
An Optimization Method for Entity Resolution in Databases:
With a Case Study on the Cleaning of
Scientific References in Patent Databases
Emiel Caron
Department of Management, Tilburg University, Warandelaan 2, Tilburg, The Netherlands
Keywords:
Entity Resolution, Data Disambiguation, Data Cleaning, Data Integration, Bibliographic Databases.
Abstract:
Many databases contain ambiguous and unstructured data which makes the information it contains difficult to
use for further analysis. In order for these databases to be a reliable point of reference, the data needs to be
cleaned. Entity resolution focuses on disambiguating records that refer to the same entity. In this paper we
propose a generic optimization method for disambiguating large databases. This method is used on a table
with scientific references from the Patstat database. The table holds ambiguous information on citations to
scientific references. The research method described is used to create clusters of records that refer to the same
bibliographic entity. The method starts by pre-cleaning the records and extracting bibliographic labels. Next,
we construct rules based on these labels and make use of the tf-idf algorithm to compute string similarities. We
create clusters by means of a rule-based scoring system. Finally, we perform precision-recall analysis using a
golden set of clusters and optimize our parameters with simulated annealing. Here we show that it is possible
to optimize the performance of a disambiguation method using a global optimization algorithm.
1 INTRODUCTION
Ambiguous data in a data set can lead to misleading,
inaccurate and incomplete results in analysis. The
problem of ambiguous and unstructured data in large-
scale databases is a common problem (Bhattacharya
and Getoor, 2007). This is also known as the record
linkage problem and entity resolution - the general
term for disambiguating records that refer to the same
entity by linking and classifying them as one group.
Since manual disambiguation is not attainable in large
databases, there is need for automatic methods. In
the past, research has been conducted on finding au-
tomatic methods for the disambiguation of records in
databases. For instance, Caron and Van Eck (2014)
presented an author name disambiguation method for
large databases, and Caron and Daniels (2016) pro-
vided a method for identifying company name vari-
ants in large databases.
In this work we want to deal with two problems
in the current entity resolution approaches. The first
problem is how to develop a generic method for dis-
ambiguating large databases that, in theory, is appli-
cable to multiple domains. The second problem is
how to optimize the parameters of a generic method.
The optimization of the method refers to finding the
model’s parameters that provide the best results, i.e.
the highest value of a certain performance measure,
typically precision-recall metrics. To the best of our
knowledge, none of the existing approaches for entity
resolution in databases use heuristics to optimize per-
formance. By making the method more efficient and
by implementing optimization techniques, we aim to
reduce computation time and obtain better results.
This in turn would open up the possibility for other
data leaning methods to be optimized as well.
1.1 Entity Resolution
In this paper, data disambiguation, record linkage
and entity resolution all refer to the same topic:
we want to determine a mapping from ambiguous
database records to real-world entities. In the last
20 years much research has been conducted on this
topic. Here we analyze a subset of past research that
is relevant for this paper. Furthermore, we briefly
identify particular methods and techniques used to
disambiguate patent databases. Entity resolution is
the problem of identifying records that refer to the
same real-world entity. For example, the names ‘J.
Doe’ and ’Doe, Jon’ may actually refer to the same
person. Without unique identifiers, this so-called
Caron, E.
An Optimization Method for Entity Resolution in Databases: With a Case Study on the Cleaning of Scientific References in Patent Databases.
DOI: 10.5220/0009972406250632
In Proceedings of the 15th International Conference on Software Technologies (ICSOFT 2020), pages 625-632
ISBN: 978-989-758-443-5
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
625

duplication problem can lead to various issues such
as erroneous computed statistics, redundant data
and data inconsistencies (Monge and Elkan, 1996;
Hern
´
andez and Stolfo, 1995). In the context of
databases, Bhattacharya and Getoor (2007) state that
the entity resolution problem includes (a) identifying
the underlying entities of a database and (b) labelling
records of the database with the entities they refer
to. Typically, these two issues cannot be solved
independently. Bhattacharya and Getoor (2007)
describe two main problems on why entity resolution
is difficult to solve. The first is the identification
problem which occurs when entities are referred to by
different name variants in a database. Second, there is
a disambiguation problem when two similar records
actually do not refer to the same entity. Failure in
disambiguation leads to lower precision; the identifi-
cation problem affects the recall of a cleaning method.
This paper is organised as follows. Related work
is presented in Section 2. In Section 3, our method
for entity resolution is explained step-by-step. After
that the method is applied on a case study involving
disambiguation of scientific references in the Patstat
database in Section 4. Here the method’s performance
is evaluated with precision-recall analysis. Finally,
we discuss conclusions and further research in Sec-
tion 5.
2 RELATED WORK
There are different existing approaches to entity res-
olution. The traditional approach is attribute-based,
where an attribute refers to a property or character-
istic of a record (Fellegi and Sunter, 1969). In these
approaches a similarity value is computed for every
pair of records based on their attributes. If a pair’s
similarity is above a certain threshold, the records are
considered to refer to the same entity. Different simi-
larity measures may be used for computing a similar-
ity value.
In general, manual disambiguation is not feasible
in large databases. Therefore, computerized meth-
ods are required. From the class of automated data
cleaning methods, we first review general data min-
ing methods, and after that we review specific work
on the large-scale cleaning of bibliographic databases
like Patstat, directly related to the case study in Sec-
tion 4.
In supervised approaches, a data set with labelled
records is used to train the learning scheme. How-
ever, large data sets with manually labelled records
would be expensive to collect and are often not avail-
able. For this reason research has focused on devel-
oping unsupervised approaches for entity resolution.
Unsupervised approaches use similarity metrics and
clustering algorithms to find clusters of name vari-
ants. While unsupervised approaches do not require
training data sets, they often perform less well than
supervised approaches (Levin et al., 2012).
Benjelloun et al. (2009) present a generic entity
resolution method and algorithms based on pairwise
decisions. Pairwise means that the method matches
two records at a time. In addition, they elaborate on
the formal properties of an efficient algorithm. How-
ever, no solution is given to find the optimal settings
for the algorithm. This paper addresses this issue.
Nguyen and Ichise (2016) describe an effective
supervised solution to entity resolution named cLink
for linking records from different data sources. cLink
is a generic method that uses a learning algorithm
to optimize the configuration of matching properties
and similarity measures. To this end, the learning al-
gorithm cLearn requires a training data set to com-
pute the F1-score of the candidates with the highest
matching score. The optimized configuration of sim-
ilarity functions is used to compute a final matching
score for every candidate pair. Only pairs with a max-
imal matching score that is above a certain threshold
are considered to be co-referent. Related to the work
of Nguyen and Ichise (2016) are the active learning
approaches RAVEN (Ngomo et al., 2011), EAGLE
(Ngonga Ngomo and Lyko, 2012) and ActiveGenLink
(Isele and Bizer, 2013). These three systems all work
similarly by using active learning to achieve accu-
rate link specifications. Active learning requires in-
teraction with the user to confirm or decline a certain
match. EAGLE and ActiveGenLink additionally ap-
ply genetic programming to achieve better efficiency.
Thoma and Torrisi (2007) compare two ap-
proaches to automatic matching techniques on linking
patent data from the PatStat database to data on firms
from the Amadeus database. Amadeus is a financial
database on European firms, maintained by Bureau
Van Dijk. Both approaches start with name standard-
ization which consists of tasks such as punctuation
cleaning, character cleaning and removal of common
terms such as ‘company’ and ‘co.’. Next, the first ap-
proach uses character-to-character comparison (per-
fect matching). The second approach uses approx-
imate string comparison based on string similarity
functions (approximate matching). To measure string
similarity Thoma and Torrisi make use of the Jaccard
index combined with tf-idf weights. Even though per-
fect matching yields high precision, the number of
matches can be quite low. Approximate matching can
increase the number of matches, but at the cost of pre-
ICSOFT 2020 - 15th International Conference on Software Technologies
626

cision. The results of the two approaches show that
approximate matching indeed yields a significant in-
crease in the number of matches, while producing a
limited loss of precision. Thoma et al. (2010) devel-
oped a matching technique that, in addition to name
similarity, compares other information such as loca-
tion and founding year to match firms in multiple
large databases. This technique uses a combination of
dictionary-based methods and rule-based approaches.
Dictionary-based methods depend on large data sets
with verified name variants. While these methods
are simple and very precise, typically the number of
matches is low in the case of firm names. Rule-based
approaches set up a collection of rules to determine
the similarity between different firm names. Thoma
et al. focus on approximate matching in their set of
rules. Following Thoma et al., Lotti and Marin (2013)
describe an improved cleaning routine to match ap-
plicants in the PatStat database to firms in the AIDA
database. AIDA is a product of Bureau Van Dijk con-
taining information on Italian companies. In addition
to name harmonization and matching, Lotti and Marin
(2013) use harmonized addresses for exact match-
ing of firms. Furthermore, approximate matches on
names were used in combination with visual checks.
Finally, the name variants contained in the data set
created by Thoma et al. (2010) were used for further
matching.
3 METHOD FOR DATA
DISAMBIGUATION
When studying past research on entity resolution,
specifically the work described in (Caron and Van
Eck, 2014; Zhao et al., 2016; Caron and Daniels,
2016), we observe that the methods on clean-
ing databases all have similar components and ap-
proaches. This common approach is in essence as fol-
lows. Features are extracted from the records in the
database and similarity rules are constructed on the
basis of these features. These rules provide evidence
that a pair of records match. Next, scores are assigned
to every rule and the total score for all pairs is com-
puted. These scores measure the similarity of a pair of
records. The pairs that score above a certain thresh-
old are linked using a clustering algorithm, e.g. con-
nected components or maximal cliques. After that the
performance of the clusters is benchmarked against
a golden verified set with precision-recall analysis.
Subsequently, the scores and threshold parameters are
updated experimentally to improve the average values
for both precision and recall. We add to this known
approach a way to optimize the method’s parameters
with a global optimization algorithm to obtain the
clusters with the highest possible average precision
and recall.
3.1 Generic Method
Here the generic method is presented. Consider N
objects or representations of entities r
1
, ..., r
N
. For i =
1, ..., N, the object r
i
has feature vector f
i
consisting
of M features f
i
= ( f
i
(1), ..., f
i
(M)). Every feature
f
i
( j) has a domain F
j
, i.e. f
i
(1) ∈ F
1
, ..., f
i
(M) ∈ F
M
which is independent of i.
We call two entities similar if their corresponding
feature vectors are similar. Stated more precisely, we
define
σ
l
: F
l
× F
l
−→ R
+
∪ {0} (1)
such that σ
l
( f
i
(l), f
j
(l)) measures the similarity of
any pair ( f
i
(l), f
j
(l)) ∈ F
l
× F
l
, where i 6= j.
To obtain a similarity score as a single number, we
define a NxN matrix S with elements s
i j
and weight
vector w = (w
1
, ..., w
M
):
s
i j
=
M
∑
k=1
w
k
· σ
k
( f
i
(k), f
j
(k)) where w
k
≥ 0, i 6= j
(2)
Objects that are similar are grouped into sets as
follows. Suppose we have Q sets Σ
1
, ..., Σ
Q
. Define a
threshold δ. Then
∪
Q
p=1
Σ
p
= {r
1
, ...r
N
}
where
r
j
∈ Σ
p
implies ∃r
i
∈ Σ
p
such that s
i j
≥ δ, i 6= j
unless
|
Σ
p
|
= 1.
The sets Σ
1
, ..., Σ
Q
are mutually exclusive:
Σ
p
∩ Σ
q
=
/
0 for p 6= q
and are chosen to be maximal:
let r
i
∈ Σ
p
. If ∃r
j
such that s
i j
≥ δ, then r
j
∈ Σ
p
.
The method’s performance is evaluated using pre-
cision and recall analysis. The F1-score is the har-
monic mean of precision and recall (Fawcett, 2006).
Since the objective is to obtain clusters with both high
precision and high recall, we maximize the F1-score.
Therefore we choose our parameters w = (w
1
, ..., w
M
)
and δ in such a way that
w
∗
, δ
∗
= arg max
w,δ
L(w, δ) (3)
where our objective function L(w, δ) is the average
F1-score of the clusters. Optimizing the method’s pa-
rameters is done using a simulated annealing algo-
rithm (Xiang et al., 1997).
An Optimization Method for Entity Resolution in Databases: With a Case Study on the Cleaning of Scientific References in Patent Databases
627

3.2 Optimization
The objective function in the optimization problem
described in Eq. (3) is nonlinear and yields many lo-
cal optima. Moreover, according to Erber and Hock-
ney (1995), the number of local optima typically in-
creases exponentially as the number of variables in-
creases. Here the variables are weights w and thresh-
olds δ used in the method described in section 3.1. We
need to use a global optimization method such as the
simulated annealing algorithm or a genetic algorithm
in order to find a global optimum instead of getting
trapped in one of the many local optima.
The problem of maximizing the F1-score with re-
spect to the weights w and thresholds δ of the model
is a combinatorial optimization problem. Research in
combinatorial optimization focuses on developing ef-
ficient techniques to minimize or maximize a func-
tion of many independent variables. Since solving
such optimization problems exactly would require a
large amount of computational power, heuristic meth-
ods are used to find an approximate optimal solution.
Heuristic methods are typically based on a ‘divide-
and-conquer’ strategy or an iterative improvement
strategy. The algorithms we focus on are based on
iterative improvement. That is, the system starts in
a known configuration of the variables. Then some
rearrangement operation is applied until a configura-
tion is found that yields a better value of the objec-
tive function. This configuration then becomes the
new configuration of the system and this process is re-
peated until no further improvements are found. Since
this method only accepts new configurations that im-
prove the objective function, the system is likely to
be trapped in a local optima. This is where simulated
annealing plays its part.
Simulated annealing is inspired by techniques of
statistical mechanics which describe the behavior of
physical systems with many degrees of freedom. The
simulated annealing process starts by optimizing the
system at a high temperature such that rearrangements
of parameters causing large changes in the objective
function are made. The “temperature”, or in general
the control parameter, is then lowered in slow stages
until the system freezes and no more changes occur.
This cooling process ensures that smaller changes
in the objective functions are made at lower tem-
peratures. The probability of accepting a configu-
ration that leads to a worse solution is lowered as
the temperature decreases (Kirkpatrick et al., 1983).
To optimize the F1-score of our method we use the
dual
annealing() function of the SciPy.optimize pack-
age in Python (SciPy.org, 2020). This dual anneal-
ing optimization is derived from the research of Xi-
ang et al. (1997) in combination with a local search
strategy (Xiang and Gong, 2000).
Next to simulated annealing, the genetic algorithm
was explored for finding a global optimum. The
genetic algorithm computes a population and pro-
duces generations by combining and altering indi-
vidual solutions of the population (Whitley, 1994).
While the genetic algorithm is often used in optimiza-
tion problems, for our problem simulated annealing
seems to be a better fit. The genetic algorithm re-
quires large generations of populations to be com-
puted which would be very time consuming consider-
ing our problem as the computation of one individual
already takes several minutes.
3.3 Method Steps
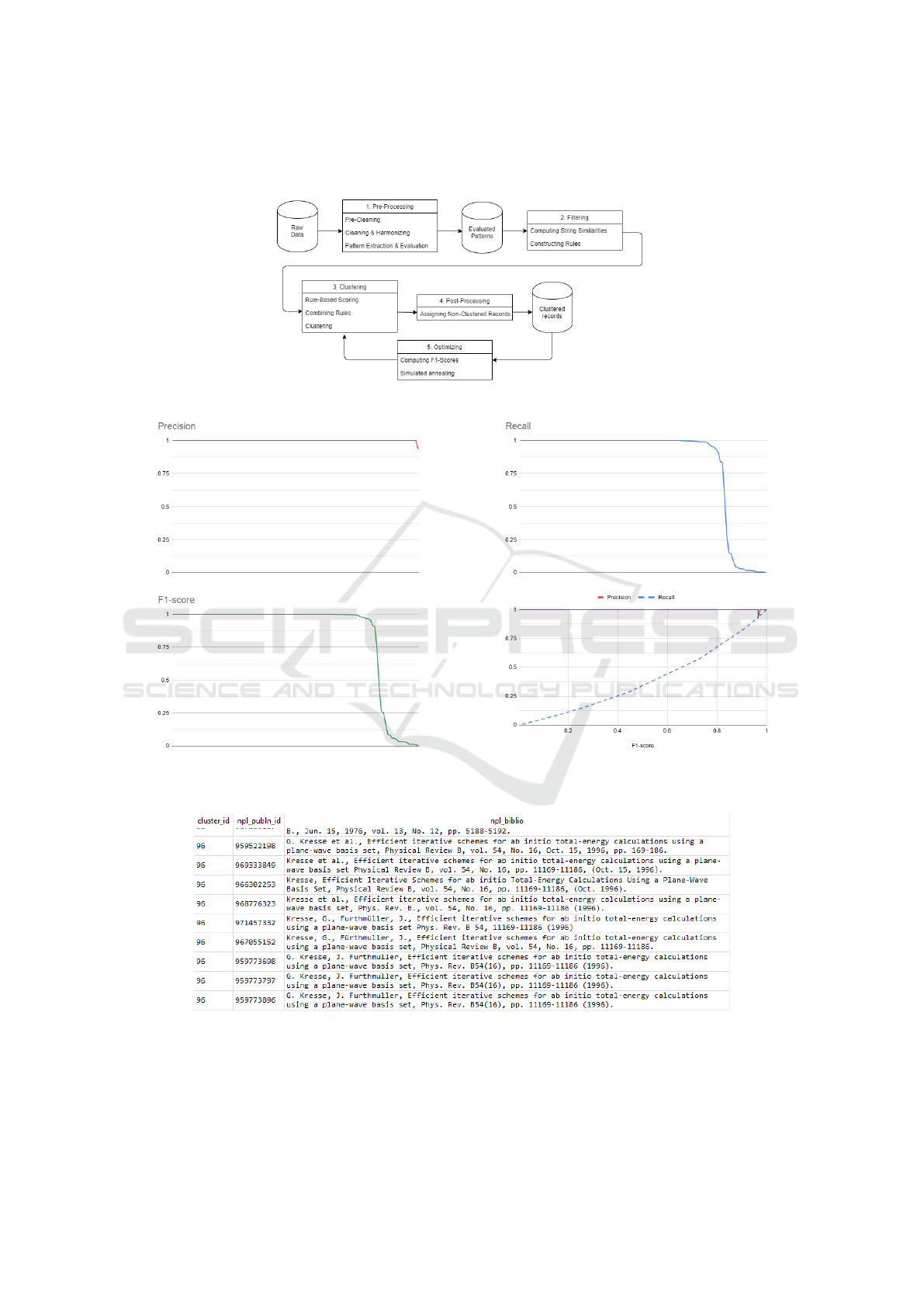
In Figure 4, an overview is given of the practical steps
in the method for entity resolution detailed in the pre-
vious section. The input of the method is raw data
that corresponds to ambiguous name variants and the
output are clusters of name variants. The method is
composed out of main 5 steps:
1. Pre-processing.
2. Filtering.
3. Rule-based clustering.
4. Post-processing.
5. Optimization.
Step 1 involves pre-cleaning the data and extract-
ing and evaluating descriptive labels. In step 2, the
tf-idf algorithm (Salton and Buckley, 1988) is used to
compute string similarities between records using Eq.
(1). By constructing rules based on the extracted la-
bels, evidence is collected for the similarity between
records and based on that candidate record pairs are
created. In step 3, rules are scored and combined us-
ing Eq. (2) and after that the connected-components
algorithm (Bondy and Murty, 1976) is used to obtain
clusters of records. In step 4, records for which no
duplicates are identified are assigned to new single-
record clusters. In step 5, the clusters are evaluated
on the golden sample using precision and recall anal-
ysis. The parameters used for constructing and scor-
ing rules, such as thresholds and scores, are adjusted
to achieve higher values in precision and recall. Using
simulated annealing (Xiang et al., 1997) we obtain the
optimal parameters. Here the average F1-score over
all clusters is used, the harmonic mean of the pre-
cision and recall measure, as our objective function
(Fawcett, 2006) as defined in Eq. (3). The method
stops when the global optimum is obtained.
ICSOFT 2020 - 15th International Conference on Software Technologies
628

3.4 Software Implementation
The generic method as described in section 3.1 is
implemented in Python
1
. In the example case of
the Patstat table with ambiguous scientific references,
the objects r
1
, ...r
N
are the records of the table and
the features of each record are described in the table
‘evaluated patterns’. The features in the case study
Section 4 are extracted bibliographic meta informa-
tion as: publication title and year, author names, jour-
nal information, etc. Eq. (1) of the generic method
refers to the rules that are constructed. These rules
provide evidence that two records are similar. In or-
der to compute the string similarities for the rules we
use an efficient implementation of tf-idf in Python.
The scores that are assigned to the rules correspond
to weight vector w in Eq. (2). The clustering of
records is done using the connected components al-
gorithm and results in the sets Σ
1
, ...Σ
Q
as described
in the generic method.
The method ‘find clusters()’, given in the
script ‘find clusters.py’, implements the whole
generic method and computes the F1-score. Its input
parameters are a configuration of variables (w, δ) and
a table containing feature vectors. We use this method
as our objective function for the dual annealing()
method (SciPy.org, 2020) described in the script
‘optimize.py’. In this way we find the optimal con-
figuration of the parameters in Eq. (3).
4 CASE STUDY ON PATSTAT
DATA
4.1 Background
PatStat (European Patent Office, 2019) is a worldwide
patent database from the European Patent Office. It
contains bibliographic information on patent applica-
tions and publications and is important source in the
field of patent statistics. One of its tables, TLS214,
holds information on scientific references cited by
patents. These references are collected from patent
application, in which patent applicants reference (sci-
entific) literature to acknowledge the contribution of
other writers and researchers to their work. In 2019,
this table contained more than 40 million records with
scientific references. Therefore, the table is an impor-
tant point of reference for research, for example, that
studies the relation between science and technology.
However, amongst the scientific references there are
1
https://emielcaron.nl/wp-content/uploads/2020/05/
entity resolution optimization code.7z
many name variants of publications caused by miss-
ing data, inconsistent input convention, different or-
der of items, typos, etc. The records in Figure 1 serve
as an example of such name variants. In order for
table TLS214 to be a reliable point of reference for
research, its records need to be disambiguated and re-
structured (Zhao et al., 2016).
4.2 Results
Here we present and analyze the results of applying
the disambiguation method on scientific references
from the Patstat database, by comparing the set of
clusters generated by the method with a golden set
of 100 clusters. The golden sample is a sample of
records with verified clusters (Caron and Van Eck,
2014), evaluated by human domain experts. We run
our algorithm and keep track of the intermediate re-
sults. After approximately 24 hours, no more im-
provements are made to the F1-score, so the algo-
rithm is stopped. The plot in Figure 2 shows the in-
crease of the F1-score against time in seconds. The
first point of the line is the F1-score of the initial clus-
ters. It is important to note that, this plot shows that
the simulated annealing algorithm indeed is able to
improve the F1-score and thus our method produces
better clusters. We observe that the algorithm makes
large improvements to the objective value in the be-
ginning and smaller improvements towards the end.
This is in accordance with the theory on simulated an-
nealing, as the smaller changes are made at low tem-
peratures near the end.
The results of our method, depicted in Table 1, are
promising in terms of precision-recall statistics. The
average values of precision, recall, and F1 are high
for all cluster categories. In comparison, the clusters
achieve significantly better precision and recall values
than the initial clusters created with the method’s ini-
tial parameters, as depicted in development of the F1-
score in Figure 2. The plots in Figure 5 show a large
positive shift to the right in the distribution of the dif-
ferent measures, compared to initial values. We also
Figure 1: A set of name variants that refer to the article
‘Protein measurement with the Folin phenol reagent’. A
selection of records from a cluster in the golden sample.
An Optimization Method for Entity Resolution in Databases: With a Case Study on the Cleaning of Scientific References in Patent Databases
629
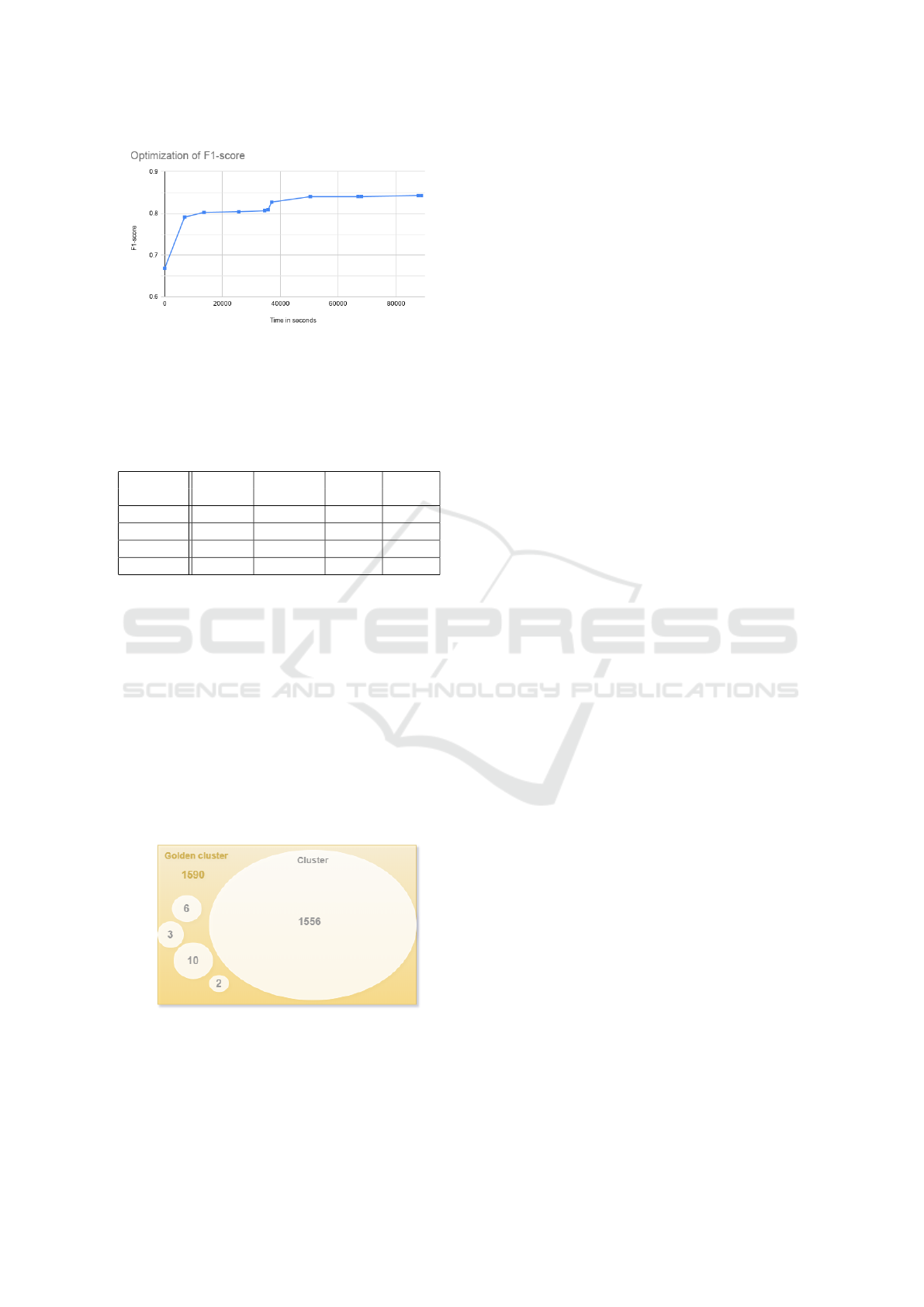
Figure 2: Optimization of the F1-score against time in sec-
onds.
see that the F1-score almost solely depends on the re-
call measure as precision is equal to 1 for almost all
clusters.
Table 1: Statistics of optimized clusters.
# of Precision Recall F1
Category clusters (Avg) (Avg) (Avg)
Large 31 0.9979 0.9893 0.9933
Medium 43 1 0.9490 0.9563
Small 41 1 0.5936 0.6114
Total 115 0.9994 0.8331 0.8433
Figure 6 shows an example cluster of correctly
classified scientific references. We observe that the
optimized method correctly identifies all records of
the cluster and achieves a precision and recall value
of 1.
In addition, when analyzing the clusters, we no-
tice that our method is slightly conservative, it val-
ues precision over recall. As a result the method is
more likely to create multiple clusters with high pre-
cision than to create one large cluster with potential
errors. Typically, the method splits the name variants
of one golden cluster into one large dominant clus-
ter and multiple small clusters. This is illustrated for
golden cluster 100 in Figure 3. The number inside
Figure 3: Illustration of a golden cluster distributed over
multiple system clusters.
each cluster is its size, i.e. the number of records in
the cluster. The remaining records of the golden clus-
ter were not paired by our method or did not score
above the threshold and were therefore not clustered.
The sizes of the golden cluster range between 2 and
1590 records. Because of this variety, we distinguish
between three cluster categories. Large clusters con-
tain more than 100 records and typically score high
on recall. These clusters contain the majority, or all,
of the records of the corresponding golden cluster.
Medium clusters contain 11 to 100 records and typ-
ically correspond to medium-sized golden clusters.
Most small clusters, containing 10 or less records, are
clusters like the small clusters in figure 8. Because of
this, recall is sometimes lower for small clusters.
5 CONCLUSIONS
Here an optimization method for entity resolution in
databases is proposed. To optimize the method, pre-
cision and recall analysis is performed using a golden
set of clusters and the F1-score is maximized using
simulated annealing. The research method is used to
disambiguate scientific references and create clusters
of records that refer to the same bibliographic entity.
The method starts by pre-cleaning the records and ex-
tracting bibliographic labels. Next, we construct rules
based on these labels and make use of the tf-idf al-
gorithm to compute a string similarity measure. We
create clusters by means of a rule-based scoring sys-
tem. Finally, we perform precision and recall analysis
using a golden set of clusters and optimize our param-
eters with a simulated annealing algorithm. Here we
show that it is possible to optimize the average F1-
score of disambiguation method using a global opti-
mization algorithm. The proposed method is generic
and applicable on similar entity resolution problems.
The results of this research are beneficial for both
academics and industry. Academics can use our
method to efficiently access information in an am-
biguous data sets. The unambiguated data in Patstat
can for example be used to study the effect of sci-
ence outputs on industry patents. Data scientists at
firms often have to deal with ambiguous data. The
disambiguation method studied provides a way to ef-
ficiently clean such data sets so that these can be used
for data analysis. To the best of our knowledge, none
of the existing approaches for entity resolution in bib-
liographic databases use heuristics to optimize the pa-
rameters for rule construction and clustering. Hence,
the results of this research provide new insights for
research on entity resolution.
In future work, we want to compare our method
with existing methods on benchmark datasets for en-
tity resolution. In addition, we could speed up the
method by incorporating parallel computing. Espe-
cially the construction of rules is suitable for parallel
ICSOFT 2020 - 15th International Conference on Software Technologies
630

computing as the rules could be split over multiple
cores of a computer and be constructed simultane-
ously. This would decrease the computation time of
the disambiguation method significantly. Moreover,
the exploration of different optimization techniques
for finding a global optimum next to simulated an-
nealing would be very interesting for comparison. A
potential candidate for comparison is Tabu search.
ACKNOWLEDGEMENTS
We kindly acknowledge Wen Xin Lin and Prof.
H.A.M. Daniels for their contribution to this work.
REFERENCES
Benjelloun, O., Garcia-Molina, H., Menestrina, D., Su, Q.,
Whang, S. E., and Widom, J. (2009). Swoosh: A
generic approach to entity resolution. The VLDB Jour-
nal, 18(1):255276.
Bhattacharya, I. and Getoor, L. (2007). Collective entity
resolution in relational data. ACM Trans. Knowl. Dis-
cov. Data, 1(1).
Bondy, J. A. and Murty, U. S. R. (1976). Graph theory with
applications, volume 290. Macmillan London.
Caron, E. and Daniels, H. (2016). Identification of organiza-
tion name variants in large databases using rule-based
scoring and clustering - with a case study on the web
of science database. In Proceedings of the 18th Inter-
national Conference on Enterprise Information Sys-
tems - Volume 1: ICEIS,, pages 182–187. INSTICC,
SciTePress.
Caron, E. and Van Eck, N.-J. (2014). Large scale author
name disambiguation using rule-based scoring and
clustering. In Noyons, E., editor, Proceedings of the
Science and Technology Indicators Conference 2014,
pages 79–86. Universiteit Leiden.
Erber, T. and Hockney, G. (1995). Comment on method
of constrained global optimization. Physical review
letters, 74(8):1482.
European Patent Office (2019). Data Catalog - PATSTAT
Global, 2019 autumn edition edition.
Fawcett, T. (2006). An introduction to roc analysis. Pattern
recognition letters, 27(8):861–874.
Fellegi, I. P. and Sunter, A. B. (1969). A theory for record
linkage. Journal of the American Statistical Associa-
tion, 64(328):1183–1210.
Hern
´
andez, M. A. and Stolfo, S. J. (1995). The merge/purge
problem for large databases. In ACM Sigmod Record,
volume 24, pages 127–138. ACM.
Isele, R. and Bizer, C. (2013). Active learning of expres-
sive linkage rules using genetic programming. Web
Semantics: Science, Services and Agents on the World
Wide Web, 23:2–15.
Kirkpatrick, S., Gelatt, C. D., and Vecchi, M. P. (1983).
Optimization by simulated annealing. science,
220(4598):671–680.
Levin, M., Krawczyk, S., Bethard, S., and Jurafsky,
D. (2012). Citation-based bootstrapping for large-
scale author disambiguation. Journal of the Ameri-
can Society for Information Science and Technology,
63(5):1030–1047.
Lotti, F. and Marin, G. (2013). Matching of patstat applica-
tions to aida firms: discussion of the methodology and
results. Bank of Italy Occasional Paper, (166).
Monge, A. E. and Elkan, C. (1996). The field matching
problem: Algorithms and applications. In KDD, vol-
ume 2, pages 267–270.
Ngomo, A.-C. N., Lehmann, J., Auer, S., and H
¨
offner, K.
(2011). Raven–active learning of link specifications.
Ontology Matching, 2011.
Ngonga Ngomo, A.-C. and Lyko, K. (2012). Eagle: Effi-
cient active learning of link specifications using ge-
netic programming. In Simperl, E., Cimiano, P.,
Polleres, A., Corcho, O., and Presutti, V., editors,
The Semantic Web: Research and Applications, pages
149–163, Berlin, Heidelberg. Springer Berlin Heidel-
berg.
Nguyen, K. and Ichise, R. (2016). Linked data entity res-
olution system enhanced by configuration learning al-
gorithm. IEICE Transactions on Information and Sys-
tems, E99.D(6):1521–1530.
Salton, G. and Buckley, C. (1988). Term-weighting ap-
proaches in automatic text retrieval. Information Pro-
cessing & Management, 24(5):513 – 523.
SciPy.org (2020). Scipy.optimize package with
dual annealing() function.
Thoma, G. and Torrisi, S. (2007). Creating powerful indica-
tors for innovation studies with approximate matching
algorithms: a test based on PATSTAT and Amadeus
databases. Universit
`
a commerciale Luigi Bocconi.
Thoma, G., Torrisi, S., Gambardella, A., Guellec, D., Hall,
B. H., and Harhoff, D. (2010). Harmonizing and
combining large datasets-an application to firm-level
patent and accounting data. Technical report, National
Bureau of Economic Research.
Whitley, D. (1994). A genetic algorithm tutorial. Statistics
and computing, 4(2):65–85.
Xiang, Y. and Gong, X. (2000). Efficiency of generalized
simulated annealing. Physical Review E, 62(3):4473.
Xiang, Y., Sun, D., Fan, W., and Gong, X. (1997). General-
ized simulated annealing algorithm and its application
to the thomson model. Physics Letters A, 233(3):216–
220.
Zhao, K., Caron, E., and Guner, S. (2016). Large
scale disambiguation of scientific references in patent
databases. In Rafols, I., Molas-Gallart, J., Castro-
Martinez, E., and Woolley, R., editors, Proceedings of
21st International Conference on Science and Tech-
nology Indicators (STI 2016), pages 1404–1410. Edi-
torial Universitat Polit
`
ecnica de Val
`
encia.
An Optimization Method for Entity Resolution in Databases: With a Case Study on the Cleaning of Scientific References in Patent Databases
631
APPENDIX
Figure 4: Overview of the disambiguation method.
Figure 5: Distribution of the precision and recall measure and the F1-score on all optimized clusters. The clusters on the
x-axis are ranked based on precision-recall-f1 values.
Figure 6: Contents of example cluster 96.
ICSOFT 2020 - 15th International Conference on Software Technologies
632
