
Morphological Classification of Heartbeats in Compressed ECG
Gennaro Laudato
1
, Francesco Picariello
2
, Simone Scalabrino
1
,
Ioan Tudosa
2
, Luca De Vito
2
and Rocco Oliveto
1
1
STAKE Lab, University of Molise, Pesche (IS), Italy
2
Department of Engineering, University of Sannio, Benevento (BN), Italy
Keywords:
ECG Analysis, Arrhythmia, Decision Support System, Compressed Sensing, Machine Learning.
Abstract:
The number of connected medical devices that are able to acquire, analyze, or transmit health data is continu-
ously increasing. This has allowed the rise of Internet of Medical Things (IoMT). IoMT-systems often need to
process a massive amount of data. On the one hand, the colossal amount of data available allows the adoption
of machine learning techniques to provide automatic diagnosis. On the other hand, it represents a problem
in terms of data storage, data transmission, computational cost, and power consumption. To mitigate such
problems, modern IoMT systems are adopting machine learning techniques with compressed sensing meth-
ods. Following this line of research, we propose a novel heartbeat morphology classifier, called RENEE, that
works on compressed ECG signals. The ECG signal compression is realized by means of 1-bit quantization.
We used several machine learning techniques to classify the heartbeats from compressed ECG signals. The
obtained results demonstrate that RENEE exhibits comparable results with respect to state-of-the-art methods
that achieve the same goal on uncompressed ECG signals.
1 INTRODUCTION
Nowadays, the use of Internet of Medical Things
(IoMT) systems for remote health monitoring is play-
ing a pivotal role in improving both the effectiveness
of medical devices and the accessibility to medical
services (Hassanien et al., 2018). Remote health mon-
itoring refers to a process where the patient’s health is
continuously checked, thus allowing the identification
and the prevention of diseases. To this aim, the use of
wearable devices for continuous monitoring is receiv-
ing increasing interest from both the health services
and the manufactures. For example, in the case of
electrocardiogram (ECG) monitoring, several IoMT
systems based on wearable devices have been pro-
posed (Balestrieri et al., 2019; Wang et al., 2019).
With the spread use of IoMT systems, the com-
plex and time-consuming steps of pre-diagnosis and
diagnosis — usually manually performed by special-
ized medical staff — can be supported or undertaken
by such systems. For this reason, in the recent years,
several methods for the automatic detection of cardiac
diseases from an ECG trace and, more specifically,
automatic classification of heartbeats have been pro-
posed (Mond
´
ejar-Guerra et al., 2019; Kandala et al.,
2019; Rajesh, 2018; Chen et al., 2017; David et al.,
2011b; David et al., 2011a; Garcia et al., 2017; Xu
et al., 2018; Mar et al., 2011). All these methods
— when applied in contexts of long-term continu-
ous monitoring — require that the physical devices
of the IoMT system continuously send the monitored
data to a gateway or directly to a server. Since wear-
able devices are battery-powered and since they use
a wireless connection to exchange data, it is very im-
portant to decrease the amount of transmitted data for
reducing the energy consumption of the device and, as
a consequence, increasing its battery life. (Balestri-
eri et al., 2020) proposed an ECG data acquisition
system that performs data compression according to
compressed sensing, a theoretical framework that ex-
ploits the sparsity of a signal in a specific domain
without computationally load the physical device that
performs the compression. The authors also showed
that — by using such kind of systems — it is possible
to reduce the number of transmitted data and increase
the battery life by ∼12 %, while keeping a good ECG
signal reconstruction quality.
In this paper, we propose RENEE (heaRtbEat
classificatioN in comprEssed ECG), a novel method
for the automatic classification of heartbeats that
works on a compressed ECG signal, through the in-
volvement of a method based on a 1-bit signal quan-
tization (Picariello et al., 2021). The advantage of us-
ing RENEE with respect to others available heartbeat
classifiers is that it allows reducing the signal data rate
and performs the heartbeat classification directly on
the quantized samples, therefore without reconstruct-
ing the signal waveform. Especially, RENEE was de-
386
Laudato, G., Picariello, F., Scalabrino, S., Tudosa, I., De Vito, L. and Oliveto, R.
Morphological Classification of Heartbeats in Compressed ECG.
DOI: 10.5220/0010236003860393
In Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2021) - Volume 5: HEALTHINF, pages 386-393
ISBN: 978-989-758-490-9
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

signed to be used in an IoMT system based on wear-
able devices, where it is needed to reduce the data
transmitted by the physical device and to automat-
ically classify the heartbeats, without analyzing the
signal waveform. In such a context, using a com-
pressed signal is beneficial both in terms of data rate
and memory occupied.
We evaluated RENEE on a public data set, the
MIT-BIH Arrhythmia Database, a reference point in
the literature. The evaluation has respected the rec-
ommended practice provided by a well-known stan-
dard, the ANSI/AAMI EC57:1998
1
. The achieved re-
sults provide evidence that RENEE allows to keep a
comparable overall accuracy — in the classification of
heartbeats — with respect to state-of-the-art methods
that works with the full (i.e., uncompressed) original
ECG signal. Indeed, one of the best approaches from
the literature shows a global accuracy of 0.947 while
RENEE achieves 0.94 in the compressed domain.
The remainder of the paper is structured as fol-
lows. Section 2 provides details about the recent state
of the art on (i) the morphological automatic classi-
fication of heartbeats and (ii) the compression algo-
rithms applied to ECG signals. Section 3 presents
RENEE, our novel approach for the morphological
classification of heartbeats in the compressed domain.
Section 4 and Section 5 reports on the design and the
results of the empirical study that we conducted to
evaluate RENEE. Finally, Section 6 concludes the pa-
per and provides suggestions for possible future re-
search directions.
2 BACKGROUND AND RELATED
WORK
This section provides details on (i) the automatic
classification of ECG through machine learning tech-
niques and (ii) approaches proposed for ECG com-
pression.
2.1 Heartbeat Classification
In the last years, several methods have been pro-
posed for the automatic classification of ECG heart-
beats (Mond
´
ejar-Guerra et al., 2019; Kandala et al.,
2019; Rajesh, 2018; Chen et al., 2017; Garcia et al.,
2017; Xu et al., 2018; Mar et al., 2011). Most of
them have shown good results. They all dealt with the
full ECG, i.e., without any type of compression. In
addition, most of them involve complex algorithms,
1
American National Standard prepared by the Association
for the Advancement of Medical Instrumentation.
i.e., that need high-computational cost for the creation
of the features. In details, these approaches classify
each heartbeat in five output categories: normal beat
(N), ventricular ectopic beat (V), supraventricular ec-
topic beat (S), fusion of a normal and a ventricular ec-
topic beat (F) and unknown beat type (Q). Also, they
have been validated on the public MIT-BIH arrhyth-
mia database
2
.
(Mond
´
ejar-Guerra et al., 2019) proposed a method
for the automatic classification of ECG based on the
combination of multiple Support Vector Machines.
The method relies on the time intervals between con-
sequent beats and their morphology for the ECG char-
acterization. Several features based on wavelets, local
binary patterns (LBP), higher order statistics (HOS)
were employed. The designed methodology approach
was tested classifying four kinds of abnormal and nor-
mal beats. The authors achieved an overall accuracy
of approximately 0.945 and, in some cases, they have
obtained better results than the related state of the art
approaches.
(Kandala et al., 2019) presented an inter-patient
heartbeat classification algorithm. The foundation of
this work is based on the consideration that the ECG
is a non-stationary, non-Gaussian signal derived from
nonlinear systems (Rajesh, 2018). Therefore, the au-
thors employed a decomposition method, namely im-
proved complete ensemble empirical mode decompo-
sition (ICEEMD), to obtain features from the ECG
beats. Then, nonlinear measures such as entropies and
higher-order statistics (HOS) were determined from
the modes obtained after ICEEMD. These were used
as features for the discrimination of the heartbeats. To
handle with class unbalance, the authors employed
a type of ensemble classification based on a major-
ity voting scheme. Finally, the authors demonstrated
good improvement — with respect to the state of the
art methods — especially for the minority classes.
(Garcia et al., 2017) proposed a heartbeat rep-
resentation, called the temporal vector-cardiogram
(TVCG), and an optimized feature extraction pro-
cess with complex networks and particle swarm op-
timization (PSO). The authors show that their method
presents an overall accuracy in the classification equal
to 0.924.
(Xu et al., 2018) proposed an end-to-end method
with a deep neural network (DNN) for both feature
extraction and classification based on aligned heart-
beats. This method avoids any further elaboration
for the features creation and produces optimized ECG
representation for heartbeat classification. The overall
working principle of this approach can be resumed as
follows: the system buffers raw single lead ECG sig-
2
https://physionet.org/content/mitdb/1.0.0/
Morphological Classification of Heartbeats in Compressed ECG
387

nals — at one end — and produces heartbeat classifi-
cation, at the other end. The pre-processing concerns
with the selection of heartbeats from continuous ECG
signals. In the proposed approach a heartbeat signal is
represented by a segment of the ECG that comprises
the consecutive sample points of a complete heartbeat
cycle, which includes not only the QRS complex but
also the P and T waves. The Neural Network (NN)
is used for both feature extraction and classification,
which are achieved by the lower part and the upper
part of the network, respectively. Two steps must be
performed in order to extract fixed-length feature vec-
tors from raw ECG signals: (i) heartbeat segmentation
and (ii) heartbeat alignment. To the best of our knowl-
edge, the approach proposed by (Xu et al., 2018) rep-
resents one of the best approaches presented in the
state of the art with an overall accuracy of 0.947 in
the 5-class classification. For this reason, we used
such an approach as baseline in the evaluation of RE-
NEE.
2.2 ECG Compression
In the literature, several compression methods of ECG
signals have been proposed with the aim of reduc-
ing the data rate of the IoMT physical device and its
energy consumption. The proposed approaches can
be classified in hardware-based methods and digital-
based methods (Picariello et al., 2021).
The hardware-based compression methods exploit
the sparsity of the ECG signal in the time domain to
design specific Analog-to-Digital Converter (ADC).
According to (Picariello et al., 2021), the digital-
based ECG compression methods can be classified in:
(i) direct methods, (ii) parameter extraction methods,
and (iii) transform domain methods.
The transform domain methods have gained sig-
nificant attention due to their good capability of rep-
resenting the ECG signal even at high compression
ratios (Picariello et al., 2021). However, most of them
require a high computational load to be implemented
in real-time on data acquisition systems having low
resources (Picariello et al., 2021).
Alternatively, Compressed Sensing (CS) has been
proposed in the literature for ECG data compression
(Picariello et al., 2021). The advantage of CS —
as compared to other methods — relies in its capa-
bility of achieving performance comparable with the
transform-domain methods, while moving the com-
putational load from the data acquisition system to the
node that receives the compressed samples. Thus, this
solution has been widely used for implementing data
compression on devices with constrained resources,
such as wearable devices.
In some cases, the data rate reduction can be
obtained by optimizing the resolution of the data,
thus introducing a controlled quantization (Jha and
Kolekar, 2018; Bera et al., 2019).
The aim of this paper is to apply a low-complexity
compression algorithm based on a 1-bit quantization
of the ECG signal and to assess the capability of some
machine learning algorithms to successfully classify
the heartbeat from the quantized samples. It is worth
noting that the proposed classification approach op-
erates directly on the compressed data and does not
require a reconstruction of the ECG waveform before
the classification. As compression algorithm, we have
chosen a simple 1-bit quantization, as proposed by
(Picariello et al., 2021). The algorithm is applied to
heartbeat signals. During this phase, (i) the data is
normalized, (ii) the dither is applied, and (iii) the 1-
bit quantization is performed. With normalization we
simply refer to the application of the formula
hbs
i
=
hbs
i
− min(hbs)
max(hbs) − min(hbs)
for each sample i of the heartbeat signal hbs; in other
words, we normalize the data between the minimum
and the maximum value for every heartbeat signal. As
a result, all the values will be in the interval [0, 1].
The application of dither consists in applying a
Gaussian dithering noise to the heartbeat signals with
power σ. Thus, let be (i) hbs a heartbeat signal,
(ii) ds the noising signal obtained by imposing ds
i
=
σ ×random(0, 1) so that |ds| = |hbs|. The noised ver-
sion of the original signal is given by nhbs = hbs +ds.
Dither can be considered as a kind of noise, but it
is typically and intentionally applied to randomize
quantization error and thus to improve the next quan-
tization step (Pohlmann, 1995).
Finally, 1-bit quantization step performs a com-
parison with a pre-defined threshold γ: if a given sam-
ple value nhbs
i
exceeds the threshold γ, 1 is assigned
to the output vector, while 0 is assigned otherwise.
Formally, for each nhbs
i
:
qhbs
i
=
(
1, if nhbs
i
≥ γ
0, otherwise
3 THE PROPOSED APPROACH
In this section, we present RENEE, a novel approach
for the classification of heartbeats in the compressed
domain. The proposed method is contextualized in
an IoMT system and in an healthcare scenario, where
a wearable device (transmitter) acquires an ECG sig-
nals and needs to send the acquired signal to a deci-
HEALTHINF 2021 - 14th International Conference on Health Informatics
388

sion support system (receiver) for its automatic anal-
ysis.
The pre-processing stage is composed by an R-
peak detection algorithm and a consecutive selection
of a complete heartbeat signal. Once executed these
steps, the further processing consists in the compres-
sion. Finally, the compressed data is provided as in-
put to the machine learning classifier. This latter is in
charge of providing the final multi-class classification
on the heartbeat types. We provide more details about
each step of our approach below.
3.1 Pre-processing of ECG Data
The pre-processing steps expected from our proposed
approach are composed of an R-Peak detector and a
heartbeat selection technique. Such steps may be per-
formed on the transmitter device.
The R-peak detector is in charge of accurately
evaluating the R-peak positioning in a single-lead
ECG signal. For our purposes, we used the R-peak
annotations available from the database but — for an
online scenario — a R-peak detection algorithm has
to be involved in RENEE, such as the Pan-Tompkins
algorithm (Pan and Tompkins, 1985; Sedghamiz,
2014).
The heartbeat selection technique needs the R-
peak positioning information provided by the previ-
ous algorithm in order to properly select the heartbeat.
According to the chosen baseline, a heartbeat signal is
defined as the samples included between two middle
points of three successive R-peaks. In other words, a
heartbeat is not computed as an ECG signal included
between two R-peaks, but as a signal composed of (i)
an individual QRS complex, and (ii) the previous and
successive dynamics.
After all these steps, we compressed the data
through the method proposed by (Picariello et al.,
2021), based on a 1-bit signal quantization.
3.2 Features Creation & Classification
In order to create informative features for the classi-
fication stage, we first defined a windowed accumu-
lation of samples by imposing a fixed window length
winLen: given the qhbs signal, we define a new sig-
nal whbs so that whbs
i
=
∑
i
j=i−winLen
qhbs
j
. In this
way, RENEE is able to represent the dynamics of the
original heartbeat signal in the compressed domain.
The classification component of RENEE—
through the use of machine learning techniques —
is in charge of providing the final classification of
the heartbeat in five types, according to the AAMI
standard (ANSI/AAMI-EC57, 1998): normal beat
(N), ventricular ectopic beat (V), supraventricular
ectopic beat (S), fusion of a normal and a ventricular
ectopic beat (F) and unknown beat type (Q).
The features we use for the automatic classifica-
tion of the heartbeats are the samples of the final sig-
nal we obtained, i.e., whbs. It is worth noting that the
number of samples may vary among different heart-
beats. However, the machine learning model, ap-
pointed for the classification of each heartbeat sig-
nal, needs the data to be aligned in terms of features
across all the instances. Therefore, we needed to se-
lect a maximum number D of whbs samples to use as
features. To do this, we used the same method used
by (Xu et al., 2018), i.e., we apply zero-padding and
truncation in case the heartbeat signal contains less
or more samples as compared to a fixed threshold D,
respectively.
Figure 1 depicts an example of how the signal
changes after each of the main stages of RENEE.
The plot in the upper row shows the original signal
waveform of a sample heartbeat. The second sub-
plot shows the signal after the application of dither-
ing noise. Then, the samples obtained by the 1-bit
quantization procedure are depicted in the third sub-
plot. Finally, the fourth line of plot shows the sig-
nal once applied the windowed accumulation of sam-
ples. Such a signal contains the features used by the
machine learning method for the classification of the
heartbeat.
4 EMPIRICAL STUDY DESIGN
The goal of this study is to evaluate the accuracy of
RENEE in classifying heartbeat types from a highly
compressed version of an ECG. The literature shows
that the uncompressed trace of an ECG allows to ob-
tain a very accurate classification (Xu et al., 2018).
Thus, the study is steered by the following research
question:
Can RENEE provide a heartbeat
classification comparable to state-of-the-art
methods based on uncompressed ECG?
The perspective of the study is both (i) of a researcher
who wants to understand if machine learning tech-
niques are able to classify heartbeat also in the com-
pressed domain, and (ii) of a practitioner who wants
to use a method in a telemedicine application that is
able to balance accuracy and data storage and trans-
mission.
Morphological Classification of Heartbeats in Compressed ECG
389
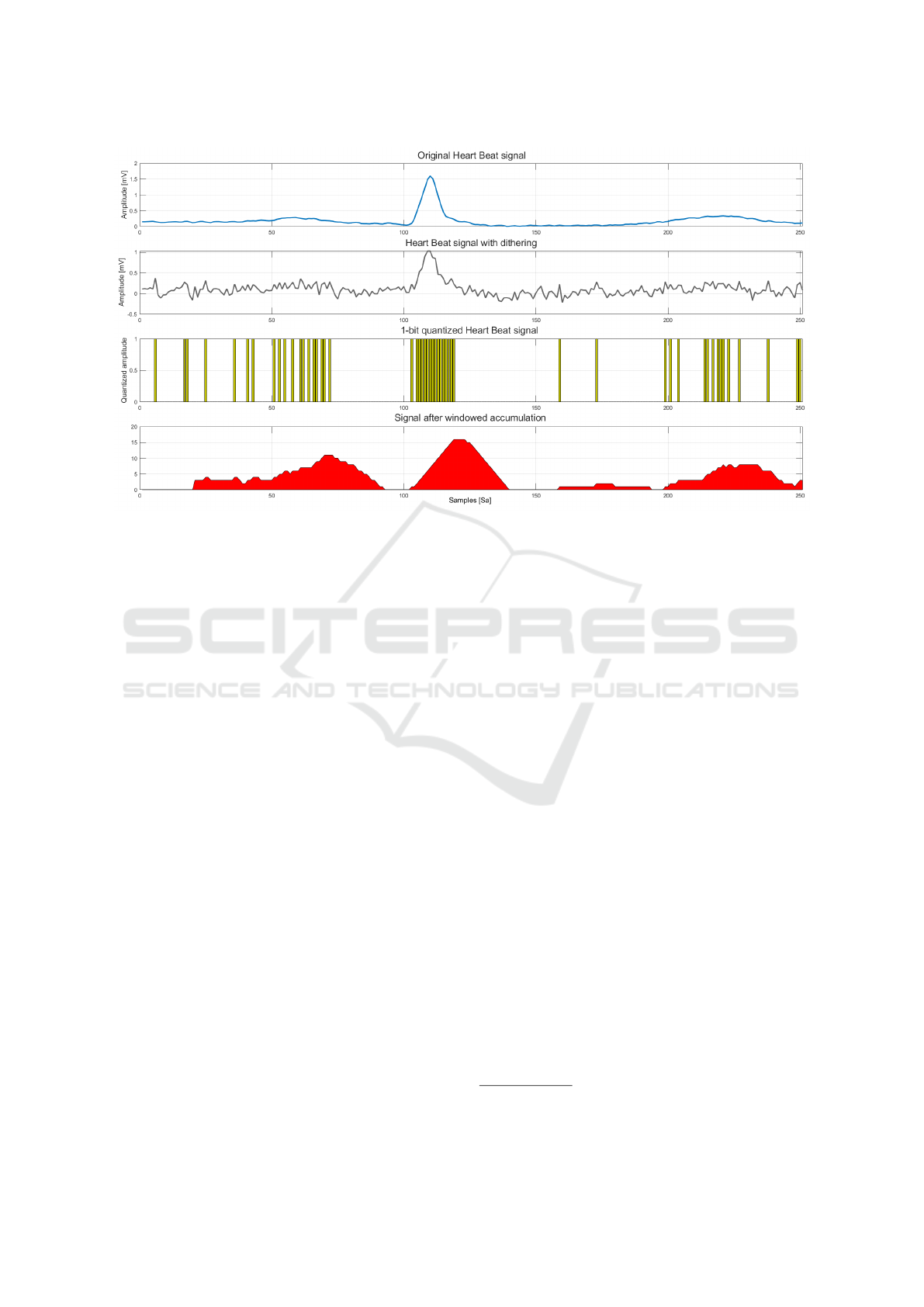
Figure 1: The four main steps performed by RENEE after the pre-processing stage: (1) the original heartbeat signal, (2) the
signal after the pre-processing and noising, (3) the compressed signal through 1-bit quantization and (4) the final elaboration
— applied to the compressed signal — that consists of a windowed accumulation of binary samples.
4.1 Context of the Study
The context of this study is represented by the MIT-
BIH Arrhythmia Database (Moody and Mark, 2001;
Goldberger et al., 2000), a commonly used bench-
mark which contains 48 half-hour two-channel am-
bulatory ECG recordings, obtained from 47 subjects.
These ECG were digitized at 360 Hz with 11-bit res-
olution over a 10 mV range. Approximately 110,000
annotations are included in the database.
Each heartbeat is classified by using 15 different
classes. These 15 types of heartbeat in the MIT-
BIH arrhythmia database have been categorized in
five classes, reported in (ANSI/AAMI-EC57, 1998).
In most of the recordings in the MIT-BIH
database, the first channel is a modified limb lead II
(MLII), and the second one is a modified lead V1. In
our experiments, only the signal from the first channel
was used for ECG classification because, typically,
QRS complexes are usually prominent (Xu et al.,
2018).
Finally, according to the AAMI recommendation
(ANSI/AAMI-EC57, 1998), we removed from the
dataset four recordings containing paced beats. The
final dataset was composed of a total of 44 records.
4.2 Experimental Procedure
We experimented a large set of machine learning
techniques to train the model embedded in RE-
NEE. In order to execute a complete experimenta-
tion, we chose at least one classifier from each cat-
egory of classifiers available from the Weka ma-
chine learning toolkit (Hall et al., 2009), i.e., J48
(Quinlan, 2014), Replication Tree (Devasena, 2014),
Random Forest (Barandiaran, 1998), Logistic regres-
sion (Cramer, 2002), AdaBoost M1 (Freund and
Schapire, 1997), BayesNet
3
, J48 (Cohen, 1995) and a
3-layer long short-term memory (LSTM) Neural Net-
work (NN) (Lang et al., 2019). We chose the Long
Short-Term Memory (LSTM) NN (instead of DNN,
for example) because ECG signals are time series data
and LSTM is capable of learning long-term depen-
dencies (Xu et al., 2018). We have also included
in our study several classifiers implemented in the
Matlab Classification Learner app
4
. Basically, they
are specific implementation of different categories
of classifiers. Examples are the k-nearest neighbors
(KNN)(Dasarathy, 1991) and the Support Vector Ma-
chine (SVM)(Noble, 2006).
The whole dataset, composed by 44 records, was
split into two datasets, i.e., DS1 and DS2: this al-
lows to perform a patient-independent classification
in which each patient appears either in the training or
in the test set, but never in both of them. Each dataset
contains approximately 50,000 beats from 22 record-
ings. We used DS1 as the training set and DS2 as the
3
https://bit.ly/3bYCFcR
4
https://bit.ly/35oMcZ9
HEALTHINF 2021 - 14th International Conference on Health Informatics
390

test set. This is a consolidated procedure from the lit-
erature. Indeed, it was used in many previous works
(De Chazal et al., 2004; Ye et al., 2012; Raj and Ray,
2018; Xu et al., 2018).
As for the parameters of RENEE, we used the con-
figurations reported in Table 1 and determined with a
trial & error approach—on a different data set—than
the one used for the classification experiment.
We compare RENEE with the chosen baseline
work, i.e., a state-of-the-art method designed to auto-
matically classify uncompressed ECG heartbeats with
high accuracy(Xu et al., 2018).
To compare the approaches, we use several met-
rics. First, we use the accuracy, i.e., the number
of correctly classified instances divided by the total
number of instances. Then, we use also some class-
level metric—designed for a given class—among the
ones we consider for our study i.e., N, S, V, F. We
list below the class-level metrics we compute for each
class:
• Precision
c
, i.e., the number of correctly classi-
fied positive instances divided by the total num-
ber of instances classified as positive, computed
as
T P
T P+FP
• Recall
c
, i.e., the number of correctly classified
positive instances divided by the total number of
instances actually positive, computed as
T P
T P+FN
• F-Measure
c
, i.e., the harmonic mean of precision
and recall, computed as
2×Precision
c
×Recall
c
Precision
c
+Recall
c
• AUC
c
, i.e., the definite integral used as a measure
of the two-dimensional area underneath the ROC
curve.
• MCC
c
(Matthews Correlation Coefficient), a very
reliable statistical rate which returns a higher
value the better are the four confusion matrix in-
dicators: true and false positives, true and false
negatives.
We report the comparison for the classes N, S, V, F.
Similarly to other studies (De Chazal et al., 2004; Ye
et al., 2012; Raj and Ray, 2018; Xu et al., 2018), we
exclude the class Q, because such a class contains
paced beats (that were excluded) and unclassifiable
beats (only 15).
Table 1: Configuration parameters of RENEE used in the
experimentation.
Parameter Description Value
σ Power of Gaussian dithering 0.1
γ Threshold for the quantization 0.2
winLen Window size for whbs 20
D Number of features 417
4.3 Threats to Validity
A limitation of this study may be represented by the
validation. Even if our kind of validation takes care
to appropriately separate the data of distinct subjects
for the training and testing phases, a more appropri-
ate validation would have been a typical L1SO-CV
(Leave 1 Subject Out Cross Validation). This impli-
cates that the data related to an individual patient will
be included once in the test data set and n-1 times in
the training data set. However, we decided to adopt
the validation method used in previous study to fa-
cilitate the comparison of the results achieved. The
replication of the study on larger data sets and with
different validation methods is part of the agenda of
our future works.
5 ANALYSIS OF THE RESULTS
We report in Table 2 the accuracy of RENEE ob-
tained by using the 10 top performing classifiers ex-
perimented in our study. The achieved results show
that RENEE allows to keep a comparable overall ac-
curacy in the classification of heartbeats when com-
pared to the baseline, which, as previously mentioned,
uses the uncompressed original ECG signal. Espe-
cially, the Random Forest, the Bagged Trees and the
Medium Gaussian classifiers achieve the highest ac-
curacy (between 0.93 and 0.94).
Table 2: Overall accuracy of RENEE by using the 10 top
performing classifiers experimented in our study. At the
bottom we also report the accuracy achieved by the ap-
proach proposed by (Xu et al., 2018).
Classifier Accuracy
Random Forest 0.940
Bagged Trees 0.937
Medium Gaussian SVM 0.932
Boosted Trees 0.928
LSTM NN 0.928
Fine Gaussian SVM 0.926
Fine Tree 0.925
Quadratic SVM 0.922
JRip 0.919
Cubic SVM 0.912
Baseline (Xu et al., 2018) 0.947
Table 3 reports the detailed results achieved by
RENEE when using the best performing classifier,
i.e., Random Forest. The results reported in Table
3 highlight a clear outcome: RENEE is able to cor-
rectly classify with a high accuracy the classes la-
Morphological Classification of Heartbeats in Compressed ECG
391

beled as N and V. In details, RENEE has correctly
classified respectively 43,978 out of 44,259 and 2,713
out of 3,221 total instances. For what concerns the
class S, RENEE still needs to improve in terms of
classification accuracy. The machine learning model
with the highest classification accuracy specific for
class S is the Quadratic SVM; such model was able
to correctly classifies 128 instances out of approxi-
mately 1,800 total instances. Finally, for the F class,
the results obtained in terms of Precision, Recall and
F-Measure are not satisfying. It is worth noting,
however, that the classification performance on these
classes has a low impact on the overall performance.
Indeed, as suggested by the standard ANSI/AAMI
EC57 (ANSI/AAMI-EC57, 1998), it is recommended
to focus the attention on the two majority arrhythmia
classes, i.e., classes S and V.
Table 3: Detailed classification evaluation of RENEE when
using the best performing classifier, i.e., Random Forest.
Class Precision Recall F-Measure AUC
N 0.945 0.994 0.969 0.966
S 0.375 0.016 0.031 0.565
V 0.907 0.842 0.873 0.987
F 0.074 0.018 0.029 0.812
Table 4 shows the MCC achieved by the two com-
pared approaches for each class. The achieved results
indicate that RENEE achieves similar results (even if
it performs slightly better) compared to the baseline
for the classes N and V. Indeed, for these classes,
the difference in terms of MCC is below 0.05. The
greatest delta has been obtained for the class S, which
touches the amount of 0.6. Thus, further improve-
ments are required for the classification in the com-
pressed domain of this particular class.
Table 4: Comparison between RENEE (Random Forest)
and the approach proposed by (Xu et al., 2018).
Class (Xu et al., 2018) RENEE Delta
N 0.69 0.67 -0.02
S 0.67 0.07 -0.60
V 0.91 0.87 -0.04
F 0.22 0.03 -0.19
6 CONCLUSION
We have presented RENEE, an automatic approach
for (i) compressing an ECG signal through 1-bit quan-
tization and (ii) classifying the heartbeats in com-
pressed ECGs using machine learning techniques. An
empirical evaluation conducted on the MIT-BIH Ar-
rhythmia Database indicates that the overall classifi-
cation accuracy of RENEE is comparable to the accu-
racy of the best state-of-the-art method for the clas-
sification of heartbeats based on uncompressed ECG
signal (0.940 vs. 0.947).
Future work will be devoted to replicate the evalu-
ation of RENEE by using more robust validation, such
as the L1SO-CV to corroborate our findings.
ACKNOWLEDGMENT
This work has been supported by the project PON
2014-2020—ARS01 00860 “ATTICUS: Ambient-
intelligent Tele-monitoring and Telemetry for
Incepting and Catering over hUman Sustainability”
funded by the Ministry of Education, University and
Research—RNA/COR 576347.
REFERENCES
ANSI/AAMI-EC57 (1998). Testing and reporting perfor-
mance results of cardiac rhythm and ST segment mea-
surement algorithms. Standard, Association for the
Advancement of Medical Instrumentation, Arlington,
VA.
Balestrieri, E., Boldi, F., Colavita, A. R., De Vito, L.,
Laudato, G., Oliveto, R., Picariello, F., Rivaldi, S.,
Scalabrino, S., Torchitti, P., and Tudosa, I. (2019).
The architecture of an innovative smart t-shirt based
on the internet of medical things paradigm. In 2019
IEEE International Symposium on Medical Measure-
ments and Applications (MeMeA), pages 1–6.
Balestrieri, E., Daponte, P., De Vito, L., Picariello, F., Ra-
puano, S., and Tudosa, I. (2020). A wi-fi iot proto-
type for ecg monitoring exploiting a novel compressed
sensing method. ACTA IMEKO, 9:38.
Barandiaran, I. (1998). The random subspace method for
constructing decision forests. IEEE Trans. Pattern
Anal. Mach. Intell, 20(8):1–22.
Bera, P., Gupta, R., and Saha, J. (2019). Preserving Abnor-
mal Beat Morphology in Long-term ECG Recording:
An Efficient Hybrid Compression Approach. IEEE
Transactions on Instrumentation and Measurement,
69(5).
Chen, S., Hua, W., Li, Z., Li, J., and Gao, X. (2017). Heart-
beat classification using projected and dynamic fea-
tures of ecg signal. Biomedical Signal Processing and
Control, 31:165–173.
Cohen, W. W. (1995). Fast effective rule induction. In
Twelfth International Conference on Machine Learn-
ing, pages 115–123. Morgan Kaufmann.
Cramer, J. S. (2002). The origins of logistic regression
(technical report). In Tinbergen Institute.
HEALTHINF 2021 - 14th International Conference on Health Informatics
392

Dasarathy, B. V. (1991). Nearest neighbor (nn) norms: Nn
pattern classification techniques. IEEE Computer So-
ciety Tutorial.
David, V., Adochiei, N., Adochiei, F., and Tudosa, I.
(2011a). ECG waves and features extraction using
Wavelet Multi-Resolution Analysis. In 2011 E-Health
and Bioengineering Conference (EHB), pages 1–4.
David, V., Adochiei, N., and Tudosa, I. (2011b). Methods
of electromagnetic interference reduction in electro-
cardiographic signal acquisition. Environmental En-
gineering and Management Journal, 10(4):553–559.
De Chazal, P., O’Dwyer, M., and Reilly, R. B. (2004). Auto-
matic classification of heartbeats using ecg morphol-
ogy and heartbeat interval features. IEEE transactions
on biomedical engineering, 51(7):1196–1206.
Devasena, C. L. (2014). Comparative analysis of random
forest rep tree and j48 classifiers for credit risk predic-
tion. In International Conference on Communication,
Computing and Information Technology (ICCCMIT-
2014).
Freund, Y. and Schapire, R. E. (1997). A decision-theoretic
generalization of on-line learning and an application
to boosting. Journal of computer and system sciences,
55(1):119–139.
Garcia, G., Moreira, G., Menotti, D., and Luz, E. (2017).
Inter-patient ecg heartbeat classification with temporal
vcg optimized by pso. Scientific Reports, 7(1):1–11.
Goldberger, A. L., Amaral, L. A., Glass, L., Hausdorff,
J. M., Ivanov, P. C., Mark, R. G., Mietus, J. E., Moody,
G. B., Peng, C.-K., and Stanley, H. E. (2000). Phys-
iobank, physiotoolkit, and physionet: components of
a new research resource for complex physiologic sig-
nals. circulation, 101(23):e215–e220.
Hall, M., Frank, E., Holmes, G., Pfahringer, B., Reutemann,
P., and Witten, I. H. (2009). The weka data min-
ing software: an update. ACM SIGKDD explorations
newsletter, 11(1):10–18.
Hassanien, A. E., Dey, N., and Borra, S. (2018). Medical
Big Data and Internet of Medical Things: Advances,
Challenges and Applications.
Jha, C. K. and Kolekar, M. H. (2018). Electrocardiogram
data compression using DCT based discrete orthogo-
nal Stockwell transform. Biomedical Signal Process-
ing and Control, 46:174–181.
Kandala, R. N., Dhuli, R., Pławiak, P., Naik, G. R., Moein-
zadeh, H., Gargiulo, G. D., and Gunnam, S. (2019).
Towards real-time heartbeat classification: evalua-
tion of nonlinear morphological features and voting
method. Sensors, 19(23):5079.
Lang, S., Bravo-Marquez, F., Beckham, C., Hall, M.,
and Frank, E. (2019). Wekadeeplearning4j: A deep
learning package for weka based on deeplearning4j.
Knowledge-Based Systems, 178:48 – 50.
Mar, T., Zaunseder, S., Mart
´
ınez, J. P., Llamedo, M., and
Poll, R. (2011). Optimization of ecg classification
by means of feature selection. IEEE transactions on
Biomedical Engineering, 58(8):2168–2177.
Mond
´
ejar-Guerra, V., Novo, J., Rouco, J., Penedo, M. G.,
and Ortega, M. (2019). Heartbeat classification fusing
temporal and morphological information of ecgs via
ensemble of classifiers. Biomedical Signal Processing
and Control, 47:41–48.
Moody, G. B. and Mark, R. G. (2001). The impact of the
mit-bih arrhythmia database. IEEE Engineering in
Medicine and Biology Magazine, 20(3):45–50.
Noble, W. S. (2006). What is a support vector machine?
Nature biotechnology, 24(12):1565–1567.
Pan, J. and Tompkins, W. J. (1985). A real-time qrs de-
tection algorithm. IEEE transactions on biomedical
engineering, (3):230–236.
Picariello, F., Iadarola, G., Balestrieri, E., Tudosa, I., and
De Vito, L. (2021). A novel compressive sampling
method for ecg wearable measurement systems. Mea-
surement, 167:108259.
Pohlmann, K. C. (1995). Principles of digital audio.
McGraw-Hill, Inc.
Quinlan, J. R. (2014). C4. 5: programs for machine learn-
ing. Elsevier.
Raj, S. and Ray, K. C. (2018). Sparse representation of ecg
signals for automated recognition of cardiac arrhyth-
mias. Expert systems with applications, 105:49–64.
Rajesh, K.N.and Dhuli, R. (2018). Classification of imbal-
anced ecg beats using re-sampling techniques and ad-
aboost ensemble classifier. Biomed. Signal Process.
Control, 41:242–254.
Sedghamiz, H. (2014). Matlab implementation of pan
tompkins ecg qrs detector. Code Available at the File
Exchange Site of MathWorks.
Wang, C., Qin, Y., Jin, H., Kim, I., Granados Vergara, J. D.,
Dong, C., Jiang, Y., Zhou, Q., Li, J., He, Z., Zou,
Z., Zheng, L., Wu, X., and Wang, Y. (2019). A low
power cardiovascular healthcare system with cross-
layer optimization from sensing patch to cloud plat-
form. IEEE Transactions on Biomedical Circuits and
Systems, 13(2):314–329.
Xu, S. S., Mak, M.-W., and Cheung, C.-C. (2018). To-
wards end-to-end ecg classification with raw signal
extraction and deep neural networks. IEEE journal of
biomedical and health informatics, 23(4):1574–1584.
Ye, C., Kumar, B. V., and Coimbra, M. T. (2012). Heartbeat
classification using morphological and dynamic fea-
tures of ecg signals. IEEE Transactions on Biomedical
Engineering, 59(10):2930–2941.
Morphological Classification of Heartbeats in Compressed ECG
393
