor, P., Rudolph, S., and Stojanovic, N.
(2011a). EP-SPARQL: a unified language for event
processing and stream reasoning. In Proc. of Int. Conf.
on World Wide Web (WWW), pages 635–644.
Anicic, D., Fodor, P., Rudolph, S., St
¨
uhmer, R., Stojanovic,
N., and Studer, R. (2011b). ETALIS: Rule-based rea-
soning in event processing. In Proc. of Reasoning in
event-based distributed systems, pages 99–124.
Baader, F., Calvanese, D., McGuinness, D., Patel-
Schneider, P., and Nardi, D. (2003). The description
logic handbook: Theory, implementation and applica-
tions. Cambridge university press.
Bienvenu, M. (2016). Ontology-mediated query answer-
ing: harnessing knowledge to get more from data. In
Proc. of Int. Join Conf. on Artificial Intelligence (IJ-
CAI), pages 4058–4061.
Cabalar, P., Otero, R. P., and Pose, S. G. (2000). Tem-
poral constraint networks in action. In Proc. of Eu-
ropean Conf. on Artificial Intelligence (ECAI), pages
543–547.
Dauxais, Y., Guyet, T., Gross-Amblard, D., and Happe, A.
(2017). Discriminant chronicles mining. In Proc. of
Conf. on Artificial Intelligence in Medicine in Europe
(AIME), pages 234–244.
Dousson, C. and Le Maigat, P. (2007). Chronicle recogni-
tion improvement using temporal focusing and hier-
archization. In Proc. of Int. Join Conf. on Artificial
Intelligence (IJCAI), pages 324–329.
Giatrakos, N., Artikis, A., Deligiannakis, A., and Garo-
falakis, M. (2017). Complex event recognition in the
big data era. In Proc. VLDB Endow., volume 10, pages
1996–1999.
Hitzler, P., Krotzsch, M., and Rudolph, S. (2009). Founda-
tions of semantic web technologies. CRC press.
Hong, N., Wen, A., Stone, D. J., Tsuji, S., Kingsbury, P. R.,
Rasmussen, L. V., Pacheco, J. A., Adekkanattu, P.,
Wang, F., Luo, Y., et al. (2019). Developing a FHIR-
based EHR phenotyping framework: A case study for
identification of patients with obesity and multiple co-
morbidities from discharge summaries. J. of Biomed-
ical Informatics, 99:103310.
Kalayci, E. G., Brandt, S., Calvanese, D., Ryzhikov, V.,
Xiao, G., and Zakharyaschev, M. (2019). Ontology-
based access to temporal data with ontop: A frame-
work proposal. Int. J. of Applied Mathematics and
Computer Science, 29(1):17–30.
O’Connor, M. J., Shankar, R. D., Parrish, D. B., and Das,
A. K. (2009). Knowledge-data integration for tem-
poral reasoning in a clinical trial system. Int. J. of
Medical Informatics, 78:77–85.
Pacaci, A., Gonul, S., Sinaci, A. A., Yuksel, M., and
Laleci Erturkmen, G. B. (2018). A semantic transfor-
mation methodology for the secondary use of observa-
tional healthcare data in postmarketing safety studies.
Frontiers in pharmacology, 9:435.
Rivault, Y., Dameron, O., and Le Meur, N. (2019).
queryMed: Semantic web functions for linking phar-
macological and medical knowledge to data. Bioin-
formatics.
Sahugu
`
ede, A., Le Corronc, E., and Le Lann, M.-V. (2018).
An ordered chronicle discovery algorithm. In 3nd
ECML/PKDD Workshop on Advanced Analytics and
Learning on Temporal Data, AALTD’18.
Sellami, C., Samet, A., and Tobji, M. A. B. (2018). Fre-
quent chronicle mining: Application on predictive
maintenance. In Proc. of Int. Conf. on Machine Learn-
ing and Applications (ICMLA), pages 1388–1393.
Snodgrass, R. T. and llsoo Ahn (1986). Temporal databases.
Computer, 19(09):35–42.
Tuppin, P., Rudant, J., Constantinou, P., Gastaldi-M
´
enager,
C., et al. (2017). Value of a national administrative
database to guide public decisions: From the syst
`
eme
national d’information interr
´
egimes de l’assurance
maladie (sniiram) to the syst
`
eme national des donn
´
ees
de sant
´
e (snds) in france. Revue d’epidemiologie et de
sante publique, 65:S149–S167.
Wang, Y., Zhu, M., Qu, L., Spaniol, M., and Weikum,
G. (2010). Timely YAGO: harvesting, querying, and
visualizing temporal knowledge from wikipedia. In
Proc. of Int. Conf. on Extending Database Technology
(EDBT), pages 697–700.
Zhang, F., Wang, K., Li, Z., and Cheng, J. (2019). Temporal
data representation and querying based on RDF. IEEE
Access, 7:85000–85023.
HEALTHINF 2021 - 14th International Conference on Health Informatics
142
