
Choosing the Appropriate QRS Detector
Justus Eilers
a
, Jonas Chromik
b
and Bert Arnrich
Hasso Plattner Institute, University of Potsdam, Rudolf-Breitscheid-Straße 187, Potsdam, Germany
Keywords:
Electrocardiography, QRS Detection, Heart Rate, Heart Rate Variability, Alarm Fatigue.
Abstract:
QRS detectors are used as the most basic processing tool for ECG signals. Thus, there are many situations and
signals with a wide range of characteristics in which they shall show great performance. Despite the expected
versatility, most of the published QRS detectors are not tested on a diverse dataset. Using 14 databases,
10,000 heartbeats for each different heartbeat type were extracted to show that there are notable performance
differences for the tested eight algorithms. Besides the analysis on heartbeat types, this paper also tests the
noise resilience regarding different noise combinations. Each of the tested QRS detectors showed significant
differences depending on heartbeat type and noise combination. This leads to the conclusion that before
choosing a QRS detector, one should consider its use case and test the detector on data representing it. For
authors of QRS detectors, this means that every algorithm evaluation should employ a dataset that is as diverse
as the one used in this paper to assess the QRS detector’s performance in an objective and unbiased manner.
1 INTRODUCTION
Monitoring the heartbeat of a patient is done in two
levels of detail. The first application is recording the
pace of the heart and the second the regularity of the
heart rate also known as heart rate variability analy-
sis. In the intensive care unit (ICU) the first use case
applies as all physiological parameters of a patient are
constantly monitored. One of these parameters is the
patient’s heart rate. Because the care personnel can-
not calculate the heart rate for every recorded elec-
trocardiogram (ECG), this task is done by an algo-
rithm. Such algorithms, called QRS detectors, need
very high accuracy as errors may lead to false alarms,
that a nurse needs to check out. If too many false
alarms are raised, this causes nurses and doctors not
responding to them anymore (Drew et al., 2014). This
phenomenon is called alarm fatigue and has to be
avoided as good as possible.
If QRS detectors are used to perform a heart rate
variability analysis (HRV) some different metrics are
needed to see if a QRS detector is suitable. Since
HRVs use the time difference between QRS com-
plexes (Cygankiewicz and Zareba, 2013) it is not only
important that a QRS detector finds all heartbeats but
also predicts them with the same offset.
Patients need to have their heart tracked for multi-
a
https://orcid.org/0000-0002-9982-3562
b
https://orcid.org/0000-0002-5709-4381
ple reasons, one reason might be that they suffer from
a heart-related illness. On one hand, cause these ill-
nesses different kinds of heartbeats (MIT-LCP, 2020),
such as bundle branch blocks or premature ventricu-
lar contractions, that have sometimes vastly different
waveforms. Such special heartbeats might not be de-
tected as well as regular, healthy heartbeats thus re-
sulting in false alarms. On the other hand, are not all
patients required to lie down in bed all the time. The
movement of the patient causes noise, which can also
cause false alarms.
This paper shows the performance differences of
QRS detectors based on the ECG signals they are
evaluated on.
The rest of this work is structured as follows: In
section 2 electrocardiography in general and QRS de-
tection in particular are introduced. In section 3 the
related work is summarized. In section 4 it is ex-
plained how the QRS detectors are compared and
which databases are used. In section 5 the results of
our comparison are shown, described, and explained.
Finally, section 6 briefly discusses the implications of
our findings.
50
Eilers, J., Chromik, J. and Arnrich, B.
Choosing the Appropriate QRS Detector.
DOI: 10.5220/0010234600500059
In Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2021) - Volume 4: BIOSIGNALS, pages 50-59
ISBN: 978-989-758-490-9
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

2 BACKGROUND
The heart beats because the sinoatrial node (Open-
Stax, 2013) propagates an electric pulse over the con-
ducting system of the heart. This electric potential can
be measured by placing electrodes on very specific
points on the human body. Based on the electrical
signal, so-called leads can be computed. These leads
may then be used to study the inner workings of the
heart and form the electrocardiogram (ECG). Medical
professionals use the ECG to diagnose illnesses for
example by looking at heartbeats with special shapes
in the ECG.
Moreover, the ECG is also used to calculate the
heart rate of a patient. To do this, algorithms called
QRS detectors locate in the ECG the largest, most
prominent spike called R-peak. The smaller spikes
before and after are called Q- and S-peak, therefore
the name QRS detector (Kohler et al., 2002).
Besides to compute the heart rate, the ECG signal
is also used for a heart rate variability analysis (Ma-
lik and Camm, 1990). Since the basis of this analy-
sis is the distribution of intervals from one R-peak to
the next, the R-peaks have to be found as precisely as
possible (Arzeno et al., 2008).
3 RELATED WORK
For medical professionals to decide for a QRS de-
tector, an evaluation of them is needed. In the ex-
isting research many such comparisons have been
performed (see (
´
Alvarez et al., 2013), (Francesca
et al., 2018), (Xiang et al., 2018), (Phukpattaranont,
2015)) but most of the time they are not compa-
rable with each other. This is caused by differing
dataset, thresholds, and preprocessing methodologies
(Elgendi et al., 2014). Additionally, all the previ-
ously listed evaluations use the MIT-BIH Arrhythmia
Database (Moody and Mark, 2001) as a resource for
the evaluation data. This has the issue that the MIT-
BIH Arrhythmia Database is so widespread, that au-
thors proposing new algorithms can optimize their al-
gorithm just for this database. Thus, the common
principle of having disjoint test and validation data
set is violated.
In (Liu et al., 2018) ten algorithms were evalu-
ated based on six different databases. As mentioned
in the paper, all the algorithms were executed on both
high-quality and low-quality ECG signals. Like the
paper mentions, the algorithms focus on speed and
not on noise resilience, their performance often does
differ a lot between high and low quality. Having
this evaluation over multiple databases, it becomes
clear how much the data quality differs in each of the
tested databases. For example, on the Telehealth ECG
Database, almost all the algorithms perform as bad as
on the ECG database explicitly labelled as poor qual-
ity.
A large listing of algorithms and their quality re-
garding robustness to noise, parameter choice and nu-
merical efficiency has been done by (Elgendi et al.,
2014). Even though the authors did not compute pos-
itive predictive value and sensitivity by themselves,
they list the values of these metrics determined by the
original publishers. Furthermore, the authors men-
tion that the robustness to noise suffers as many pa-
pers only use record from the MIT-BIH Arrhyth-
mia Database. Sometimes not even papers using
the MIT-BIT Arrhythmia Database are comparable
as they exclude certain unfavourable records or seg-
ments. (Arzeno et al., 2008) excludes a period at atrial
flutter in record 204 and (Elgendi et al., 2010) does
not exclude anything. If different databases are used,
this also causes incomparable results. Thus, a stan-
dard database would be needed that contains already
prepared evaluation data. Furthermore, this database
would need to contain such diverse ECG recording
that makes it difficult for authors to optimize their al-
gorithms just for this database as this is a case of over-
fitting.
4 METHODS AND MATERIALS
This section gives a quick overview of the databases
used for the evaluation process presented in this paper.
For the latter one includes the data preparation, used
metrics and algorithms.
4.1 Databases
To not get biased towards a single database, especially
the MIT-BIH Arrhythmia Database, many databases
available on PhysioNet (Goldberger et al., 2000) were
inspected. It follows a list of the databases that got
into closer consideration. The most occurring exclu-
sion criteria where very special recordings for exam-
ple fetal ECG, automated/missing annotations or only
very short, hand-selected recordings. In Figure 1 the
decision process behind the exclusion of databases is
shown. Only databases with manual annotations can
be used as otherwise a QRS detector would be judged
based on the performance of another QRS detector.
The MIT-BIH Arrhythmia Database was excluded as
well in an attempt to reduce bias. Fetal and infant
ECG recordings are performed differently than their
adult counterparts and thus yield different ECG sig-
Choosing the Appropriate QRS Detector
51

ECG databases available
on Physionet (n=41)
Databases with manual annotation
from a medical professional (n=25)
Excluding fetal and
infant databases (n=19)
Excluding heart failure and
endocardial databases (n=17)
Excluding databases designed
for noise stress testing (n=15)
Databases with valid annotation
data format (n=14)
Figure 1: Decision process for including or excluding ECG
databases.
nals. Hence, this work focuses on adult ECG record-
ings. All databases containing heart failure were ex-
cluded as this is a very special scenario. Because this
scenario is so rarely recorded, even in the databases
containing heart failures there is not a representa-
tive number of these instances. Without reaching the
threshold of having a representative amount, all find-
ings are not statistically significant. Also, this work
focuses on externally measured ECG data and thus
the Intracardiac Atrial Fibrillation Database had to be
dropped. Left with 17 databases two more were de-
signed for testing noise resilience. Those contained
recordings with noise but as this paper wants to test
the noise influence it is important to know exactly
which and how much noise is added. Finally, one
database had to be dropped as it contained too many
files with invalid annotations or not processible anno-
tation files.
This leaves the following databases for evaluating
QRS detectors: ANSI/AAMI EC13 Test Waveforms
1
,
MIT-BIH Atrial Fibrillation Database
2
, CiPA ECG
Validation Study (Vicente et al., 2019), ECG Effects
of Dofetilide, Moxifloxacin, Dofetilide+Mexiletine,
Dofetilide+Lidocaine and Moxifloxacin+Diltiazem
(Johannesen et al., 2016), ECG Effects of Ranolazine,
Dofetilide, Verapamil, and Quinidine (Johannesen
et al., 2014), European ST-T Database (Taddei et al.,
1
https://physionet.org/content/aami-ec13/1.0.0/
2
https://physionet.org/content/afdb/1.0.0/
1992), St Petersburg INCART 12-lead Arrhythmia
Database, Long Term ST Database (Jager et al.,
2003), MIT-BIH Normal Sinus Rhythm Database
(Moody and Mark, 2001), MIT-BIH Noise Stress
Test Database (only for the noise recordings) (Moody
et al., 1984), QT Database (Laguna et al., 1997),
MIT-BIH Polysomnographic Database (Ichimaru and
Moody, 1999), MIT-BIH Supraventricular Arrhyth-
mia Database (Greenwald, 1990), MIT-BIH Malig-
nant Ventricular Ectopy Database (Greenwald, 1986)
Especially valuable are the databases with special
focus on certain illnesses or ECG recordings of pa-
tients under the influence of certain drugs. This is
because in the ICU people are often very sick, thus a
diverse set of drugs is used and QRS detectors need
to work regardless of these circumstances.
4.2 QRS Detectors
In this paper, eight algorithms for QRS detection are
evaluated. A brief description of them is given in the
following:
Engelse-Zeelenberg (Engelse and Zeelenberg,
1979) (Zeelenberg and Engelse, 1975): is the
oldest algorithm in the ones presented here with
a publishing date ranging back to 1979, which
is even earlier than the Pan-Tompkins algorithm.
It works with basic waveform comparison and
analysis techniques.
Pan-Tompkins (Pan and Tompkins, 1985): uses
two adaptive thresholds and bandpass filtering
to detect the QRS-complexes. This means that
the algorithm performance on a specific QRS-
complex depends on the previously seen signal,
which influences the adaptive thresholds.
Hamilton (Hamilton and Tompkins, 1986): is very
similar to the Pan-Tompkins algorithm. It also
uses a low followed by a high-pass filter, calcu-
lates the derivative, computes the moving aver-
age, and finds the QRS complexes by peak detec-
tion and applying detection rules. The main dif-
ference between Hamilton and the Pan-Tompkins
algorithm are the two last stages.
GQRS:
3
has not been published yet but is distributed
in the wfdb software package, which is authored
by George B. Moody. Although this algorithm
has been optimized for sensitivity, the software
package comes with a post-processing algorithm,
called gqpost, that should increase the positive
predictions at cost of sensitivity. Nevertheless,
gqpost will not be used in the paper.
XQRS:
4
has not been published in any available pa-
per, just like the GQRS algorithm. But unlike with
BIOSIGNALS 2021 - 14th International Conference on Bio-inspired Systems and Signal Processing
52

GQRS, the exact QRS-detection algorithm is ex-
plained in the documentation. After initialization
and bandpass filtering, moving wave integration
and Ricker wavelet are applied to the signal. The
next step is unique as the algorithm tries to learn
parameters for noise and the QRS amplitudes, the
QRS-detection threshold and recent R-R intervals.
The final output is produced with the previously
processed signal and the learned parameters.
Christov (Christov, 2004): starts to process the sig-
nal by applying multiple moving average filters.
Then adaptive steep-slope thresholding is per-
formed. The initial parameters for that are kept for
the first 5s, where the author expects at least two
QRS complexes to occur. Additionally, an adap-
tive integrating threshold, that should explicitly
remove muscle movement artefacts, is computed.
Also having adaptive beat expectation threshold-
ing shall then compensate for heartbeats with nor-
mal amplitude followed by beats with smaller am-
plitude. It generated the final output called com-
bined adaptive thresholding with is the sum of the
previous thresholding approaches.
Two Moving Average (Elgendi et al., 2010): also
starts with a bandpass filter to remove unwanted
noise, such as power line noise. It follows
the generation of potential blocks containing
QRS-complexes. In this part, the authors assume
a duration of 100 ± 20 ms as the duration of the
QRS complex but also mention, that this is the
average for healthy adults. Thus this algorithm
might struggle for children or adults with severe
illnesses. Finally, the R-peaks are determined
using thresholding based on the statistics of a
healthy adult. This might face the same issues as
the previous step.
Stationary Wavelet Transform (Kalidas and
Tamil, 2017): uses the first ten seconds of the
provided sample as the learning phase. The
signal is then split into three-second segments, on
which the detection is performed. The algorithm
begins with resampling to 80 Hz to reduce noise
and improve the computation speed. After that,
the stationary wavelet transform is computed on
which a squaring and moving window averaging
is performed. The final step is the R-peak detec-
tion, consisting of initialization, threshold-based
peak detection, missed beat detection, threshold
updating, and finally R-peak localization.
4.3 Evaluation Procedure
To see how certain beat types influence the perfor-
mance of algorithms, enough examples of such beats
need to be extracted from the databases. In (Kohler
et al., 2002) the majority of the inspected algorithms
is listed as having more than 99% Sensitivity and Pos-
itive Predictive Value. Because QRS detectors per-
form so well, they need to be evaluated on a suf-
ficiently large dataset. As an adequate size at least
10,000 heartbeats has been chosen. This number al-
lows calculating the classification metrics with five
decimal places. At the same time, this number can
be derived for a real-world consideration. When rec-
ognizing that the normal resting heart rate for an adult
lies at about 60 to 100 bpm (OpenStax, 2013), this re-
sults in 86,000 to 144,000 beats per day. The time
of 24 hours is chosen because it represents the com-
mon time for long-term ECG monitoring for exam-
ple to perform an HRV analysis. Usually, metrics
are recorded with two to four decimal places (such
as 99% or 99.99%). At the example of the Sensitivity
using a resolution of 10
−5
means that the algorithm
misses 8.6 to 14.4 beats per day with every decrease
of the Sensitivity by 0.0001. This is about ten ex-
tra seconds of missed beats. Using only one decimal
place fewer would mean that an algorithm only miss-
ing ten seconds or heartbeats is just as bad as an algo-
rithm missing one and a half minute.
After selecting 10,000 beats at random from the
available databases of each beat types they are ex-
tracted from the original recording. This is done to
reduce the computational effort as it is not needed to
run an algorithm on a half-hour recording if just the
prediction of a couple of beats is of interest. Thus,
each selected beat gets sliced with 10 beats before and
after. This length is chosen as 10 beats is the longest
any of the used algorithms needs to learn parameters.
The heartbeat types that were chosen for this eval-
uation are the ones that occur most often in the used
databases:
N normal beat
S supra-ventricular premature or ectopic beat
V premature ventricular contraction
R right bundle branch block
B unspecified bundle branch block
L left bundle branch block
A atrial premature beat (Goldberger et al., 2000)
To assess the noise resilience of the algorithms,
more or less noise-free ECG recordings need to be
induced with noise. The MIT-BIH Noise Stress Test
Database (Moody et al., 1984) is used as a source for
noise. The noise types contained in this database are
baseline wander, muscle artefacts, and electrode mo-
tion artefacts. Each combination of these three noises
was then added to each slice of the noise-free ECG
Choosing the Appropriate QRS Detector
53
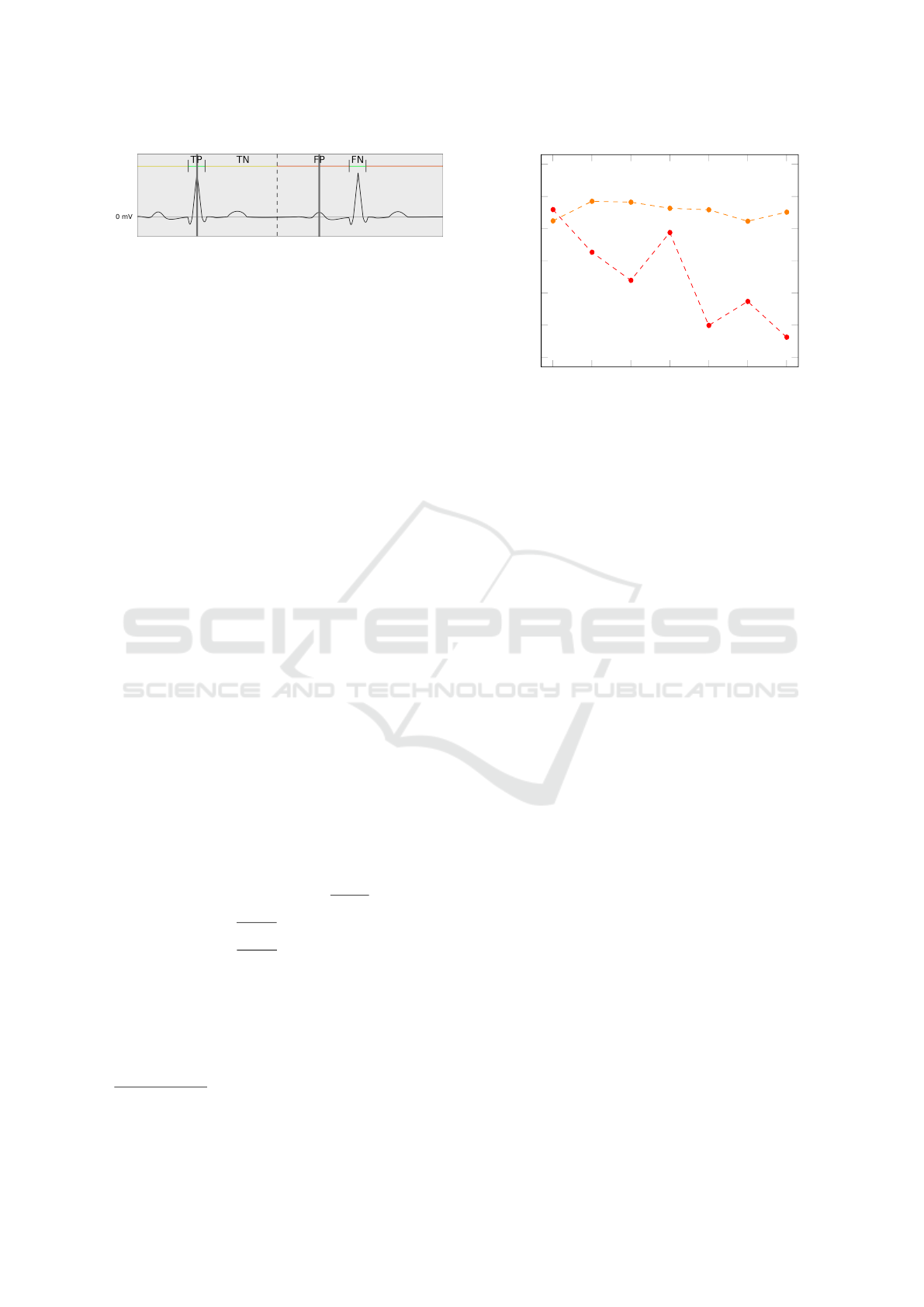
Figure 2: Exactly one example for each: True Positive (TP),
True Negative (TN), False Positive (FP), and False Negative
(FN). The green intervals represent the time frame in which
predictions are accepted as correct. In the yellow and red in-
tervals, no predictions are expected. The grey vertical lines
show predictions. As dashed line you see the mid-point be-
tween two QRS complexes, marking the end and beginning
of an interval that may be a TN or FP.
signal. This creates eight versions for each slice: One
with no noise, three with only one noise type added,
another three with each possible pair, and one with all
three noise types combined.
Just as many of the presented works, this paper
will also use the Positive Predictive Value and Sensi-
tivity as algorithm performance measures. Addition-
ally, the Specificity will be used to show how well
the algorithms are able to recognize periods without
heartbeats, and the Mean Error to show the prediction
offsets.
Detections within the range of 100 ms to the left
and right of each QRS complex are accepted as a valid
prediction. Each QRS complex interval containing a
valid prediction is counted as True Positive. If the
prediction is farther away than 100 ms from the QRS
complex, it is associated with the QRS complex that is
closest and counted as False Positive. For every 200
ms interval around a QRS complex not containing a
prediction, a False Negative is counted. When an in-
terval between two QRS complexes minus their 100
ms thresholds does not contain any prediction, this
counts as a True Negative. Visually are these metrics
explained in Figure 2. The green intervals represent
the threshold of 100 ms seconds around each QRS
complex. Grey vertical lines mark algorithm predic-
tions. Based on these metrics, the following three ag-
gregated metrics can be computed:
Positive Predictive Value (PPV)
T P
T P+FP
Sensitivity (Sens)
T P
T P+FN
Specificity (Spec)
T N
T N+FP
The Mean Error (ME)
5
gets calculated by averag-
ing the time differences between a prediction and the
closest actual QRS complex. This also means that if
no prediction is closest to a QRS complex, this is not
reflected in the metric.
5
here, Error is equivalent to Prediction Offset or Time Dif-
ference, respectively
N
S
V R B L A
0.4
0.5
0.6
0.7
0.8
0.9
1
Beat type
Sensitivity
Figure 3: Shows the problem of evaluating on record-
ings where normal beats (N) occur more often than other
heartbeat types. Orange is Hamilton and in red Engelse-
Zeelenberg, just as in Figure 5.
5 RESULTS AND DISCUSSION
In this section, the influence of different heartbeat
types, as well as noise on the performance of QRS
detectors, is shown. To have a baseline for comparing
the performance to, Figure 4 displays the performance
of algorithms on the MIT-BIH Arrhythmia Database.
5.1 Heartbeat Type Influence
When looking into the heartbeat dependent perfor-
mance for QRS detectors, each of the seven heartbeat
types in Figure 5 has their individual impact on the
algorithms. But before diving into the general trends
for the heartbeat types and the four metrics, looking
at Figure 3 shows that evaluating algorithm on whole
ECG recordings does not encapsulate the true QRS
detector performance.
When whole real-world ECG recordings are used,
the vast majority of the occurring heartbeat types are
normal beats. This means that any computed met-
ric based on these evaluations will have a strong bias
towards normal beats. In the example of Figure 3
this would mean that here Engelse-Zeelenberg (red,
0.85905) would show a higher Sensitivity than Hamil-
ton (orange, 0.82392). Although, when all heartbeat
types are tested Hamilton shows a better average per-
formance with 0.85524 over 0.64961.
Following the principle of this evaluation, all the
other algorithms can be compared for the four metrics
Positive Predictive Value, Sensitivity, Specificity, and
Mean Error. For each of these metrics follows a de-
tailed analysis. The corresponding plots can be seen
in Figure 5.
BIOSIGNALS 2021 - 14th International Conference on Bio-inspired Systems and Signal Processing
54
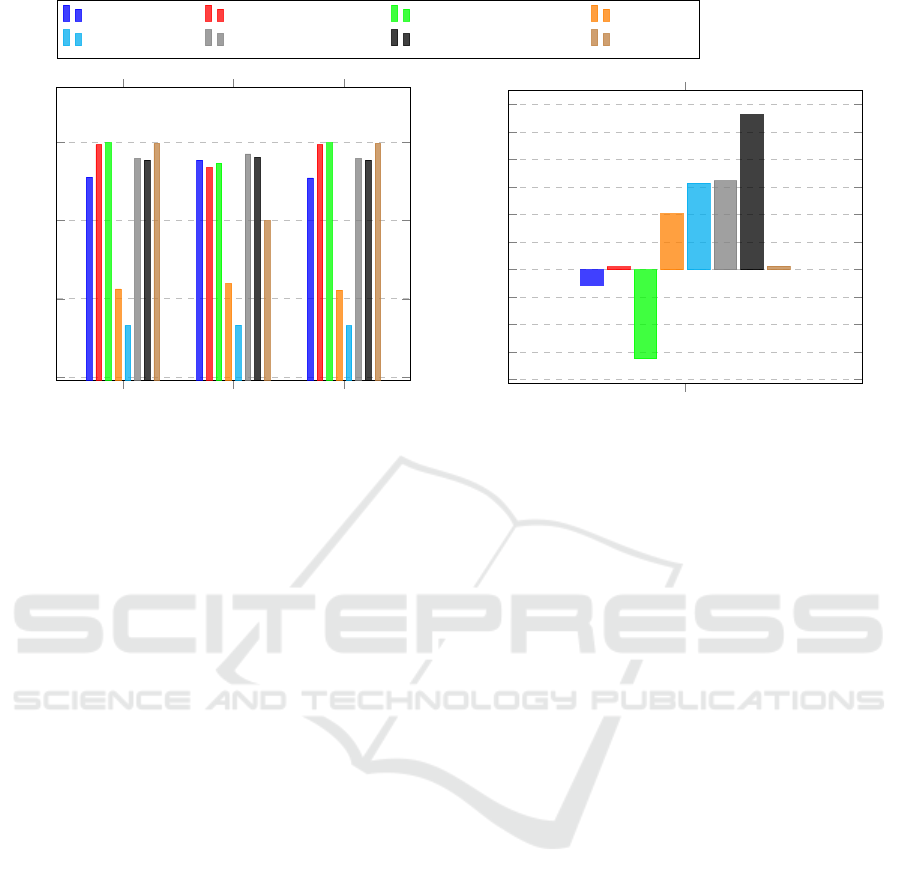
PPV
Sens Spec
0.7
0.8
0.9
1
Christov Engelse-Zeelenberg GQRS Hamilton
Pan-Tompkins Stat. Wavel. Transf. Two Moving Average XQRS
Mean Error in Seconds
−4
−3
−2
−1
0
1
2
3
4
5
6
·10
−2
Figure 4: Algorithm results on the MIT-BIH Arrhythmia Database with our evaluation method.
5.1.1 Positive Predictive Value
As one of the two most used evaluation metrics, the
PPV shows consistent values for each of the heart-
beat types in Figure 5. Ignoring the outliers from Two
Moving Average, the values per type do not deviate
more than 10% to the mean of them. An exception to
this is the values for normal heartbeats. On a per algo-
rithm basis, Stationary Wavelet Transform shows that
even if an algorithm is in the good-performing cluster,
it can still show significant value deviations (compare
type B at 0.66 with S at 0.86).
5.1.2 Sensitivity
As the other commonly used evaluation metric, the
Sensitivity shows a larger fluctuation of the results
than the PPV. Mainly Engelse-Zeelenberg and again
Two Moving Average seem to struggle with many
False Negatives. The other algorithms show consis-
tently good results with again varying performance
for each of the heartbeat types. As for the PPV,
here again, atrial premature beats (A) and left bun-
dle branch blocks (L) are the worst two. The effect of
different heartbeat types is shown in this metric, for
example by Pan-Tompkins (see type B at 0.901 and L
at 0.742).
5.1.3 Specificity
Is not usually used and shows a wider spread of value
for every heartbeat type. Excluding normal beats (N)
that again have a higher deviation than all the other
types, the algorithms differ by about 15% around the
mean. Compared to the PPV the algorithms Speci-
ficity values roughly share their performance i.e. a
high PPV leads to a high Specificity. Despite the con-
stant performance in PPV, Christov shows large dif-
ferences namely in types S at 0.816 and A at 0.622.
5.1.4 Mean Error
As the Mean Error can only be computed if a pre-
diction for a heartbeat was made, this metric is not
significant for judging the False Negatives. This can
be seen in both the figure for Specificity as well as
Sensitivity, where left bundle branch blocks (L) do
not have the worst performances, even though all al-
gorithm struggle most with predicting this type. Fur-
thermore, the Mean Error shows how different the al-
gorithm predicts for each heartbeat type. Even though
normal beats showed one of the largest metric devi-
ations for each metric, this figure shows that it has
the most accurate predictions overall. Only right bun-
dle branch blocks (R) and unspecified bundle branch
blocks (B) have similarly accurate predictions.
5.2 Noise Resilience
For evaluating the impact of different noise types and
their combinations, in Figure 6 all possible combi-
nations of electrode movement, muscle artefacts, and
baseline wander are examined. Just like for the heart-
beat type-specific analysis, a more detailed evaluation
of the results follows.
Choosing the Appropriate QRS Detector
55
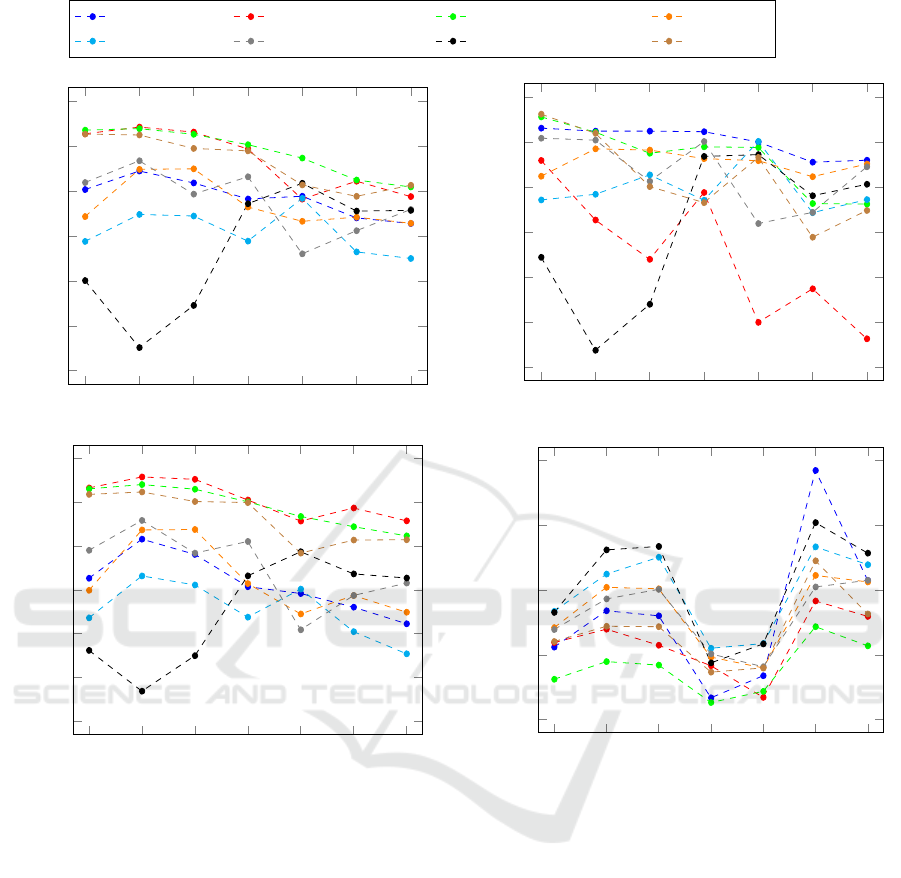
N
S
V R B L A
0.4
0.5
0.6
0.7
0.8
0.9
1
Beat type
PPV
Christov Engelse-Zeelenberg GQRS Hamilton
Pan-Tompkins Stat. Wavel. Transf. Two Moving Average XQRS
N
S
V R B L A
0.4
0.5
0.6
0.7
0.8
0.9
1
Beat type
Sensitivity
N
S
V R B L A
0.4
0.5
0.6
0.7
0.8
0.9
1
Beat type
Specificity
N
S
V R B L A
−0.05
0
0.05
0.1
0.15
Beat type
Mean Error in Seconds
Figure 5: The algorithm results for Positive Predictive Value (PPV), Sensitivity, Specificity, and Mean Error show largely
different results for each beat type.
5.2.1 Positive Prediction Value
Figure 6 shows clearly that electrode movement re-
sults in noise that is harder for algorithms to distin-
guish from the clean ECG data than other types of
noise. The combinations with electrode movement
noise (em-noise) do not only show on average the
lowest values but also a higher spread among the algo-
rithms. For the non-em-noise combinations, the PPV-
values range from 0.6746 to 1.0 and for the em-noises
from 0.5076 to 0.9126.
Although all algorithms decreased in PPV from
the non-em noises to the em-noises, large differences
can be observed on a per algorithm basis. While the
Pan-Tompkins algorithm decreased from 0.7 (none)
to 0.6 (em), Christov dropped from almost 1 to 0.65.
5.2.2 Sensitivity
Generally, the same results can be observed as for
the PPV. The electrode movement noise combinations
show a worse Sensitivity than the other noise com-
binations. However, the decrease is not as large as
for the PPV. Comparing the em-noise to the no-noise
variant of the data, the biggest drop in Sensitivity
happens for the Engelse-Zeelenberg algorithm from
0.9517 to 0.7590. Nevertheless, there are algorithms
like Hamilton or Pan-Tompkins that even show an im-
provement.
Overall the Hamilton algorithm is very interest-
ing as it shows its best performance for the combi-
nation of muscle artefacts with baseline wander at
0.883. This is only slightly over the pure muscle arte-
fact variant which is at 0.877. The worst performance
BIOSIGNALS 2021 - 14th International Conference on Bio-inspired Systems and Signal Processing
56
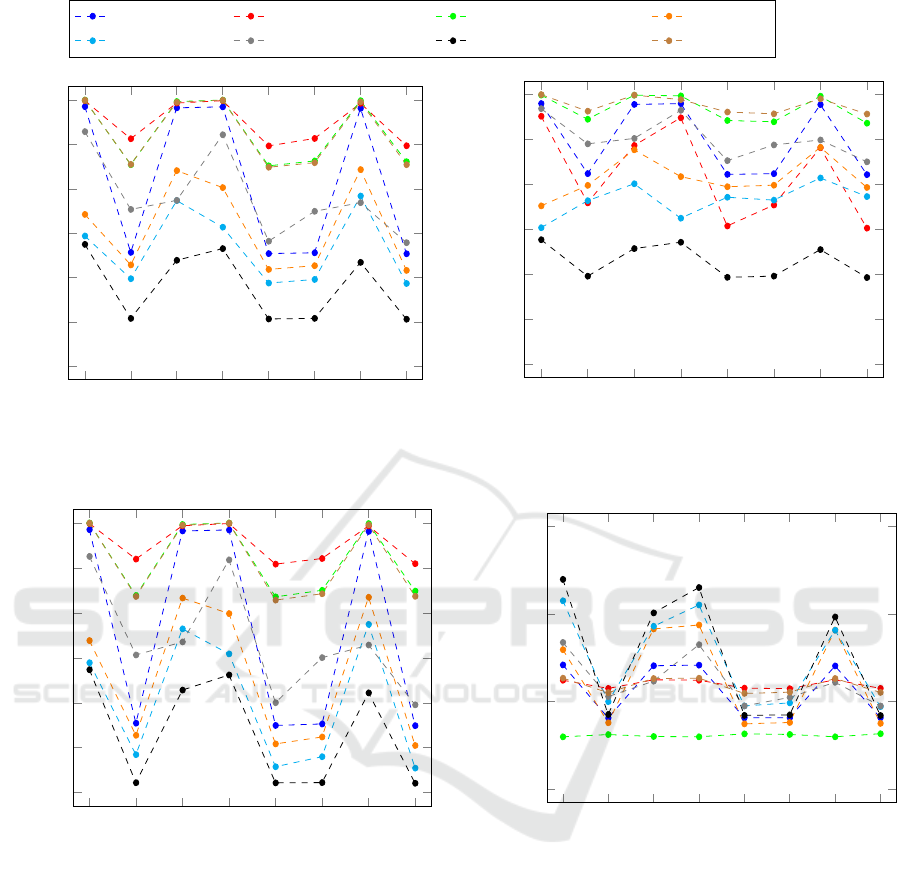
none
em
ma
bw
em-ma
em-bw
ma-bw
em-ma-bw
0.4
0.5
0.6
0.7
0.8
0.9
1
Noise combination
PPV
Christov Engelse-Zeelenberg GQRS Hamilton
Pan-Tompkins Stat. Wavel. Transf. Two Moving Average XQRS
none
em
ma
bw
em-ma
em-bw
ma-bw
em-ma-bw
0.4
0.5
0.6
0.7
0.8
0.9
1
Noise combination
Sensitivity
none
em
ma
bw
em-ma
em-bw
ma-bw
em-ma-bw
0.4
0.5
0.6
0.7
0.8
0.9
1
Noise combination
Specificity
none
em
ma
bw
em-ma
em-bw
ma-bw
em-ma-bw
−0.05
0
0.05
0.1
Noise combination
Mean Error in Seconds
Figure 6: Large algorithm specific differences can be observed. Even each individual algorithm shows a wide range of results
depending on the noise combination from muscle artefacts (ma), electrode movement (em), and baseline wander (bw).
for Hamilton is actually for no noise all with a Sen-
sitivity of 0.752. The same pattern of Sensitivity in-
creasing in the presence of noise can also be observed
with the Pan-Tompkins algorithm. This is caused by
noise spikes that are detected as QRS complexes and
are close to actual QRS complexes so that these de-
tections are recognised as true positives. The drop for
Positive Predictive Value also supports this explana-
tion as it would mean that generally False Positives
are increasing and more heartbeats are detected.
5.2.3 Specificity
The trend of electrode movement noise showing the
worse results is also confirmed by the Specificity.
All algorithms show almost identical results for all
the em-noise combinations. While XQRS, Engelse-
Zeelenberg, Christov, and GQRS give similar re-
sults for all the other noise combinations, Stationary
Wavelet Transform and Hamilton are not consistent at
all.
From all the classification metrics, the Speci-
Choosing the Appropriate QRS Detector
57

ficity shows the largest deviation of values. For pure
em-noise, the values range from 0.91946 (Engelse-
Zeelenbeerg) to 0.42204 (Two Moving Average).
5.2.4 Mean Error
At first sight, it seems counter-intuitive that for the
recordings with no noise the Mean Error shows the
largest value deviation. However, the Mean Error can
only be computed if a prediction for a QRS com-
plex has been made. If an algorithm has not pre-
dicted anything, this will not count into the Mean
Error. The consequence of this is, that for very dif-
ficult noise combinations, here the electrode move-
ment noise, only very obvious heartbeats get detected.
Such obvious or easy to detect heartbeats are also the
ones, that all algorithms can accurately locate.
When comparing the Mean Error per noise type
to the Mean Error per heartbeat type from Figure 5 it
shows that the predictions are on average more accu-
rate.
6 CONCLUSION
It has been shown that QRS detector evaluations
should be executed with more care and on diverse
datasets. Especially the large gap between the al-
gorithm performance on the MIT-BIH Arrhythmia
Database and a more diverse dataset justifies the ques-
tion if some algorithms were optimized specifically
for this database and were not tested for rare beat
types or noise resilience. Furthermore, the results
showed that different heartbeat types and noise com-
binations have two different effects on the algorithms.
While different heartbeat types cause algorithms to
make more inaccurate predictions, noise has the ef-
fect of obfuscating heartbeats such that they are not
found at all.
The evaluated algorithms have a wide range of
noise resilience as well as the ability to deal with other
than normal heartbeat types. Especially for electrode
movement noise and almost all heartbeat types the
PPV, Sensitivity, and Specificity values that most of
the time were not even close to 99% in harsh con-
trast to what the authors state. Algorithms with 99%
Sensitivity would miss 864 heartbeats per day and all
of the evaluated ones showed worse performances. If
the wrong algorithms are used in clinical practice, it
will cause an unnecessary amount of false alarms. Be-
cause these false alarms result in alarm fatigue and in
the end, might result in a patient not receiving needed
help, it is important to allow medical practitioners to
choose the right QRS detector for each use case.
PPV
Sens
Spec
1-10*abs(ME)
Figure 7: Deciding for an algorithm based on the perfor-
mance on premature ventricular contractions (V).
For that to be possible, the algorithm authors need
to test their algorithms on a dataset that is as diverse as
possible and contains equal amounts of heartbeats for
each type and is evaluated with different noise com-
binations.
For medical practitioners there needs to be an easy
way of understanding which algorithm performs best
in the use case at hand. In a first attempt to visual-
ize how such a decision aid may look like Figure 7
shows the eight algorithms of this paper for the four
metrics Positive Predictive Value (PPV), Sensitivity
(Sens), Specificity (Spec) and Mean Error (ME). The
Mean Error had to be transformed to keep that the
more area a curve spans the better the algorithm per-
formance.
REFERENCES
Arzeno, N. M., Deng, Z.-D., and Poon, C.-S. (2008). Anal-
ysis of First-Derivative Based QRS Detection Algo-
rithms. IEEE transactions on bio-medical engineer-
ing, 55(2):478–484.
Christov, I. I. (2004). Real time electrocardiogram QRS de-
tection using combined adaptive threshold. BioMedi-
cal Engineering OnLine, 3:28. Publisher: Citeseer.
Cygankiewicz, I. and Zareba, W. (2013). Chapter 31 - Heart
rate variability. In Buijs, R. M. and Swaab, D. F., edi-
tors, Handbook of Clinical Neurology, volume 117 of
Autonomic Nervous System, pages 379–393. Elsevier.
Drew, B. J., Harris, P., Z
`
egre-Hemsey, J. K., Mammone,
T., Schindler, D., Salas-Boni, R., Bai, Y., Tinoco, A.,
Ding, Q., and Hu, X. (2014). Insights into the Prob-
lem of Alarm Fatigue with Physiologic Monitor De-
vices: A Comprehensive Observational Study of Con-
secutive Intensive Care Unit Patients. PLoS ONE,
9(10):e110274.
Elgendi, M., Eskofier, B., Dokos, S., and Abbott, D.
(2014). Revisiting QRS Detection Methodologies for
BIOSIGNALS 2021 - 14th International Conference on Bio-inspired Systems and Signal Processing
58

Portable, Wearable, Battery-Operated, and Wireless
ECG Systems. PLoS ONE, 9(1):e84018.
Elgendi, M., Jonkman, M., and De Boer, F. (2010). Fre-
quency Bands Effects on QRS Detection. BIOSIG-
NALS, 2003:2002.
Engelse, W. A. H. and Zeelenberg, C. (1979). A single scan
algorithm for QRS-detection and feature extraction.
Computers in cardiology, 6(1979):37–42. Publisher:
IEEE Computer Society Press.
Francesca, S., Carlo, C. G., Nunzio, L. D., Rocco, F., and
Marco, R. (2018). Comparison of Low-Complexity
Algorithms for Real-Time QRS Detection using Stan-
dard ECG Database. Inter. Journal on Advanced Sci-
ence Engineering Information Technology, 8(2).
Goldberger, A. L., Amaral, L. A., Glass, L., Hausdorff,
J. M., Ivanov, P. C., Mark, R. G., Mietus, J. E., Moody,
G. B., Peng, C.-K., and Stanley, H. E. (2000). Phys-
ioBank, PhysioToolkit, and PhysioNet: components
of a new research resource for complex physiologic
signals. circulation, 101(23):e215–e220. Publisher:
Am Heart Assoc.
Greenwald, S. D. (1986). The development and analysis
of a ventricular fibrillation detector. Thesis, Mas-
sachusetts Institute of Technology. Accepted: 2015-
01-20T17:51:30Z.
Greenwald, S. D. (1990). Improved detection and classi-
fication of arrhythmias in noise-corrupted electrocar-
diograms using contextual information. Thesis, Mas-
sachusetts Institute of Technology. Accepted: 2005-
10-07T20:45:22Z.
Hamilton, P. S. and Tompkins, W. J. (1986). Quantitative in-
vestigation of QRS detection rules using the MIT/BIH
arrhythmia database. IEEE transactions on biomedi-
cal engineering, BME-33(12):1157–1165. Publisher:
IEEE.
Ichimaru, Y. and Moody, G. B. (1999). Development of the
polysomnographic database on CD-ROM. Psychiatry
and Clinical Neurosciences, 53(2):175–177. eprint:
https://onlinelibrary.wiley.com/doi/pdf/10.1046/j.144
0-1819.1999.00527.x.
Jager, F., Taddei, A., Moody, G. B., Emdin, M., Antoli
ˇ
c, G.,
Dorn, R., Smrdel, A., Marchesi, C., and Mark, R. G.
(2003). Long-term ST database: A reference for the
development and evaluation of automated ischaemia
detectors and for the study of the dynamics of myocar-
dial ischaemia. Medical & Biological Engineering &
Computing, 41(2):172–182.
Johannesen, L., Vicente, J., Mason, J. W., Erato, C.,
Sanabria, C., Waite-Labott, K., Hong, M., Lin, J.,
Guo, P., Mutlib, A., Wang, J., Crumb, W. J., Bli-
nova, K., Chan, D., Stohlman, J., Florian, J., Ugander,
M., Stockbridge, N., and Strauss, D. G. (2016). Late
sodium current block for drug-induced long QT syn-
drome: Results from a prospective clinical trial. Clin-
ical Pharmacology and Therapeutics, 99(2):214–223.
Johannesen, L., Vicente, J., Mason, J. W., Sanabria,
C., Waite-Labott, K., Hong, M., Guo, P., Lin, J.,
Sørensen, J. S., Galeotti, L., Florian, J., Ugander, M.,
Stockbridge, N., and Strauss, D. G. (2014). Differen-
tiating drug-induced multichannel block on the elec-
trocardiogram: randomized study of dofetilide, quini-
dine, ranolazine, and verapamil. Clinical Pharmacol-
ogy and Therapeutics, 96(5):549–558.
Kalidas, V. and Tamil, L. S. (2017). Real-time QRS de-
tector using Stationary Wavelet Transform for Auto-
mated ECG Analysis. In 2017 IEEE 17th Inter. Conf.
on Bioinformatics and Bioengineering (BIBE). IEEE.
Kohler, B.-U., Hennig, C., and Orglmeister, R. (2002). The
principles of software QRS detection. IEEE Engineer-
ing in Medicine and Biology Magazine, 21(1):42–57.
Laguna, P., Mark23, R. G., Goldberg, A., and Moody23,
G. B. (1997). A Database for Evaluation of Algo-
rithms for Measurement ofQT and Other Waveform
Intervals in the ECG. Computers in cardiology.
Liu, F., Liu, C., Jiang, X., Zhang, Z., Zhang, Y., Li, J., and
Wei, S. (2018). Performance Analysis of Ten Com-
mon QRS Detectors on Different ECG Application
Cases. Journal of Healthcare Engineering,2018:1–8.
´
Alvarez, R. A., Pen
´
ın, A. J. M., and Sobrino, X. A. V.
(2013). A comparison of three QRS detection algo-
rithms over a public database. Procedia Technology,
9:1159–1165. Publisher: Elsevier.
Malik, M. and Camm, A. J. (1990). Heart rate variability.
Clinical Cardiology, 13(8):570–576.
MIT-LCP (2020). PhysioBank Annotations.
Moody, G. and Mark, R. (2001). The impact of the MIT-
BIH Arrhythmia Database. IEEE Engineering in
Medicine and Biology Magazine, 20(3):45–50.
Moody, G. B., Muldrow, W., and Mark, R. G. (1984). A
noise stress test for arrhythmia detectors. Computers
in cardiology, 11(3):381–384.
OpenStax, C. (2013). Anatomy & Physiology. OpenStax
College.
Pan, J. and Tompkins, W. J. (1985). A Real-Time QRS De-
tection Algorithm. IEEE Transactions on Biomedical
Engineering, 32(3).
Phukpattaranont, P. (2015). Comparisons of wavelet func-
tions in QRS signal to noise ratio enhancement and
detection accuracy. arXiv:1504.03834 [cs]. arXiv:
1504.03834.
Taddei, A., Distante, G., Emdin, M., Pisani, P., Moody,
G. B., Zeelenberg, C., and Marchesi, C. (1992). The
European ST-T database: standard for evaluating sys-
tems for the analysis of ST-T changes in ambula-
tory electrocardiography. European Heart Journal,
13(9):1164–1172.
Vicente, J., Zusterzeel, R., Johannesen, L., Ochoa-Jimenez,
R., Mason, J. W., Sanabria, C., Kemp, S., Sager, P. T.,
Patel, V., Matta, M. K., Liu, J., Florian, J., Garnett, C.,
Stockbridge, N., and Strauss, D. G. (2019). Assess-
ment of Multi-Ion Channel Block in a Phase I Ran-
domized Study Design: Results of the CiPA Phase I
ECG Biomarker Validation Study. Clinical Pharma-
cology & Therapeutics, 105(4):943–953.
Xiang, Y., Lin, Z., and Meng, J. (2018). Automatic
QRS complex detection using two-level convolutional
neural network. BioMedical Engineering OnLine,
17(1):13.
Zeelenberg, C. and Engelse, W. (1975). On-Line Analysis
of Exercise Electrocardiograms.Computers in Cardi-
ology, 8(7).
Choosing the Appropriate QRS Detector
59
