
What do You Mean, Doctor? A Knowledge-based Approach for Word
Sense Disambiguation of Medical Terminology
Erick Velazquez Godinez, Zolt
´
an Szl
´
avik, Edeline Contempr
´
e and Robert-Jan Sips
myTomorrows, Anthony Fokkerweg 61 1059CP, Amsterdam, The Netherlands
Keywords:
Medical Word Sense Disambiguation, Knowledge-based, Semantic Similarity, Word Embeddings, Data
Understanding.
Abstract:
Word Sense Disambiguation (WSD) is an essential step for any NLP system; it can improve the performance
of a more complex task, like information extraction, named entity linking, among others. Consequently,
any error, while disambiguating a term, spreads to later stages with a snowball effect. Knowledge-based
strategies for WSD offer the advantage of wider coverage of medical terminology than supervised algorithms.
In this research, we present a knowledge-based approach for word sense disambiguation that can use different
semantic similarity measures to determine the correct sense of a term in a given context. Our experiments show
that when our approach used WordNet-based similarity measures, it achieved a very close performance when
using the semantic measures based on word embeddings. We also constructed a small dataset from real-world
data, where the feedback received from the annotators made us distinguish between true ambiguous terms
and vague terms. This distinction needs to be considered for future research for WSD algorithms and dataset
construction. Finally, we analyzed a state-of-the-art dataset with linguistic variables that helped to explain our
approach’s performance. Our analysis revealed that texts containing a high score of lexical richness and a high
ratio of nouns and adjectives lead to better WSD performance.
1 INTRODUCTION
One of the challenges that a BioNLP system still faces
is to decide the correct sense of the ambiguous med-
ical term. E.g., cold can have at least two meanings,
one to refer to the absence of heat and a second that
refers to the common cold. The task of determining
the sense of a given word, in its context, is called
Word Sense Disambiguation (WSD) (Navigli, 2009).
WSD in medical language faces different chal-
lenges than in layperson language, which stems
from the frequent use of specialized terminology,
acronyms, and abbreviations. Although these chal-
lenges have been addressed before (Zhang et al.,
2019; Antunes and Matos, 2017a), no propositions
have incorporated knowledge-type data to keep the
understandability of the system’s output. We are in-
terested in a solution with good performance that is
also reasonably transparent to interpretation. We be-
lieve this can be achieved if we limit the use of word-
embeddings for specific sub-steps within the WSD
pipeline.
Compared to supervised algorithms, knowledge-
based strategies cover a wider range of terminol-
ogy (Navigli, 2009) for WSD; this is an advantage for
situations of real-world scenarios. Knowledge-based
strategies can rely on similarity measures that exploit
the concept network of lexicons. The basic idea is to
compute the semantic similarity between the context
of the target word and its definitions. With this simi-
larity value, the system will determine which sense to
select (Navigli, 2009).
Our contributions are:
• proposing a new knowledge-based WSD ap-
proach that uses a semantic similarity measure
based on the concept of information coverage. We
compare term definitions and segments of text in
which target terms appear;
• creating a small dataset
1
from a real-world use
case, in addition to evaluating our approach on a
standard dataset (i.e., the MeSH corpus (Jimeno-
Yepes et al., 2011));
• analysing the (re)source data and results by utilis-
ing various linguistic features commonly used in
corpus linguistics (e.g., token type ratio (TTR)),
1
See https://research.mytomorrows.com/datasets
Godinez, E., Szlávik, Z., Contempré, E. and Sips, R.
What do You Mean, Doctor? A Knowledge-based Approach for Word Sense Disambiguation of Medical Terminology.
DOI: 10.5220/0010180502730280
In Proceedings of the 14th International Joint Conference on Biomedical Engineer ing Systems and Technologies (BIOSTEC 2021) - Volume 5: HEALTHINF, pages 273-280
ISBN: 978-989-758-490-9
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
273

which provides a profile of the characteristics of
texts leading to accurate disambiguation;
• distinguishing vague vs. ambiguous terms that
may offer new opportunities to improve WSD al-
gorithms.
2 BACKGROUND
While WordNet (Miller, 1995) is a knowledge-
based resource used to assist WSD in layperson lan-
guage (Navigli, 2009), the Unified Medical Language
System, UMLS, plays a similar role in the medical
domain. UMLS integrates taxonomies and ontolo-
gies of the medical domain (Bodenreider, 2004). Re-
sources like WordNet and UMLS encode semantic re-
lationships (synonymy, hypernymy, hyponymy, etc.)
that give a graph-like structure. Several semantic
measures exploit these semantic relations and graph
structure to compute semantic similarity among con-
cepts (Jiang and Conrath, 1997; Lin et al., 1998; Lesk,
1986).
However, semantic measures have a drawback as
well; they depend on how complete the knowledge
source is. As alternatives, word embeddings are able
to capture relational meanings, which make them suit-
able to compute semantic similarity between words.
Since word embeddings are vector space representa-
tions, the cosine similarity measure is commonly used
to express how similar two word-embedding are (Ju-
rafsky and Martin, 2020, ch. 6). Word embeddings
are created from unlabeled data (Jurafsky and Martin,
2020, ch.6), this makes it possible to have a greater
vocabulary and reduce human intervention. Particu-
larly, definitions of concepts in UMLS have an essen-
tial role for WSD and word embeddings construction
for the medical domain. Pesaranghader et al. (2019)
used UMLS definitions to create word embeddings
before initializing a neural network for a supervised
WSD.
Several authors have worked on WSD for the
medical domain (Zhang et al., 2019; Pesaranghader
et al., 2019; Wang et al., 2018). While these studies
have made significant contributions to the WSD task,
there is a need to evaluate such systems with real-
world scenarios and human experts, which we also
address in this paper.
For our research, we focused on previous works
that used knowledge-based approaches to address
WSD in the medical domain. For instance, Jimeno-
Yepes and Aronson (2010) compared three methods
on the NLM WSD data set (Weeber et al., 2001). The
first method, presented in (McInnes, 2008), is very
similar to the Lesk algorithm (Lesk, 1986); it com-
pares the overlaps of the ambiguous terms to the rep-
resentations made out of the definitions of the can-
didates’ senses. The second one is an adaptation of
the PageRank algorithm. Presented by Agirre and
Soroa (2009), this adapted version treats UMLS as
a directed graph where the PageRank value is com-
puted after the ambiguous terms and their contexts
are integrated into the graph. The third algorithm is
the Journal Descriptor Indexing (JDI), originally pre-
sented by Humphrey et al. (2006). It is based on sta-
tistical associations between ambiguous concepts and
their semantic types that are mapped to a set of jour-
nal descriptions. In their comparison, Jimeno-Yepes
and Aronson (2010) found that the JDI algorithm per-
forms the best among the three methods compared.
However, these methods rely entirely on UMLS, and
they do not integrate any other source, e.g. WordNet
or word-embbedings, that could improve their perfor-
mance.
In more recent research, Antunes and Matos
(2017b,a) presented a knowledge-based approach that
they applied to resolve ambiguities in the MeSH cor-
pus (Jimeno-Yepes et al., 2011). Antunes and Matos
used the cosine similarity measure to assess the se-
mantic similarity of two terms and the pairwise mu-
tual information value of these terms. For the sim-
ilarity computation, the two terms were represented
by word embeddings. For the pairwise mutual in-
formation calculation, they used the MEDLINE Co-
Occurrences (MRCOC) Files
2
. The final score was
then used to determine the sense of the ambiguous
term. The sense that had the highest score was se-
lected. In the same way, we select the sense with the
higher score for the ambiguous term.
As mentioned in the previous paragraphs, the text
similarity is used to compare senses and their con-
texts to select the right sense. It is important to notice
that both elements (definitions and context) are fun-
damentally different texts. Until now, this difference
has not been considered in WSD. We believe the dif-
ference between definitions and context text needs to
be addressed.
With this regard, Velazquez et al. (2016) posed the
problem of comparison of two segments of texts as
a coverage information task on students’ texts. Ve-
lazquez et al. (2016) compared two segments of text,
R and S, to determine to which extent S covers the in-
formation of R. The two segments of text hold a dif-
ferent role, the referent R and the subject of compari-
son S, as Tversky (1977) stated in his model of com-
parison. R is the object holding the most prominent
features, and S is the object with less salient features.
Doing an extrapolation of this definition, Velazquez
2
See https://ii.nlm.nih.gov/MRCOC.shtml
HEALTHINF 2021 - 14th International Conference on Health Informatics
274

et al. see syllabus documents as the referent R, which
contains essential concepts that students should dis-
cuss in the final dissertation. The final dissertation
is considered as S, since it contains a discussion and
paraphrases of the concepts in R.
In our case, we could define R as the set of def-
initions of an ambiguous term. Each of them is
considered to hold prominent features/words that can
help disambiguate the meaning of an ambiguous term.
Then we could see S as the segment of text where an
ambiguous term appears. Its features/words could dif-
fer from the actual definition of the ambiguous terms
since they reflect the use and context words of the am-
biguous terms. Thus, these contextual words share
some semantic information.
3 METHODOLOGY
3.1 Methods
We tackle the problem of word sense disambiguation
with a strategy based on the principle of coverage of
information (Velazquez et al., 2016). To disambiguate
an ambiguous term, we compute the coverage be-
tween the definitions and the segments of texts where
the ambiguous term appears. The definition with the
highest coverage value is considered the final sense.
The coverage of the information is computed using
the following formula:
coverage(R,S) =
∑
w∈{R}
maxSim(w, S) ∗ id f (w)
∑
w∈{R}
id f (w)
(1)
Where R is the referent, and S is the subject of com-
parison; both are segments of texts. However, the
referent R is a definition of ambiguous terms, and S
is the segment of text where the ambiguous term ap-
pears. The maxSim is a function where w is the word
that belongs to R, and it is being compared with each
word in S using a semantic similarity measure. The
function, then selects the word w from R that has the
greater similarity value with the words in S.
As baseline, we used the First Sense Baseline
(FSB); it is solely based on the frequency of occur-
rence of a given sense. The frequency corresponds to
the senses of ambiguous terms that have been manu-
ally annotated in a corpus. In a real-life application,
this approach tends to lead to the long tail getting for-
gotten, which in the medical domain may lead to fur-
ther isolation of people with rare diseases.
3.2 Data-sets and Data Preparation
3.2.1 Data for Evaluation
We used the MeSH WSD corpus (Jimeno-Yepes et al.,
2011), consisting of 203 ambiguous terms, where 106
terms are abbreviations, 88 terms are word-terms, and
nine terms that can be a combination of both. For
each term, there are 100 instances per sense obtained
from MEDLINE. The ambiguous terms come from
the medical subset headings (MeSH) of UMLS.
In addition to this dataset, we manually an-
notated the sense of three ambiguous terms from
UMLS, i.e., ACS, albumin, and basal cell carci-
noma, in 129 clinical trials that we collected from
https://clinicaltrials.gov. We conducted an annotation
task using a group of five experts with a medical back-
ground. For the ACS term, we collected 36 clini-
cal trials. The ACS term contains six definitions or
senses. The term albumine contains only two defini-
tions, and we retrieved 47 clinical trials. Finally, the
term basal cell carcinoma has three senses, and we
collected 47 clinical trials. This dataset is available at
https://research.mytomorrows.com/datasets.
For the annotation process, we presented a docu-
ment with the definitions of the ambiguous terms and
the clinical trials that contained the ambiguous terms.
We asked the annotators to select, from a list of defini-
tions, the sense that corresponds to the actual clinical
trial context.
Regarding the definitions of the ambiguous terms,
we first extracted all of them. Since UMLS incor-
porates multiple data sources, there may be duplicate
concepts – and consequently, definitions – present for
the same term. With medical experts’ help, we dedu-
plicated concept definitions that were in the scope of
our experiments. This is a starting point of a project
that aims to incorporate more ambiguous text and
enrich the MeSH corpus. We computed the inter-
annotator agreement for the annotations, resulting in
a value of 0.484, which indicates a moderate agree-
ment (Pustejovsky and Stubbs, 2012).
3.2.2 Data Sources
First, for the semantic similarity, we used two
different word embedding representations, a)
from (Pyysalo et al., 2013), that was trained on
biomedical data and are publicly available
3
, and
b) word embeddings corresponds to the model
en core sci lg in sci-spacy (Neumann et al., 2019).
3
http://bio.nlplab.org.
What do You Mean, Doctor? A Knowledge-based Approach for Word Sense Disambiguation of Medical Terminology
275

3.3 Data Analysis
The purpose of this analysis is twofold: on the first
hand, it helps us with understanding the nature of in-
put data, i.e., definitions and context texts. On the
other hand, it could give insights into our method’s
performance. For that reason, we decided to use sev-
eral linguistic features that are commonly used to de-
scribe the variation of texts in corpus linguistics stud-
ies, see (Biber, 2006, p. 221). The selected linguistic
features are meant to explain how informative texts
are, their vocabulary concentration, and vocabulary
distribution. Each variable can give a different dimen-
sion of the characteristics of the text. Thus, we used
the token type ratio (TTR) to measure the lexical di-
versity of texts in a corpus; its value goes from 0 to
1; one means that the vocabulary is varied It has been
used to assess the difference between written and oral
language (Biber, 2006). We also used the number of
tokens per document to see the impact of the size of
texts. Besides, we evaluated the distribution of nouns,
verbs, adjectives, and adverbs using a normalized fre-
quency per 100 token-words. For instance, adverbs
and adjectives seem to expand and elaborate on the
information presented in the text (Biber, 2006). A
high concentration of nouns may indicate a high in-
formational focus on the text (Biber, 2006). We com-
pute these linguistic features for the text of the defini-
tions and the text instances of the MeSH corpus. We
will refer to each kind of text as definitions and MeSH
texts, respectively.
Finally, we built two multivariate models to an-
alyze the correlation between these features and our
approach’s performance.
• The vocabulary variation model verifies the rela-
tionship between the number of tokens, the lexi-
cal diversity (TTR), and the accuracy of our ap-
proach.
• The informativeness model that verifies the rela-
tionship of the number of nouns, verbs, adverbs,
and adjectives and the accuracy of our approach.
We took the accuracy as the independent variable
in our model, since it is the standard measure used to
discuss results in NLP.
The models were built using the Ordinary Least
Squares (OLS) method in the statsmodels package of
python.
4 RESULTS AND DISCUSSION
4.1 WSD Results
Table 1 shows the results for the MeSH dataset in
terms of accuracy, precision, recall and F1-measure.
Since the dataset contains different terms, the results
correspond to the weighted average values. In the re-
sults, we can see that all configurations outperformed
the baseline. Regarding the WordNet-based seman-
tic measures, the best performance is for the JCN’s
measure with 70.01 of F1 score, representing a differ-
ence of 36.38 with the baseline. Then, we have 61.38
of F1-measure value for Res’ measure and 59.58 for
Lin’s measure. Previous research reported lower per-
formance of these measures. According to Navigli
(2009), the performance of the WordNet-based mea-
sures for WSD tasks in layperson language is 29.5 for
Res’ measure, 39.0 for JCN’s measure, and 33.1 for
Lin’s measure. From our experiment, our proposi-
tion seems to enhance the performance of knowledge-
based measures. In order to confirm this claim, we
need to conduct more experiments with layperson
language dataset. However, our results show that
WordNet-based measures perform satisfactory even
for medical language.
Regarding the word embeddings’ performance,
we see that the use of id f did not presented an im-
provement. We remark a slight decline in the perfor-
mance of 0.77 but we did not find this to be of statis-
tical significance. However, this difference of perfor-
mance is probably because word embeddings are ini-
tially trained on a frequency-based matrix (Jurafsky
and Martin, 2020, ch. 6), thus any lexical information
and words distribution is already captured. Originally,
Velazquez et al. (2016)’s work (see formula 1 mixes a
WordNet semantic measure with the lexical informa-
tion of the term, id f to calculate the similarity value.
Thus, when we adapted Velazquez et al. (2016)’s for-
mula to use a word-embedding similarity measure, we
expected that the id f may not be necessary. We then
run experiments with both options to see what the im-
pact is in practice.
Considering our case study data, the results are
slightly different. The baseline performed the best
with a 77.89 of F1-score. It is followed by Res’
measure with 76.59 and the three word-embedding
strategies, with 72.65 for Embeddings-IDF, 74.01 for
Embeddings-no idf, and 73.6 for Embeddings-spacy.
In the bottom rank, we find Lin’s measure with 71.13
and JCN’s measure with 64.04. Despite not having
the scale to trust in statistics, we looked at the per-
formance. We found the following: we attribute the
baseline performance to a disparity in the distribution
HEALTHINF 2021 - 14th International Conference on Health Informatics
276

Table 1: Results for the MeSH dataset.
Semantic measure Accuracy Precision Recall F1-score
Baseline 48.06 73.07 48.06 33.62
Embeddings-idf 74.69 74.98 74.69 74.65
Embeddings-no idf 75.46 75.75 75.46 75.45
Embeddings-spacy 73.42 73.66 73.42 73.42
Lin 59.65 59.79 59.65 59.58
Res 61.53 61.54 61.53 61.38
JCN 70.00 70.15 70.00 70.01
of instances for all senses in the dataset we annotated.
For instance, the term ACS has six different senses in
UMLS; in our dataset, 34 instances correspond to the
sense of acute coronary syndrome (CUI
4
C0948089)
and only one instance for the sense acute chest syn-
drome (C0742343). Thus, when building a dataset,
we need to put a special effort into keeping an equal
distribution among each ambiguous term’s instances.
This will lead to a more robust baseline for the evalu-
ation and a better representation of the senses that the
dataset intent to cover.
After observing our results, we decided to analyse
the characteristics of the dataset and give an explana-
tion on the performance of our approach.
4.2 Understanding Our Data
Regarding the definitions, we found out that a high
lexical diversity (TTR) has a positive impact when
disambiguating a term p< 0.05. TTR and the ac-
curacy have a Pearson coefficient value of 0.15, see
fig 1-A. With the number of tokens and the TTR as
the independent variables, the vocabulary variation
model explains 91.4 percent of the data (0.914 R-
square value). The TTR has a t value of 50.07 vs.
7.31 for the number of tokens. Thus, TTR is the more
significant of the two variables. In practice, a lexically
diverse definition allows for higher WSD accuracy.
For example, in table 2, we see that the defini-
tion of the term plaque has a 99.0 TTR value, which
means that almost every token-word is unique in the
segment of text. Its counterpart is the definition of the
term sodium that has a 63.63 TTR value and an accu-
racy of 48.19 . We can remark that in this definition,
the word sodium is repeated four times, and the words
used, compounds, and food are repeated twice. Hav-
ing repeated words, or a low lexical diversity in a def-
inition reduce the context that an algorithm can use to
disambiguate terms. Indeed, WSD on sodium shows
a lower accuracy than plaque on the MeSH dataset.
Furthermore, this demonstrates the influence of XAI,
where the quality of the explanation with high TTR
increases clarity of the term.
4
Concept Unique Identifier in UMLS
Regarding the informativeness model, we found
that the number of nouns also has a determining role
in resolving ambiguity more accurately p < 0.05. In
this model, the number of nouns, adjectives, verbs,
and adverbs explains 91.4 % of the dataset. The num-
ber of nouns has a t value of 17.28; thus, a higher
ratio of nouns leads to higher accuracy in WSD. In
the second place of importance, we found the number
of adjectives with a t value of 8.86. This could be ex-
plained by the fact that adjectives modify nouns; thus,
next to a high number of nouns, a high ratio of adjec-
tives ensures higher accuracy. This also has an expla-
nation from a linguistic perspective; a high ratio of
nouns indicates a focus on information, and a high ra-
tio of adjectives expands and elaborates the informa-
tion of texts (Biber, 2006). In table 2, we observe that
plaque has a ratio of 30.43 vs 45.45 for sodium, but
plaque presents a ratio of 8.69 for the adjectives and
sodium has no adjectives at all. In the case of plaque,
the nouns and the adjectives ensure a higher degree of
informativeness, and consequently, a higher accuracy.
Thus, more nouns are associated with higher WSD
accuracy.
Regarding the text where the ambiguous terms ap-
pear in the MeSH dataset, the vocabulary variation
model found a slight difference in the importance be-
tween the number of tokens and TTR; their t value
is 7.08 and 6.59, respectively. Thus, when it comes
to the accuracy of WSD, both variables seem to con-
tribute to high accuracy. In practical terms, a text that
has high values of TTR and number of tokens leads to
more accurate disambiguation.
For the informativeness model, we found that the
number of verbs is determinant for high accuracy of
p < 0.05. The model is able to explain 92.5% of the
dataset. When testing the correlation between the ac-
curacy and the number of verbs, we found a Pearson
coefficient of 0.22 p < 0.05, see 1-B. Thus, for the
MeSH dataset, a high ration of verbs leads to a higher
accuracy.
The difference in the two models (the vocabulary
variation and the informativeness) for UMLS defi-
nitions and the texts in the MeSH dataset confirms
that each text has different linguistic characteristics.
Knowing the linguistic characteristics of texts or sec-
What do You Mean, Doctor? A Knowledge-based Approach for Word Sense Disambiguation of Medical Terminology
277
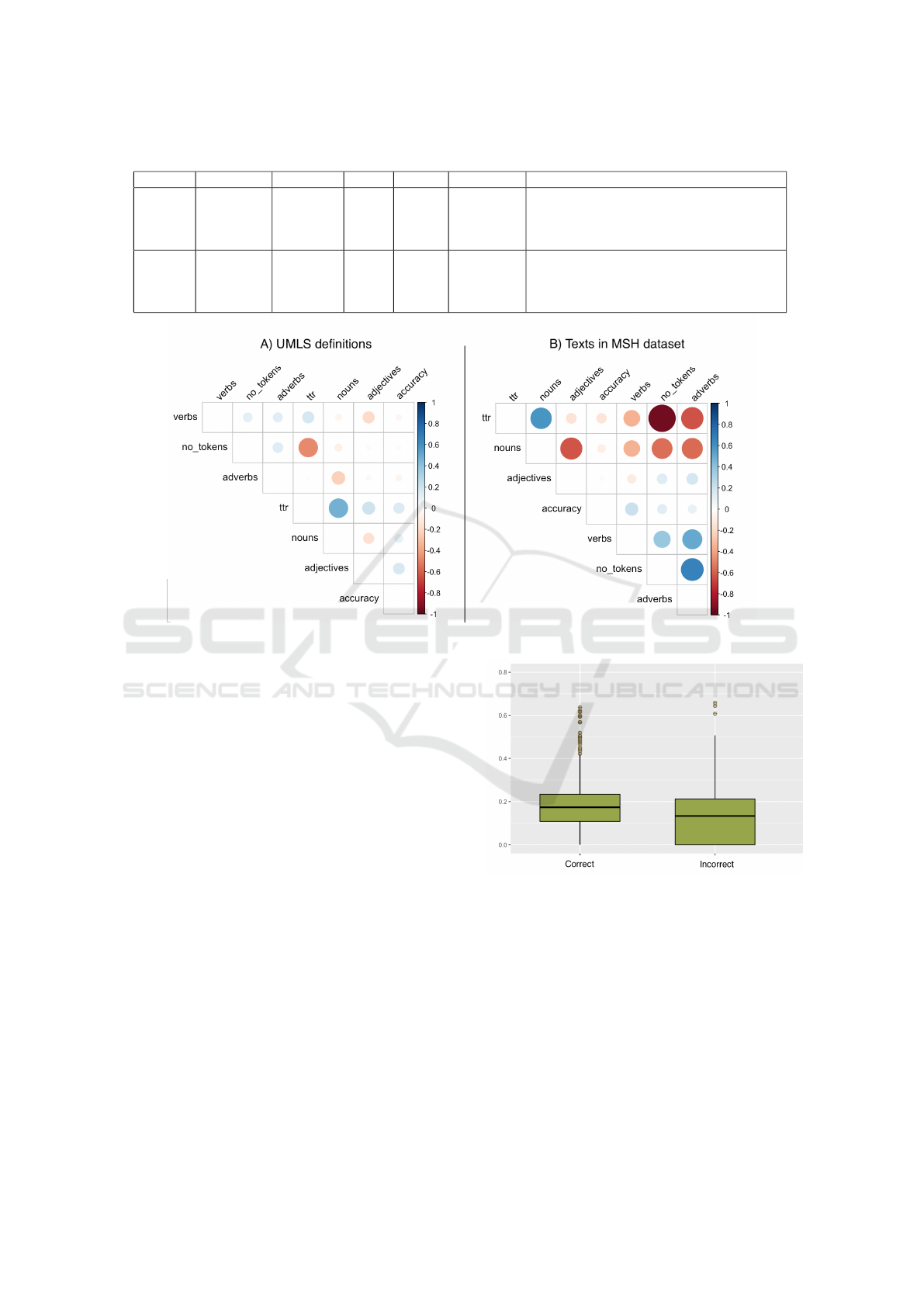
Table 2: Example of high and low TTR for the terms plaque and sodium.
Term CUI Accuracy TTR Nouns Adjectives Definition
Plaque C0011389 95.65 99.0 30.43 8.69 A film that attaches to teeth, often causing
DENTAL CARIES and GINGIVITIS. It is
composed of MUCINS, secreted from sali-
vary glands, and microorganisms.
Sodium C0037570 48.19 63.63 45.45 0.00 Sodium or sodium compounds used in
foods or as a food. The most frequently
used compounds are sodium chloride or
sodium glutamate.
Figure 1: Correlation matrix of the linguistic features and the accuracy.
tions of a document has a direct application to real-
world scenarios. For example, clinical trial docu-
ments are composed of an official title, a summary, in-
clusion, exclusion criteria sections; ambiguous terms
can appear in any of these sections. In the case of the
official title section, it may not have sufficient infor-
mation to disambiguate the term. Thus, picking the
section that has the linguistic profile that our models
describe will ensure accurate disambiguation.
In formula 1, the function maxSim(w, S) selects
the word w from the referent that has the maximum
similarity value with the subject of comparison. We
collected these words, which are very similar to a se-
mantic field. A semantic field is “a set of semantically
related lexical items whose meaning are mutually in-
terdependent and which together provide a concep-
tual structure for a certain domain of reality” (Geer-
aerts, 2010, p.52). E.g. a semantic field for “school”
would be composed by teacher, student, blackboard,
book, notebook.
To evaluate the quality of the automatically cre-
ated semantic fields, we measure the semantic simi-
larity among the group of words. This strategy has
been used to measure the quality of topic modeling
(Koren
ˇ
ci
´
c et al., 2018), which is also somehow similar
Figure 2: Boxplots for the semantic cohesion of the seman-
tic fields for the classes that have been correctly classified
(left) and the class incorrectly classified (right).
to a semantic field. In table 3, we can see an example
of the ambiguous term “coffee”, its definitions, and
the semantic field created by all the good classified in-
stances of each sense. In previous research (Gui et al.,
2019), the evaluation of topic modeling has been used
to reinforce the learning process of a deep neural net-
work model. Similarly, WSD with supervised and un-
supervised methods could benefit from this kind of
feedback to increase their performance.
HEALTHINF 2021 - 14th International Conference on Health Informatics
278

Table 3: Meanings and lexical fields for the ambiguous term “coffee”.
CUI Coherence Definition Semantic field
C0085952 0.3016 A plant genus of the family RUBIACEAE. It is best known for
the COFFEE beverage prepared from the beans (SEEDS).
’coffee’, ’genus’,
’family’, ’tree’,
’plant’
C0009237 0.4859 A beverage made from ground COFFEA beans (SEEDS) in-
fused in hot water. It generally contains CAFFEINE and
THEOPHYLLINE unless it is decaffeinated.
’consume’, ’bever-
age’, ’coffee’, ’caf-
feine’, ’roast’
4.3 Feedback from the Annotation
Process
At the end of the annotation process, we received
feedback from the annotators. One of them remarked
that for the term basal cell carcinoma with CUIs
C3540686, C2984322, and C0007117, the definitions
are vague and difficult to make a distinction. Consid-
ering these comments, we decided to investigate some
terms in the MeSH dataset to see if there were similar
cases. Our assumption was the following: in theoreti-
cal semantics, we deal with vague terms and ambigu-
ous terms. We talk of a vague term when the contexts
where it appears gives information not specified in the
definition. In the sentences he is our publicist and she
is our publicist, the term publicist is vague for gen-
der
5
. Their contexts only give us more details that do
not appear in their definition. In ambiguous terms,
their contexts will cause one of the senses to be se-
lected (Saeed, 2008, p. 61). In dictionaries, for the
ambiguous terms, lexicographers separate senses by
domain. In UMLS, the semantic types could have a
similar role. Thus, for an ambiguous term, if its def-
initions are associated with different semantic types,
we will most probably deal with a truly ambiguous
term. On the other hand, if the definitions of an am-
biguous term share the same semantic type, we will
more probably deal with vague terms.
We found 25 terms out of 203 (12.31%) under
this assumption, i.e., where the definitions have the
same semantic type. For example, for the term B-
Cell Leukemia associated both with CUIs C2004493
and C0023434, the CUIs have the same semantic type
T191 (Neoplastic Process). However, the definition
of C2004493 gives a general description of the dis-
ease, while the definition of C0023434 gives more
details and offers a classification of the disease. In-
specting the MRREL table from UMLS, we see that
CUI C2004493 has a parent-child relationship with
CUI C0023434. Thus, these two definitions are not
ambiguous but vague. In another example, we can
find with the term milk with CUIs C0026131 and
C0026140, where the last one has a parent-child re-
5
These examples were extracted from (Saeed, 2008, p.62).
lationship similar to the B-Cell Leukemia term. This
observation has two implications for future research:
first, researchers seeking to improve WSD systems
should consider the difference between ambiguous vs.
vague terms. Both terms need to be tackled differ-
ently. Second, for those seeking to build datasets for
WSD, they need to be aware that ambiguity is more
potential than real (Saeed, 2008, p.61). The possi-
ble ambiguous terms need to undergo ambiguity tests
to determine if they are vague or ambiguous. Such
a test could be automated by checking the semantic
relationships between the candidates in the MRREL
table. This practice will ensure that the dataset will
help to answer the question of WSD.
The presence of vague terms in the MeSH dataset
could mislead the research of WSD since vagueness
and ambiguity are two different problems. Solving
vagueness could be a different NLP problem where
the aim is to retrieve as much information as possi-
ble to make it less vague. Thus, we recommend that
terms in the MeSH dataset be enriched with labels on
vagueness and ambiguity.
5 CONCLUSIONS
In this paper, we presented a knowledge-based ap-
proach for word sense disambiguation for medical ter-
minology that uses an asymmetrical strategy. Our ap-
proach can be configured to use any semantic measure
on WordNet or a semantic measure based on word
embeddings. In our experiments, we found that the
WordNet-based measures performed very closely to
those based on word embeddings. Such performance
puts our strategy in advantage to others when there is
no specialized domain resource to tackle ambiguity.
We conducted statistical analysis of the texts in the
MeSH corpus and a small clinical trial based dataset
we constructed, using linguistic variables commonly
used in corpus linguistics studies. This analysis
helped us understand the characteristics of the input
texts and their impact on our models’ performance.
Our results suggest that definitions need to be lexi-
cally diverse and informative to ensure better accu-
racy.
What do You Mean, Doctor? A Knowledge-based Approach for Word Sense Disambiguation of Medical Terminology
279

During our data analysis we have also identified
the need for differentiation between vague and am-
biguous terms, which we believe has implications for
the use of test corpora – such as MeSH – for WSD
research, even beyond the medical domain.
REFERENCES
Agirre, E. and Soroa, A. (2009). Personalizing pagerank
for word sense disambiguation. In Proceedings of the
EACL 2009, pages 33–41.
Antunes, R. and Matos, S. (2017a). Biomedical word sense
disambiguation with word embeddings. In Interna-
tional Conference on Practical Applications of Com-
putational Biology & Bioinformatics, pages 273–279.
Springer.
Antunes, R. and Matos, S. (2017b). Supervised learning
and knowledge-based approaches applied to biomedi-
cal word sense disambiguation. Journal of integrative
bioinformatics, 14(4).
Biber, D. (2006). University language: A corpus-based
study of spoken and written discourse. Amsterdam:
John Benjamin.
Bodenreider, O. (2004). The unified medical language sys-
tem (umls): integrating biomedical terminology. Nu-
cleic acids research, 32(suppl 1):D267–D270.
Geeraerts, D. (2010). Theories of lexical semantics. Oxford
University Press.
Gui, L., Leng, J., Pergola, G., Xu, R., He, Y., et al. (2019).
Neural topic model with reinforcement learning. In
Proceedings of the 2019 Conference on EMNLP-
IJCNLP, pages 3469–3474.
Humphrey, S. M., Rogers, W. J., Kilicoglu, H., Demner-
Fushman, D., and Rindflesch, T. C. (2006). Word
sense disambiguation by selecting the best semantic
type based on journal descriptor indexing: Prelimi-
nary experiment. JASIST, 57(1):96–113.
Jiang, J. J. and Conrath, D. W. (1997). Semantic similarity
based on corpus statistics and lexical taxonomy. arXiv
preprint cmp-lg/9709008.
Jimeno-Yepes, A. J. and Aronson, A. R. (2010).
Knowledge-based biomedical word sense disam-
biguation: comparison of approaches. BMC bioinfor-
matics, 11(1):569.
Jimeno-Yepes, A. J., McInnes, B. T., and Aronson, A. R.
(2011). Exploiting mesh indexing in medline to gen-
erate a data set for word sense disambiguation. BMC
bioinformatics, 12(1):223.
Jurafsky, D. and Martin, J. H. (2020). Speech
& language processing [Book in preparation].
https://web.stanford.edu/ jurafsky/slp3/.
Koren
ˇ
ci
´
c, D., Ristov, S., and
ˇ
Snajder, J. (2018). Document-
based topic coherence measures for news media text.
Expert Systems with Applications, 114:357–373.
Lesk, M. (1986). Automatic sense disambiguation using
machine readable dictionaries: how to tell a pine cone
from an ice cream cone. In Proceedings of the 5th an-
nual international conference on Systems documenta-
tion, pages 24–26.
Lin, D. et al. (1998). An information-theoretic definition of
similarity. In Icml, volume 98, pages 296–304.
McInnes, B. (2008). An unsupervised vector approach to
biomedical term disambiguation: integrating umls and
medline. In Proceedings of the ACL-08: HLT Student
Research Workshop, pages 49–54.
Miller, G. A. (1995). Wordnet: a lexical database for en-
glish. Communications of the ACM, 38(11):39–41.
Navigli, R. (2009). Word sense disambiguation: A survey.
ACM computing surveys (CSUR), 41(2):1–69.
Neumann, M., King, D., Beltagy, I., and Ammar, W.
(2019). Scispacy: Fast and robust models for biomed-
ical natural language processing. arXiv preprint
arXiv:1902.07669.
Pesaranghader, A., Matwin, S., Sokolova, M., and Pe-
saranghader, A. (2019). deepbiowsd: effective deep
neural word sense disambiguation of biomedical text
data. JAMIA, 26(5):438–446.
Pustejovsky, J. and Stubbs, A. (2012). Natural Language
Annotation for Machine Learning: A guide to corpus-
building for applications. ” O’Reilly Media, Inc.”.
Pyysalo, S., Ginter, F., Tapio, S., and Sophia, A. (2013).
Distributional semantics resources for biomedical text
processing. Proceedings of LBM, pages 39–44.
Saeed, J. I. (2008). Semantics. Wiley-Blackwell.
Tversky, A. (1977). Features of similarity. Psychological
review, 84(4):327.
Velazquez, E., Ratt
´
e, S., and de Jong, F. (2016). Analyz-
ing students’ knowledge building skills by compar-
ing their written production to syllabus. In Interna-
tional Conference on Interactive Collaborative Learn-
ing, pages 345–352. Springer.
Wang, Y., Zheng, K., Xu, H., and Mei, Q. (2018). Inter-
active medical word sense disambiguation through in-
formed learning. JAMIA, 25(7):800–808.
Weeber, M., Mork, J. G., and Aronson, A. R. (2001). Devel-
oping a test collection for biomedical word sense dis-
ambiguation. In Proceedings of the AMIA Symposium,
page 746. American Medical Informatics Association.
Zhang, C., Bi
´
s, D., Liu, X., and He, Z. (2019). Biomedi-
cal word sense disambiguation with bidirectional long
short-term memory and attention-based neural net-
works. BMC bioinformatics, 20(16):502.
HEALTHINF 2021 - 14th International Conference on Health Informatics
280
