
Deep-Learning-based Segmentation of Organs-at-Risk in the Head
for MR-assisted Radiation Therapy Planning
László Ruskó
1
, Marta E. Capala
3
, Vanda Czipczer
1
, Bernadett Kolozsvári
1
, Borbála Deák-Karancsi
1
,
Renáta Czabány
2
, Bence Gyalai
2
, Tao Tan
1
, Zoltán Végváry
5
, Emőke Borzasi
5
, Zsófia Együd
5
,
Renáta Kószó
5
, Viktor Paczona
5
, Emese Fodor
5
, Chad Bobb
8
, Cristina Cozzini
7
, Sandeep Kaushik
7
,
Barbara Darázs
2
, Gerda M. Verduijn
3
, Rachel Pearson
6
, Ross Maxwell
6
,
Hazel Mccallum
6
,
Juan A. Hernandez Tamames
4
, Katalin Hideghéty
5
, Steven F. Petit
3
and Florian Wiesinger
7
1
GE Healthcare, Budapest, Hungary
2
GE Healthcare, Szeged, Hungary
3
Erasmus MC Cancer Institute, Department of Radiation Oncology, Rotterdam, The Netherlands
4
Erasmus MC, Department of Radiology and Nuclear Medicine, Rotterdam, The Netherlands
5
University of Szeged, Department of Oncotherapy, Szeged, Hungary
6
Newcastle University, Northern Institute for Cancer Research, Newcastle, U.K.
7
GE Healthcare, Munich, Germany
8
GE Healthcare, Milwaukee, U.S.A.
Keywords: Organ-at-Risk, Head, Radiation Therapy, MRI, Segmentation, Deep Learning, U-Net.
Abstract: Segmentation of organs-at-risk (OAR) in MR images has several clinical applications; including radiation
therapy (RT) planning. This paper presents a deep-learning-based method to segment 15 structures in the head
region. The proposed method first applies 2D U-Net models to each of the three planes (axial, coronal,
sagittal) to roughly segment the structure. Then, the results of the 2D models are combined into a fused
prediction to localize the 3D bounding box of the structure. Finally, a 3D U-Net is applied to the volume of
the bounding box to determine the precise contour of the structure. The model was trained on a public dataset
and evaluated on both public and private datasets that contain T2-weighted MR scans of the head-and-neck
region. For all cases the contour of each structure was defined by operators trained by expert clinical
delineators. The evaluation demonstrated that various structures can be accurately and efficiently localized
and segmented using the presented framework. The contours generated by the proposed method were also
qualitatively evaluated. The majority (92%) of the segmented OARs was rated as clinically useful for radiation
therapy.
1 INTRODUCTION
Head-and-neck cancers are one of the most common
cancers worldwide causing more than 200 000 deaths
per year (Tong et al., 2018). Radiation therapy (RT)
is an important treatment option for head-and-neck
cancer. In a state-of-the-art radiation therapy
treatment plan, radiation dose is shaped precisely to
the tumor. In that way, high energy photon/particle
beams can eradicate cancer cells while sparing as
much healthy tissues as possible. In the treatment
preparatory phase accurate definition of the target
volumes and organs-at-risk is essential. In the current
clinical practice, the planning phase of radiation
therapy is highly dependent on computer tomography
(CT) scans, hence it provides the electron density data
for the dose calculation algorithms. Therefore, it is
common practice that the structure contouring takes
place on the CT scans using information of further,
more sensitive imaging modalities either separately
or in fusion to the planning CT scans. Magnetic
resonance (MR) is becoming more and more
widespread as an additional imaging modality, due to
its high contrast for soft tissues, high spatial
resolution, non-ionizing radiation and non-invasive
nature. These properties render MR imaging superior
to the CT for cancer diagnosis and treatment
planning, as precise detection and localization of the
tumorous growth and surrounding organs-at-risk
Ruskó, L., Capala, M., Czipczer, V., Kolozsvári, B., Deák-Karancsi, B., Czabány, R., Gyalai, B., Tan, T., Végváry, Z., Borzasi, E., Együd, Z., Kószó, R., Paczona, V., Fodor, E., Bobb, C.,
Cozzini, C., Kaushik, S., Darázs, B., Verduijn, G., Pearson, R., Maxwell, R., Mccallum, H., Hernandez Tamames, J., Hideghéty, K., Petit, S. and Wiesinger, F.
Deep-Learning-based Segmentation of Organs-at-Risk in the Head for MR-assisted Radiation Therapy Planning.
DOI: 10.5220/0010235000310043
In Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2021) - Volume 2: BIOIMAGING, pages 31-43
ISBN: 978-989-758-490-9
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
31

(OARs) is crucial. For this reason, MR-only RT
planning solutions are under heavy research, where
the required electron density data is derived from a
pseudo-CT created from the MR scan (Wiesinger et
al., 2018). This workflow would be more convenient
for the patient, as one single MR examination would
deem enough for RT planning, as opposed to current
practice where the patient is scanned on different
imaging devices, often several times. Moreover, the
additional radiation dose from the CT scan could also
be avoided.
Currently, the standard clinical practice often
consists of manual contouring of OARs, which is
performed for various structures slice-by-slice by
experienced clinicians. This process is time
consuming (takes usually several hours per patient),
expensive and introduces inconsistencies due to both
intra- and inter-observer variabilities (Chlebus et al.,
2019). The unmet need for precise and automated
segmentation tools is unquestionable. Deep-learning-
based methods for image segmentation can bridge the
limitations of traditional atlas and machine learning
algorithms which are less suited to generalize for
unseen patient anatomies. Convolutional neural
networks (CNNs) are frequently used in medical
image analysis. In 2018, a model-based segmentation
method was proposed (Orasanu et al., 2018) applying
a CNN-based boundary detector to get better results
compared to the boundary detector using classic
gradient-based features. In the past few years U-Net
architectures (Ronneberger et al., 2015) became the
new state-of-the-art for image segmentation. For
example, a method proposed in (Mlynarski et al.,
2019) uses a 2D U-Net for multi-class segmentation
of 11 head organs and applies a graph-based
algorithm that forces the connectivity between
neighbouring organs. Both, Lei et al. (2020) and Chen
et al. (2019) developed a framework that first
localizes and then segments 8 and 6 head-and-neck
OARs, respectively. The method proposed in (Lei et
al., 2020) utilizes 3D Faster R-CNN to detect the
locations of OARs and uses attention U-Net to
segment them, while the algorithm in (Chen et al.,
2019) uses standard 3D U-Nets (Çiçek et al., 2016) in
a cascade manner in a way that it uses prior
segmentations (e.g. brainstem and eyes) to determine
the bounding box of the next target OAR (e.g. optic
nerves). Our proposed method utilizes a similar
approach, where the target OAR is first roughly
localized and then fine-segmented.
The presented method describes a U-Net deep
neural network architecture to segment various OARs
in the head region, crucial for radiotherapy planning.
As an initial step for head-and-neck OAR
segmentation, we aim to segment a total of 15
relevant structures in this region, including structures
of the optic system (eyeballs, lenses, lacrimal glands,
optical nerves, chiasm), as well as the brain,
brainstem, pituitary gland, cochleas, and patient body
contour. Examples of these structures are shown in
Figure 1.
Figure 1: Manual annotations of organs-at-risk in head
region. Upper row: T2-weighted MR images in axial,
coronal and sagittal directions, respectively. Lower row:
manual annotations of the 15 organs-at-risk.
The difficulties in segmenting organs within the
head region, originates from suboptimal image
resolution and low contrast between neighbouring
tissues. For better segmentation, we developed a two-
stage framework for separately locating the target
OAR with 3 slice-based 2D models and segmenting it
with a 3D model. Additionally, for larger structures
(body, brain), which might not be completely covered
on the scan, only one 2D model is utilized for
segmentation.
The proposed method differs from the algorithm
in (Chen et al., 2019) as it is not a cascaded approach,
so the segmentation of OARs does not depend on the
segmentation of other OARs, similarly to the method
in (Lei et al., 2020). Additionally, different models
are trained independently for each organ unlike in
(Mlynarski et al., 2019). Another main difference
from the previously mentioned state-of-the-art
methods is that the proposed method is applied to
segment more OARs in the head region. Furthermore,
the models’ accuracy is also evaluated on an unseen
dataset with different MR image acquisition settings
from a separate source than the source of the training
dataset.
2 METHODS
The proposed method is based on deep learning
image segmentation. The algorithm starts with a
localization step that involves training 2D U-Net
BIOIMAGING 2021 - 8th International Conference on Bioimaging
32

models on axial, coronal and sagittal planes, fusing
their prediction maps, and using it to determine the
location of each organ by a surrounding 3D bounding
box. After this step, the proper image part with a
safety margin is cropped from the 3D image and a 3D
U-Net model is used to segment this smaller area.
This approach results in significantly less false
positive voxels in the segmentation and allows
considerably faster model training and inferencing.
This section describes the image datasets used in
this study, the model architecture, the details of the
model training, and the applied pre- and post-
processing methods.
2.1 Image Dataset
The image dataset incorporated in this work is a
combination of a publicly available dataset and a
private database. The public dataset is from the RT-
MAC (Radiation Therapy – MRI Auto-Contouring)
challenge hosted by the American Association of
Physicists in Medicine (AAPM), referred to as
AAPM dataset (Cardenas et al., 2019). The AAPM
dataset (available at The Cancer Imaging Archive
(TCIA) website (Clark et al., 2013)) includes 55 T2-
weighted images of the head-and-neck region with 2
mm slice thickness and 0.5 mm pixel spacing,
acquired with Siemens Magnetom Aera 1.5T scanner.
All scans have a matrix size of 512x512x120 points
and a squared 256 mm field of view. In most of the
scans, the top of the head is missing.
The private database (acquired by our clinical
partners) consists of 24 T2-weighted MR images
depicting the head-and-neck area, scanned using
different T2-weighted MR sequences (2D
PROPELLER, 2D FRFSE, 3D CUBE), with slice
thickness between 0.5 and 3 mm. These scans were
acquired on volunteers, using GE scanners
(MR750w, SIGNA (PET/MR, Artist, Architect)).
These scans, unlike the ones in the AAPM dataset, are
not uniform, and differ in MR image parameter
settings (e.g. resolution, pixel spacing). The models
in this study were trained solely on the publicly
available dataset and evaluated on both the public and
private databases.
Manual labelling for the AAPM dataset was done
by medical students under the supervision of a
medical doctor experienced in clinical delineation,
according to the RTOG and DAHANCA guidelines
(Brouwer et al., 2015). The list of structures was
defined together with radiation oncology specialists,
prioritizing those structures that are important for RT
planning of head-and-neck tumors. Irradiating these
structures above dose constraints would cause severe
side effects. For example if the anterior visual
pathways – optic nerves and chiasm – are exposed to
excessive radiation, it may lead to radiation-induced
optic neuropathy, which is defined as a sudden,
painless, irreversible visual loss in one or both eyes
occurring up to years after radiation treatment
(Akagunduz et al., 2017). Excessive radiation
exposure can be avoided by optimizing irradiation
parameters, like dose, beam shape and direction.
As not all scans included every organ (e.g. in
some scans only half of the eyeballs were present),
the number of manual segmentations varies for each
structure. The number of contours per structure can
be found in Table 1, where positive slices refer to the
(axial) slices in every MR image in the AAPM dataset
that contain the structure and negative slices are the
ones that does not contain the structure.
Table 1: Number of contours drawn in the AAPM datasets
and the count of positive and negative (axial) slices per
organ (left side/right side). (g.:gland).
Structure No. of
contours
Pos.
slices
Neg. slices
e
y
e
(
L/R
)
22/22 290/287 2350/2353
lens (L/R) 22/22 113/115 2527/2525
lacrimal
g
.
(
L/R
)
22/22 179/175 2461/2465
optic nerve (L/R) 36/36 158/162 4162/4158
chias
m
35 97 4103
b
rain 31 1244 2476
b
rainste
m
30 975 2625
p
ituitary g. 28 108 3252
cochlea
(
L/R
)
31/31 79/73 3641/3647
b
ody 29 3480 0
The contoured cases were separated into 3 subsets
(training, validation, and testing) using 60:20:20
ratio. The number of the training and validation
samples varied organ by organ, however, the test set
included the same 5 cases for all organs. The train-
validation- and test samples were used to optimize the
model, to select the best model, and to evaluate the
best model, respectively. For each organ the same
train/validation/test separation was used for both 2D
and 3D segmentation models.
From the private dataset (that was only used
during evaluation) 5 out of 24 scans were selected for
quantitative evaluation. The annotations for all 15
structures on these scans were defined by radiation
oncologists. In all of the private cases the
segmentation result was qualitatively evaluated. The
manual contour is referred to as gold standard in the
rest of the paper.
Deep-Learning-based Segmentation of Organs-at-Risk in the Head for MR-assisted Radiation Therapy Planning
33
2.2 Preprocessing
The following preprocessing was applied to the image
dataset before model training and inferencing. First,
the voxel size was normalized to be nearly equal to
1x1x1 mm (using integer factor for up or down-
sampling). Then, the images were cropped or padded
with zero voxels to have 256x256x256 resolution.
Finally, min-max normalization was applied to
intensity values, such that the intensity belonging to
99.9 histogram percentile was used instead of the
global intensity maximum.
Additional preprocessing was applied to the
image in case of body segmentation, which attempts
to eliminate background noise using multilevel
thresholding of Otsu’s method and morphological
operations (including closing, dilation, and removal
of air objects connected to the edge of the image).
2.2.1 Harmonization
The scans in the private database have different
intensity range compared to the AAPM dataset.
Therefore, an image harmonization step was
introduced prior to the data preprocessing, which
changes the intensity of the image to be statistically
similar to the reference image chosen from the
training samples.
For image harmonization, each MR volume 𝐼 was
decomposed into images belonging to different
energy band images:
𝐿
,𝑖1,…,B
using:
𝐿
𝑥
𝐼
𝑥
, 𝐿
𝑥
𝐿
𝑥
∗𝐺
𝑥;𝜎
(1)
where 𝐺 is a Gaussian kernel and 𝜎
is randomly
selected increasing number. Furthermore, let
𝐼
𝑥
𝐿
𝑥
𝐿
𝑥
,
𝑓
𝑜𝑟 i1,…,B1
(2)
𝐼
𝑥
𝐿
𝑥
(3)
The above energy bands are computed for a reference
image (of the AAPM dataset). The aim is to make the
statistics (i.e. mean and standard deviation) of the
energy bands belonging to an input (𝑖𝑛) image similar
to that of the reference (𝑟𝑒𝑓) image:
𝐼
𝐼
𝑚𝑒𝑎𝑛
𝐼
∙
𝑠𝑡𝑑
𝐼
𝑠𝑡𝑑
𝐼
𝑚𝑒𝑎𝑛
𝐼
(4)
The harmonized image is computed by adding the
modified energy band images 𝐼
,𝑖1,…,B.
An example of the input, output and reference
image and their histograms is shown in Figure 2. The
motivation for this preprocessing step is to reduce
unwanted variability introduced by MR parameter
settings and to adjust the images of the private dataset
used for evaluation such as to be similar to the
training examples from the AAPM dataset. The
harmonization was only applied to the private dataset
before the evaluation.
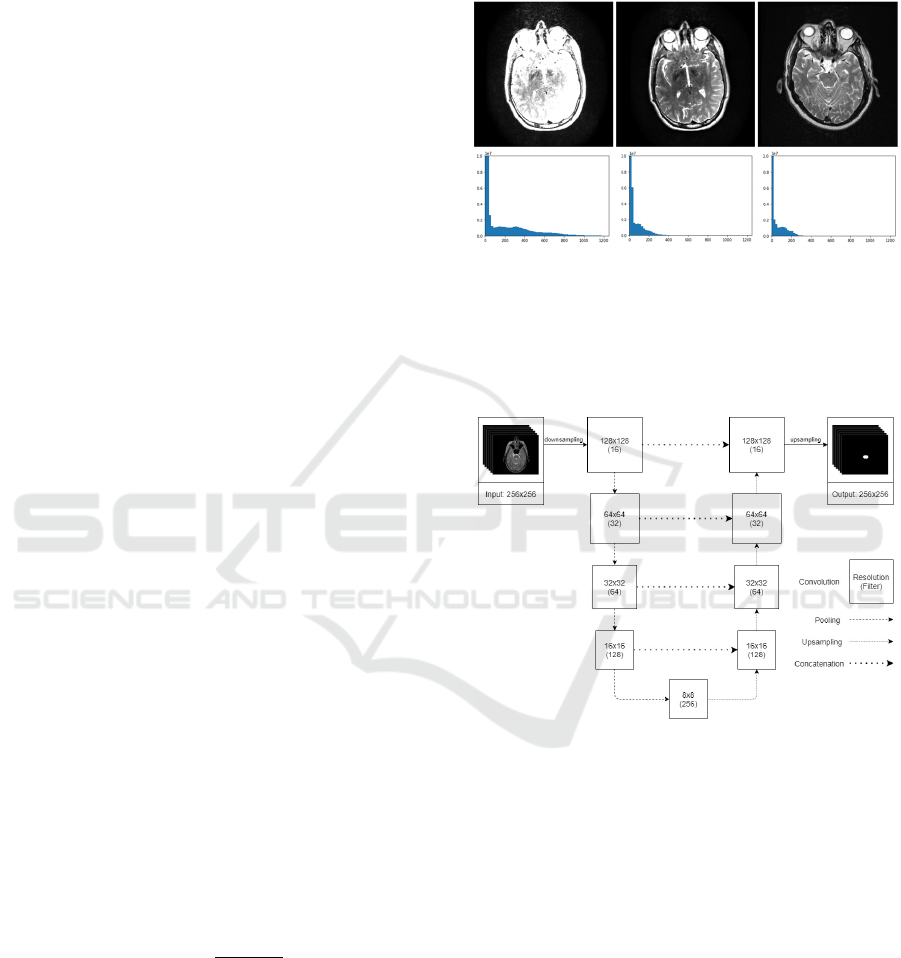
Figure 2: Image harmonization. In the upper row, the input,
the harmonized and the reference image is shown,
respectively. The lower row shows their histograms in the
same order.
2.3 2D Model
Figure 3: Architecture of the 2D segmentation model used
for localization. The axial model is depicted on the figure,
but the same architecture is used for coronal and sagittal
models, as well.
The 2D model’s architecture (shown in Figure 3) is a
state-of-the-art U-Net, which is used to segment
structures on axial, coronal, or sagittal slices
independently (i.e. not using 3D information). The
size of the input is a 256x256 single-channel matrix
representing one slice of the MR image which
resolution is halved to 128x128 with a voxel size of
2x2x2 mm. The output is a 128x128 matrix with
prediction values, where 1 is the highest probability
indicating the presence of the organ, and 0 is the
lowest. The size of the output is increased to 256x256
using upsampling as the last layer.
BIOIMAGING 2021 - 8th International Conference on Bioimaging
34

The model has 4 levels, at each level there are 2
consecutive convolution filters (with 3x3 kernel) with
batch normalization before the ReLU activation
layers. The number of filters is equal to 16 at the input
resolution and doubles after each pooling layer. The
model has 4 pooling layers (with 2x2 pool size), so
the resolution decreases to 8x8 (with 256 filters) at
the “bottom” of the network. Subsequently, the image
is gradually upsampled to the original resolution
using skip connections at each resolution level.
Each 2D model was trained for 75 epochs, except
the brain and body model, where the number of
epochs was set to 30. In each epoch 100% of positive
(including the organ) and the same number of
negative (not including the organ) slices were used.
Due to randomization, most of the negative slices are
used for training. For example, chiasm is detected in
97 positive slices (vide in Table 1), thus only 97 out
of 4103 negative slices are selected randomly in each
epoch. Note that less samples were used during
training than reported in Table 1, since that includes
all (train/validation/test) cases. This approach
accelerates and stabilizes the learning process and
increases the accuracy of the final model. Adam
optimizer was used with 8 batch size. The initial
learning rate was 0.001, and it was halved after every
25 epochs over the training process. During training,
accuracy and loss were calculated based on Dice. At
the end of each epoch the actual model was evaluated,
and the final model was selected based on the
validation loss.
Separate model was trained for each (axial,
coronal, sagittal) orientation. In case of paired organs,
both left and right structures were included in the
positive samples to increase the sagittal model’s
accuracy. For the two largest structures (brain and
body), which were partially covered in the input
images, only the 2D axial model was trained. In these
cases, the input resolution was equal to the original
512x512, the model architecture included 2 more (6
in total) pooling layers, and the model was trained for
30 epochs.
Although the accuracy of neither 2D model is
outstanding (except for the body and brain
segmentation), the combination of the 3 model
outputs is a good basis for the localization of the
organ. After applying each model (slice-by-slice) to a
3D volume, the 3 predictions are combined in the
following way. First, the predictions were binarized.
Then, those voxels were taken, where at least 2 of the
3 models predicted the organ. Finally, the largest
connected component was taken. This combination of
2D models is referred to as fused 2D model in the rest
of the paper.
2.4 3D Model with the Fused 2D
Localization
Figure 4: Architecture of the 3D segmentation model. The
input resolution varies by organ. The size of brainstem's
bounding box was used as an example.
The main advantage of the bounding box localization
for the 3D models is to speed up the training process
and increase the segmentation accuracy. The size of
the bounding box (encompasses the to-be-segmented
organ) was pre-defined and calculated based on the
whole training dataset. The centre of the bounding
box was computed from the gold standard during the
training, and it was computed from the fused 2D
model during the inferencing.
During the training of the 3D model, the bounding
box of the organ is cut from the preprocessed image
and fed into the CNN, thus the input of the network is
considerably smaller than the original resolution
(256x256x256). To account for possible inaccuracies
of the fused 2D model, the centre of the bounding box
was shifted with a random 3D vector before cutting
(using enough safety margin to include all voxels of
the organ) as an augmentation. In contrast to the 2D
model training, the histogram-based intensity
normalization as well as the additional mean/std
normalization was applied only to the bounding box
instead of the whole scan.
The architecture of the 3D model (shown in Figure
4) is created by changing the 2D model’s architecture
to accommodate the 3D input. The 2D layers are
replaced with 3D layers (convolution, pooling,
upsampling). The number of pooling layers is
decreased to 3 (using 2x2x2 pool size). The
convolutional layers use 3x3x3 kernel size. The
number of filters was increased to 24 at the input
resolution (and doubled after each pooling layer).
The 3D model was trained for 100 epochs. In each
epoch all training samples were used. The batch size
was reduced to 4 due to the increased memory needs
of the network. The same (Adam) optimizer and flat
(0.001) learning rate was used with best model
Deep-Learning-based Segmentation of Organs-at-Risk in the Head for MR-assisted Radiation Therapy Planning
35

selection based on validation loss. Training and
validation loss were defined with the Dice metric.
Separate 3D models were trained for each OAR.
During model inferencing the centre of bounding box
was calculated automatically (note that the size is an
organ specific constant) by taking the centre of the
bounding box of nonzero pixels in the result of the
fused 2D model.
2.5 Postprocessing
The prediction of the models was binarized using 0.5
threshold and resized to the original resolution.
Additional postprocessing was applied to body and
brain segmentations that involved filling the holes
and taking the largest connected component.
2.6 Evaluation
For each organ, all models (2D axial, 2D coronal, 2D
sagittal, fused 2D, 3D) were quantitatively evaluated
using the AAPM test set. Furthermore, the 3D models
were (qualitatively and quantitatively) evaluated on
the private cases. The following subsections describe
the evaluation methods.
2.6.1 Quantitative Evaluation
The segmentation results were compared with the
gold standard using Dice, and Surface Dice metrics.
The Dice similarity coefficient is the most
commonly used metric in validating medical image
segmentations by direct comparison between
automatic and manual segmentations. the formula for
calculating Dice is the following:
𝐷𝑖𝑐𝑒2
|
𝑋∩𝑌
|
|
𝑋
|
|
𝑌
|
, (5)
where
|
𝑋
|
and
|
𝑌
|
are the number of voxels in the
automatic and manual segmentations, respectively
and
|
𝑋∩ 𝑌
|
is the number of overlapping voxels
between the two segmentations.
The limitation of this metric is that it weights all
inappropriately segmented voxels equally and
independently of their distance from the surface.
Thus, when comparing two segmentations, assessing
how well the surfaces of the contours are aligned can
provide useful information about the segmentation
accuracy. Surface Dice measures deviations in border
placement by computing the closest distances
between all surface points on one segmentation
relative to the surface points on the other (Wang et
al., 2019). The Surface Dice value represents the
percentage of surface points that lies within a defined
tolerance (as tolerance we used 1 and 2 mm).
To be able to compare the quantitative results to
the state-of-the-art, mean distance (MD) between
manual and automatic segmentation ( 𝑋 and 𝑌,
respectively) is defined as follow:
𝑀𝐷
𝑋,𝑌
1
|
𝑋
|
|
𝑌
|
Inf
∈
𝑑
𝑥,𝑦
Inf
∈
𝑑
𝑦,𝑥
∈∈
(6)
Where 𝑑 is Euclidean distance.
2.6.2 Qualitative Evaluation
The segmentations were qualitatively evaluated on
the private dataset by a radiation oncologist to assess
the usability of the segmented contours for
radiotherapy treatment planning (RTP). Each
segmentation result was classified into one of the
following 4 categories:
1. The contour is missing, or it can be used for
RTP after major corrections that would take
similar time as re-contouring.
2. The contour can be used for RTP after some
corrections that would take less time than re-
contouring.
3. The contour can be used for RTP after minor
corrections that would take significantly less
time than re-contouring.
4. The contour can be used for RTP without any
corrections.
2.7 Implementation Details
The deep learning training and inferencing
frameworks were implemented using Keras 2.3 with
Tensorflow 2.1 backend in Python 3.6 platform. The
2D and 3D models were trained and tested on an HP
Z440 workstation with 32 GB RAM, 12 core, 3.6
GHz CPU and GTX 1080, 8 GB RAM, 2560 CUDA
cores GPU.
3 RESULTS
3.1 Evaluation on AAPM Dataset
Figure 5 demonstrates the best and the worst 3D
results for all anatomy structures from the 5 test cases
of AAPM dataset. In the images the gold standard is
represented with red overlay, while the model
prediction is shown with green outline. All results are
displayed in the axial views.
BIOIMAGING 2021 - 8th International Conference on Bioimaging
36
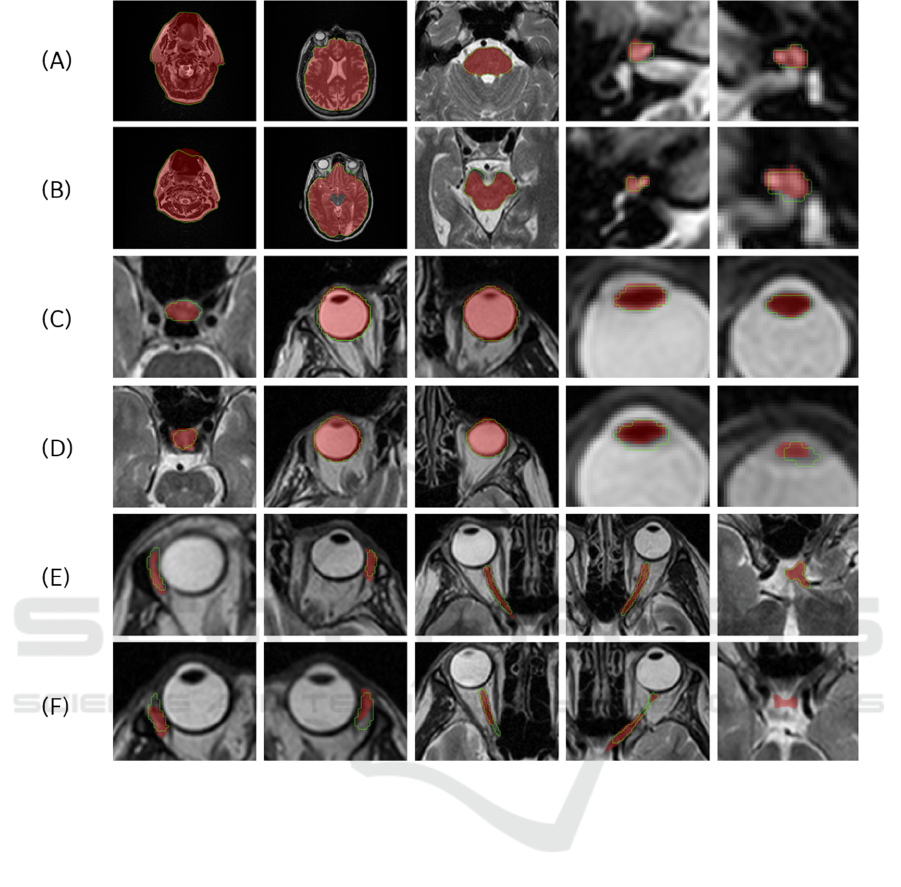
Figure 5: The best ((A), (C) and (E)) and worst ((B), (D) and (F)) segmentation results in the axial direction for the 15 OARs
((A) and (B) shows body, brain, brainstem, right and left cochlea, (c) and (D) shows the pituitary gland, right and left eyeball,
right and left lens, (E) and (F) shows right and left lacrimal gland, right and left optic nerve and chiasm). The predicted result
is depicted in green, and the gold standard is shown as a red overlay.
Table 2
demonstrates the Dice metric reflecting the
overall accuracy of the models and the Surface Dice
metric, which provides the surface accuracy within a
tolerated distance. The paired organs were trained and
tested separately (left and right part) except for the 2D
sagittal model, where both left and right parts were
used since location information is not included in
sagittal images (note that different accuracy of the left
and right sagittal models is due to randomization
during model training). According to the 2D results
the accuracy of the models is similar, and none of
them is significantly better than the other. It is
remarkable that the fused 2D model outperforms most
of the 2D models. The 3D model has an overall
outstanding performance compared to the other
models, except for the chiasm and the cochlea (R)
structures where the 3D model was only the second
best based on Dice accuracy.
3.2 Evaluation on Private Dataset
3.2.1 Qualitative Evaluation on the Private
Database
The proposed method was qualitatively evaluated on
the set of 24 T2-weighted MR images of the private
database for the 15 OAR structures. The results are
summarized in
Table 3
. Our models were able to
provide useful contour (i.e. rating ≥ 2) for 92% of the
segmentation tasks, and only 8% of the contours was
useless (including a few failed segmentations
indicated as bold numbers in the table).
Deep-Learning-based Segmentation of Organs-at-Risk in the Head for MR-assisted Radiation Therapy Planning
37
Table 2: 2D (axial (ax.), coronal (cor.), sagittal (sag.)), Fused 2D (F. 2D) and 3D model accuracy for 5 test cases of the public
dataset. (BS: brainstem, co.: cochlea, lac.:lacrimal gland, ON: optic nerve, pituit.:pituitary gland).
Dice (%) Surface Dice (%) (1 mm) Surface Dice (%) (2 mm)
2D
ax.
2D
cor.
2D
sag.
F. 2D 3D
2D
ax.
2D
cor.
2D
sag.
F.
2D
3D
2D
ax.
2D
cor.
2D
sag.
F.
2D
3D
body 99.4 - - - - 98.1 - - - - 99.4 - - - -
brain 97.8 - - - - 94.5 - - - - 97.5 - - - -
BS 89.9 89.8 88.8 90.9 92.1 90.8 88.5 88.4 91.7 97.0 96.5 95.3 95.0 97.0 97.8
chiasm 63.5 56.9 54.3 62.6 59.2 86.0 81.8 84.9 87.5 91.9 89.3 87.6 89.8 91.9 93.5
co. (L) 62.8 65.7 67.5 72.9 82.3 90.7 85.8 86.1 94.9 97.3 95.0 88.8 89.0 97.3 100.0
co. (R) 61.9 60.7 63.1 76.2 72.1 95.1 74.1 80.4 98.0 99.9 98.9 76.3 82.7 99.9 99.8
eye (L) 92.6 89.1 93.7 93.7 93.9 98.9 92.0 98.0 98.7 99.7 99.7 97.0 99.3 99.7 100.0
eye (R) 92.3 93.7 93.5 94.2 94.6 96.8 98.3 98.0 99.2 99.9 98.5 99.8 99.0 99.9 100.0
lac. (L) 56.4 50.8 43.1 53.8 59.9 77.1 68.1 60.0 70.8 83.8 89.4 83.4 73.6 83.8 91.7
lac. (R) 44.8 49.5 49.1 50.3 57.5 70.5 66.7 64.7 71.0 83.9 84.2 79.9 75.5 83.9 88.7
lens (L) 74.2 75.5 74.5 77.7 79.7 93.6 96.3 98.0 98.3 99.4 97.3 98.1 99.7 99.4 99.9
lens (R) 75.6 73.6 75.0 76.9 81.5 96.1 95.0 97.5 97.3 99.5 99.0 97.6 99.3 99.5 99.7
ON (L) 68.1 61.3 66.0 71.6 73.7 88.4 82.0 87.9 89.8 93.9 93.4 88.1 93.0 93.9 95.8
ON (R) 65.7 63.9 65.5 69.3 71.5 88.7 84.7 87.2 89.1 92.5 93.7 90.0 91.7 92.5 95.7
pituit. 59.4 63.0 67.8 69.2 73.2 75.4 85.0 87.2 87.8 93.7 87.8 91.3 93.0 93.7 94.9
3.2.2 Quantitative Evaluation on the Private
Database
The models were also evaluated quantitatively on 5
annotated MR scans from the private database. The
summarized results of the 3D models are found in
Table 4 and Table 5. Compared to the results on the
AAPM test examples, the accuracy metrics are
somewhat lower. The models were able to provide
contour for all organs on each scan, except for Exam
14, where the proposed method for left lens failed and
segmented the right lens (more details in discussion).
3.3 Training and Segmentation
Efficiency
In this study, the training of a 2D model took 10-20
minutes (per image orientation) except for body and
brain, where it took several hours. The training of the
3D model took ~10 minutes per organ. Note that
during training no online augmentation was applied
to the images in addition to the random shift of the
bounding box. The average segmentation time (using
GPU) including the preprocessing, inferencing of
three 2D models, the computation of the bounding
box, and the inferencing of the 3D model, and the
post-processing took 30 seconds per organ per case.
4 DISCUSSION
Based on the quantitative evaluation metrics in Table
2
and Table 4, the average Dice score of the 3D and
2D axial models (for body and brain segmentation)
indicates that the proposed method was able to
segment the large anatomical structures with higher
than 90% Dice score on the AAPM dataset. However,
Dice metric was generally considerably lower for
BIOIMAGING 2021 - 8th International Conference on Bioimaging
38
Table 3: Qualitative evaluation of the private database by a radiation oncologist (MFS: Magnetic Field Strength, BS:
brainstem, pituit.: pituitary gland, cochl.: cochlea, ON: optic nerve, lacr.:lacrimal gland, PROP: Axial Propeller sequence) –
Bold numbers indicate the cases where the proposed method failed to segment the given organ. The exams that were used for
quantitative evaluation are highlighted in bold.
Sequence
(MFS)
body brain BS pituit.
cochl.
left
cochl.
right
chiasm
ON
left
ON
right
eye
left
eye
right
lacr.
left
lacr.
right
lens
left
lens
right
Mean
Exam 1 PROP (1.5T) 4 3 3 3 3 3 2 3 2 4 4 4 4 4 4 3.3
Exam 2 PROP (1.5T) 4 3 3 2 3 3 2 2 2 4 4 4 3 4 4 3.1
Exam 3 PROP (1.5T) 2 2 3 3 3 4 3 1 1 3 3 3 1 4 4 2.7
Exam 4 PROP (1.5T) 2 3 3 3 3 3 2 1 1 3 3 4 4 1 1 2.5
Exam 5 PROP (1.5T) 4 2 3 2 4 4 1 1 1 4 4 4 4 3 4 3.0
Exam 6 PROP (1.5T) 2 2 3 2 3 3 2 3 2 4 4 3 3 1 4 2.7
Exam 7 CUBE (3T) 2 1 2 1 3 3 2 2 2 3 3 3 3 4 3 2.5
Exam 8 CUBE (1.5T) 3 2 3 3 3 3 2 1 1 4 3 4 4 3 3 2.8
Exam 9
CUBE (3T) 4 2 3 2 3 3 2 2 2 3 3 3 4 3 2 2.7
FRFSE (3T) 3 3 3 2 2 2 3 2 2 3 3 3 3 3 3 2.7
Exam 10
PROP (3T) 4 2 3 2 4 4 2 3 3 3 3 3 1 4 4 3.0
FRFSE (3T) 3 2 4 2 3 3 2 3 3 3 3 3 4 4 3 3.0
Exam 11
CUBE (1.5T) 3 3 2 3 3 3 1 1 1 4 4 3 3 3 3 2.7
PROP (1.5T) 4 2 4 2 3 3 2 2 2 3 3 3 3 3 3 2.8
Exam 12
CUBE (1.5T) 3 2 3 3 2 3 2 2 2 3 4 4 4 3 3 2.9
PROP (1.5T) 4 2 3 3 4 4 2 3 4 3 4 3 4 3 4 3.3
Exam 13
CUBE (3T) 3 2 3 2 3 3 2 2 2 3 4 4 4 3 4 2.9
PROP (3T) 4 2 3 3 2 4 3 2 2 3 3 3 3 3 3 2.9
FRFSE (3T) 3 3 3 3 3 4 2 3 2 3 3 3 4 3 4 3.1
Exam 14
CUBE (1.5T) 2 2 3 2 3 3 2 1 2 3 3 3 3 1 3 2.4
PROP (1.5T) 4 3 4 1 3 3 3 2 3 4 3 3 3 1 4 2.9
Exam 15 CUBE (1.5T) 3 2 3 2 3 3 2 1 1 3 3 1 1 3 3 2.3
Exam 16
PROP (3T) 4 2 3 2 3 3 2 2 2 3 3 3 4 4 4 2.9
FRFSE (3T) 3 1 3 3 3 3 2 2 2 3 3 4 4 3 2 2.7
Mean 3.2 2.2 3.0 2.3 3.0 3.2 2.1 2.0 2.0 3.3 3.3 3.3 3.3 3.0 3.3
small structures (50-90%), where a slight mismatch
(that might be clinically irrelevant in terms of their
effect on radiotherapy treatment) can decrease the
accuracy significantly. The average Surface Dice in
Table 2 shows that at least 90% of segmented
structures’ surface was properly outlined within a
defined tolerance of 2 mm, meaning that only a small
fraction (maximum of 10%) of the surface needed to
be corrected compared to the gold standard surface.
These results indicate that the proposed method can
accurately segment various structures in the head
region.
Based on the qualitative evaluation on the private
dataset in
Table 3, the proposed method failed only on
small organs, such as pituitary gland (1), optic nerves
(3), lacrimal glands (3) and lenses (4). The reason
behind this might be that their 2D models (trained on
low number of positive slices) could not generalize
well. Therefore, there was no overlap between any
two of the 2D results when taking the majority vote.
This resulted in an empty fused 2D result, and as a
consequence, the bounding box for the 3D
inferencing couldn’t be generated. This can be
Deep-Learning-based Segmentation of Organs-at-Risk in the Head for MR-assisted Radiation Therapy Planning
39

Table 4: Dice accuracy for 5 annotated case of private database. (BS: brainstem, pituit.: pituitary gland, cochl.: cochlea, ON:
optic nerve, lacr.:lacrimal gland).
Protocol body brain BS pituit.
cochl.
left
cochl.
right
chiasm
ON
left
ON
right
eye
left
eye
right
lacr.
left
lacr.
right
lens
left
lens
right
Mean
Exam 1 PROP 99.1 90.7 90.5 42.0 77.8 77.3 24.4 71.8 54.4 91.1 90.7 37.6 61.1 79.6 82.6 71.4
Exam 2 PROP 97.5 92.2 89.7 57.4 49.0 57.3 69.1 41.2 42.3 93.1 90.7 37.0 56.9 51.8 49.4 65.0
Exam 13 PROP 98.5 90.4 90.2 61.3 55.3 75.8 59.1 5.3 35.0 86.3 88.8 67.2 68.3 63.0 55.6 66.7
Exam 14 PROP 99.2 89.4 87.2 1.3 54.0 59.2 34.7 15.1 47.4 91.7 90.7 38.4 28.7 0.0 53.4 52.7
Exam 16 PROP 98.2 90.3 90.3 60.8 64.3 79.7 71.7 7.4 47.2 90.1 87.5 47.0 59.4 75.5 48.3 67.8
Mean 98.5 90.6 89.6 44.6 60.1 69.8 51.8 28.2 45.2 90.5 89.7 45.4 54.9 54.0 57.9
Table 5: Surface Dice (2 mm) accuracy for 5 annotated case of private database. (BS: brainstem, pituit.: pituitary gland,
cochl.: cochlea, ON: optic nerve, lacr.:lacrimal gland).
Protocol body brain BS pituit.
cochl.
left
cochl.
right
chiasm
ON
left
ON
right
eye
left
eye
right
lacr.
left
lacr.
right
lens
left
lens
right
Mean
Exam 1 PROP 99.4 81.4 91.4 96.0 95.1 99.6 92.2 95.2 84.9 100.0 99.1 79.5 95.5 98.9 99.2 93.8
Exam 2 PROP 98.6 81.4 96.4 96.5 79.6 86.5 96.1 75.0 91.9 99.9 99.2 76.0 88.5 90.0 87.7 89.6
Exam 13 PROP 98.6 77.5 96.0 92.5 97.4 91.0 81.6 22.6 72.0 99.1 99.5 90.9 88.1 92.1 86.2 85.7
Exam 14 PROP 99.0 75.2 87.9 20.5 97.0 99.1 84.5 49.4 88.7 99.4 98.5 84.0 80.4 0.0 86.9 76.7
Exam 16 PROP 96.7 89.3 96.6 92.0 93.9 98.1 96.3 27.0 82.0 99.3 99.2 73.6 94.1 98.7 94.2 88.7
Mean 98.5 81.0 93.7 79.5 92.6 94.9 90.2 53.8 83.9 99.5 99.1 80.8 89.3 76.0 90.8
later fixed by detecting these small structures in
connection with a larger organ located nearby.
In some cases, the failed results were due to image
properties that differed greatly from the training
dataset. In case of the optic nerve segmentation, in
two exams the low image quality resulted in poor
visibility of optic nerves that would also have
hampered the manual segmentation. An example is
shown in Figure 6.(A). In exam 14, the left lens
segmentation has failed since the structure was hardly
visible in the preprocessed image (shown in Figure
6.(B) lower image). The left lens was barely
observable in the original MR image (shown in
Figure 6.(B) upper image), which might have caused
its disappearance after preprocessing. Figure 7. shows
a case, where the right lens was segmented instead of
the left one. In this case the coronal model predicted
only false positive voxels within the right lens and the
sagittal model correctly segmented the right lens (as
it learns on both, left and right structure), so the 2D
model fusion involved the poorly segmented right
lens voxels, thus the bounding box was cut on the
wrong side. The lacrimal glands are usually
distinguishable from the eye, but in two cases the
lacrimal gland has disappeared into the surrounding
tissue, which led to the unsuccessful segmentation
(shown in Figure 6.(C)).
Figure 6: Examples for failed segmentation. (A) shows the
optic nerve which the model failed to segment. (B) depicts
the (upper) original and (lower) preprocessed MR image,
where the left lens disappears after preprocessing. (C)
represents the lacrimal gland which is barely
distinguishable from the neighbouring tissues.
The qualitative evaluation shows that 92% of the
models’ results achieved 2 or above qualitative score,
which means most of these segmentations were
clinically useful and only 8% of the segmentations
were classified into the first category which implies
that they can’t be utilized for radiotherapy planning.
BIOIMAGING 2021 - 8th International Conference on Bioimaging
40
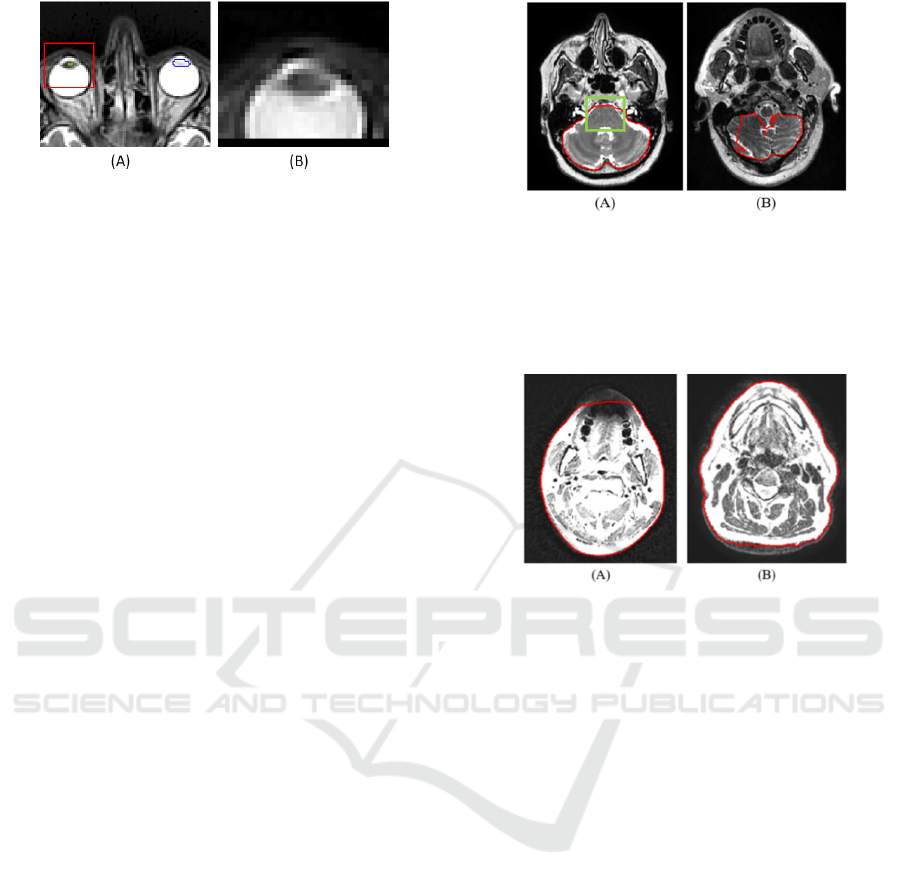
Figure 7: Faulty left lens segmentation. (A) Blue outline is
the gold standard; green outline marks the fused 2D result.
The red rectangle is the bounding box that is cut from the
image. (B) The bounding box of the left lens encompasses
the right lens due to the poor fused 2D result.
It is important to mention that the brain model was
trained on images where the top of the head was
missing (unlike most of the private cases), thus it was
not capable of properly segmenting the brain above
the ventricles. The qualitative evaluation did not,
while the quantitative evaluation did take this area
into account. Therefore, the average Dice score of the
brain model decreased compared to the result on the
AAPM test set significantly (from 97.8 to 90.6). The
2D brain model got an average of 2.2 qualitative
score, which means that most of the time it is still
useful for the radiotherapy planning, since the
correction of the results would require less time than
recontouring the whole organ. In the majority of the
results, that were rated as 2, the brain model failed to
differentiate between brainstem and brain tissues
resulting in over-segmentation. This might be caused
by the lack of spatial information due to only using a
2D model or due to the similar intensity of the upper
slices of brainstem and the white matter (shown in
Figure 8.(A)). Additionally, it was observed that in
certain cases the caudal part of the brain was under-
segmented. An example is presented in Figure 8.(B).
Although the 2D body model achieved relatively
high mean qualitative (3.3 out of 4) and quantitative
(Dice: 98.5) score on the private dataset, the radiation
oncologist noticed some typical faults in the
segmentation results during evaluation. Firstly, due to
some artefacts, the mouth area appeared as blurred in
the original MR image, and thus this part of the body
was under-segmented (represented in Figure 9.(A)).
The second observation was that, occasionally, the
caudal and in few cases, the cranial parts of the
contour were incomplete. An example of the under-
segmentation of the caudal part is depicted in Figure
9.(B).These two might be the results of the additional
preprocessing step for the body segmentation.
However, if we omit this step from the segmentation
process, large over-segmentation could occur around
the body due to background noise.
Figure 8: Segmentation faults of the brain model. Red
contour shows the brain segmentation. (A) depicts that the
brainstem (inside the green bounding box) is hardly
distinguishable from the white matter. (B) represents the
under-segmentation that occurred on the caudal part of the
brain.
Figure 9: Segmentation faults of body model. Red contour
shows the body segmentation. The images depict the under-
segmentation of (A) mouth and (B) caudal part. The image
intensity was changed to highlight the under-segmented
parts that fade into the background.
Out of the results of the 3D models, the pituitary
gland, optic nerves and chiasm got the lowest scores
during both the qualitative and quantitative
evaluation. These organs are the most difficult to
segment as they are small structures, only appearing
in 2-5 slices and their visibility is dependent on MR
image parameter settings. In case of chiasm, if the
image has been acquired with an appropriate angle, it
is visible in 1-3 slices as an X-shaped structure,
slightly darker than its neighbouring tissues.
However, if the angle is different, the chiasm
becomes too fragmented, and hard to distinguish from
the surrounding brain matter. An example of an over-
segmented chiasm is shown in Figure 10.(A). With
regard to the optic nerves, in some cases they were
not clearly defined, and hard to distinguish from the
surrounding tissue. Such small structures are hard to
segment on scans with relatively large slice
thicknesses (3 mm or above), like the ones in the
private dataset. Consequently, this structure is
typically under-segmented (Figure 10.(B)).
Additionally, optic nerves can be hard to segment,
since they might consist of separate disconnected
Deep-Learning-based Segmentation of Organs-at-Risk in the Head for MR-assisted Radiation Therapy Planning
41

components on the image slices, as a result of the
slight curvature of the organ. The pituitary gland is
usually easy to contour, since it is a little orb situated
under the chiasmatic cross. However, the pituitary
gland is often inhomogeneous, thus the results can be
under-segmented. A poorly detected pituitary gland is
depicted in Figure 10.(C).
Figure 10: Examples of segmentation faults of the proposed
method that achieved the lowest Dice scores. Gold standard
is shown as red overlay, the prediction of the 3D model is
marked with green outline (A) depicts an over-segmented
chiasm. (B) shows an example of an under-segmented optic
nerve. (C) The 3D model was unable to localize the
pituitary gland inside the bounding box.
Table 6: Comparison of state-of-the-art Dice (%) and mean
distance (MD) (mm) results. (BS: brainstem, ONC: optic
nerves and chiasm, pitu.: pituitary gland). Under proposed
method, the metrics calculated for the AAPM test set are
shown.
Orasanu
et al.
(2018)
Mlynarski et al.
(2019)
Chen
et al.
(2019)
Proposed
method
MD Dice MD Dice Dice MD
brain
96.8 0.08
97.8 0.78
BS 0.56 88.6 0.26 90.6 92.1 0.62
ONC 0.80 67.4 0.48 75.7 65.9 0.61
eye 0.53 89.6 0.11 94.2 94.3 0.28
lens 0.67 0.63 58.8 80.6 0.22
pitu. 58.0 0.69 73.2 0.43
To the best of our knowledge, there are only three
prior publications on deep-learning-based OAR
segmentation of the head from MR images ( (Orasanu
et al., 2018), (Mlynarski et al., 2019) and (Chen et al.,
2019)) that reported Dice and/or mean distance (MD)
results (Mlynarski et al., 2019). To be able to compare
the results, the paired organs’ and optic nerves’ and
chiasm’s quantitative results were averaged and an
additional metric, the mean distance results were
calculated. Based on Table 6, the proposed method
outperforms the state-of-the art Dice scores for most
structures, while our mean distance results are
comparable to the state-of-the-art. However, the
comparison is difficult due to different datasets.
Orasanu et al. validated their results on 16 T2-
weighted MR images using 5-fold cross-validation,
Mlynarski et al. reported cross-validated results on a
dataset including 44 contrast-enhanced T1-weighted
MR images, Chen et al. used a dataset of 80 T1-
weighted MR images from which 20 was used during
testing, while in this work, a maximum of 26 T2-
weighted image were available for training.
The advantages of the proposed approach over the
standard 2D or 3D UNET solutions are the robust
localization that is based on more 2D models, and the
precise 3D segmentation that is efficient to train and
infer due to the reduced 3D domain. The
disadvantages are the need for maintaining more
models and the limited capability of segmenting
organs which are partially covered in the image.
5 CONCLUSION
In this paper, the segmentation of 15 head structures
was presented using the well-known deep-learning
architectures. Separate 2D models were trained to
segment various structures on axial, coronal, and
sagittal slices of MR images. For the body and brain,
a 2D axial model alone could provide accurate 3D
segmentation. For the other (smaller) organs, the
combination of the 2D models was used for accurate
localization of the organ’s bounding box that was
accurately segmented with a 3D model.
The proposed models were trained on a public
dataset and evaluated on both public and private
image database. Based on the quantitative results, the
presented approach was able to provide precise
segmentation of various structures in the head region
despite the limited size of the training database
(maximum of 26 data from the AAPM dataset) and
different challenges introduced by the private
database (in particular, different MR parameter
settings such as larger slice thickness, no head
fixation as in the AAPM dataset thus more artifact is
present). The qualitative evaluation given by a
radiation oncologist on the set of 24 MR images of
the private dataset demonstrated that the majority
(92%) of the segmentations were found clinically
useful for radiation therapy treatment planning. It also
showed that the proposed method was not sensitive to
different T2 sequences, which indicates its ability to
generalize. The presented approach demonstrates
competitive performance compared to the prior state-
of-the art in terms of Dice scores and mean distance.
In the future, we aim to improve organ models to
segment structures more accurately by increasing the
BIOIMAGING 2021 - 8th International Conference on Bioimaging
42

training dataset and utilizing more augmentations
during training. Furthermore, to decrease the
inferencing time, we intend to develop a multiclass
segmentation method based on the proposed
approach, which can also improve the robustness of
the localization. Finally, the presented approach can
be extended to other organs in the neck region.
ACKNOWLEDGEMENT
This research is part of the Deep MR-only Radiation
Therapy activity (project numbers: 19037, 20648)
that has received funding from EIT Health. EIT
Health is supported by the European Institute of
Innovation and Technology (EIT), a body of the
European Union receives support from the European
Union´s Horizon 2020 Research and innovation
programme.
REFERENCES
Akagunduz, O. O., Yilmaz, S. G., Yalman, D., Yuce, B.,
Biler, E. D., Afrashi, F., and Esassolak, M. (2017).
Evaluation of the Radiation Dose–Volume Effects of
Optic Nerves and Chiasm by Psychophysical,
Electrophysiologic Tests, and Optical Coherence
Tomography in Nasopharyngeal Carcinoma.
Technology in cancer research & treatment 16.6
(2017), 16(6), 969-977.
Brouwer, C. L., Steenbakkers, R. J., Bourhis, J., Budach,
W., Grau, C., Grégoire, V., . . . Langendijk, J. A. (2015).
CT-based delineation of organs at risk in the head and
neck region: DAHANCA, EORTC, GORTEC,
HKNPCSG, NCIC CTG, NCRI, NRG Oncology and
TROG consensus guidelines. Radiotherapy and
Oncology, 117(1), 83-90.
Cardenas, C. E., Mohamed, A. S., Sharp, G., Gooding, M.,
Veeraraghavan, H., and Yang, J. (2019). Data from
AAPM RT-MAC Grand Challenge 2019. The Cancer
Imaging Archive.
doi:https://doi.org/10.7937/tcia.2019.bcfjqfqb
Chen, H., Lu, W., Chen, M., Zhou, L., Timmerman, R., Tu,
D., . . . Gu, X. (2019). A recursive ensemble organ
segmentation (REOS) framework: application in brain
radiotherapy. Physics in Medicine & Biology, 2, 64.
Chlebus, G., Meine, H., Thoduka, S., Abolmaali, N., van
Ginneken, B., Hahn, H. K., and Schenk, A. (2019).
Reducing inter-observer variability and interaction time
of MR liver volumetry by combining automatic CNN-
based liver segmentation and manual corrections. PloS
one, 14(5), e0217228.
Çiçek, Ö., Abdulkadir, A., Lienkamp, S. S., Brox, T., and
Ronneberger, O. (2016). 3D U-Net: learning dense
volumetric segmentation from sparse annotation.
International conference on medical image computing
and computer-assisted intervention, 424-432.
Clark, K., Vendt, B., Smith, K., Freymann, J., Kirby, J.,
Koppel, P., Prior, F. (2013). The Cancer Imaging
Archive (TCIA): Maintaining and Operating a Public
Information Repository, Journal of Digital Imaging.
Journal of digital imaging, 26(6), 1045-1057.
https://doi.org/10.1007/s10278-013-9622-7.
Lei, Y., Zhou, J., Dong, X., Wang, T., Mao, H., McDonald,
M., Yang, X. (2020). Multi-organ segmentation in head
and neck MRI using U-Faster-RCNN. In Medical
Imaging 2020: Image Processing. International Society
for Optics and Photonics.
Mlynarski, P., Delingette, H., Alghamdi, H., Bondiau, P.-
Y., and Ayache, N. (2019). Anatomically Consistent
Segmentation of Organs at Risk in MRI with
Convolutional Neural Networks. arXiv preprint
arXiv:1907.02003.
Orasanu, E., Brosch, T., Glide-Hurst, C., and Renisch, S.
(2018). Organ-at-risk segmentation in brain MRI using
model-based segmentation: benefits of deep learning-
based boundary detectors. International Workshop on
Shape in Medical Imaging, Springer(Cham), 291-299.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-net:
Convolutional networks for biomedical image
segmentation. International Conference on Medical
image computing and computer-assisted intervention.
Tong, N., Gou, S., Yang, S., Ruan, D., and Sheng, K.
(2018). Fully automatic multi-organ segmentation for
head and neck cancerradiotherapy using shape
representation model constrained fullyconvolutional
neural networks. Medical physics, 45(10), 4558-4567.
Wang, Y., Zhao, L., Wang, M., and Song, Z. (2019). Organ
at risk segmentation in head and neck ct images using a
two-stage segmentation framework based on 3D U-Net.
IEEE Access 7
, 144591-144602.
Wiesinger, F., Bylund, M., Yang, J., Kaushik, S.,
Shanbhag, D., Ahn, S., Cozzini, C. (2018). Zero TE-
based pseudo-CT image conversion in the head and its
application in PET/MR attenuation correction and MR-
guided radiation therapy planning. Magnetic
Resonance in Medicine, 80(4), 1440-1451.
Deep-Learning-based Segmentation of Organs-at-Risk in the Head for MR-assisted Radiation Therapy Planning
43
