
Using Conditional Generative Adversarial Networks to Boost the
Performance of Machine Learning in Microbiome Datasets
Derek Reiman
a
and Yang Dai
b
Department of Bioengineering, University of Illinois at Chicago, 851 S Morgan St., Chicago, IL 60607, U.S.A.
Keywords: Microbiome, Metagenomics, Generative Adversarial Networks, Data Generation, Data Augmentation.
Abstract: The microbiome of the human body has been shown to have profound effects on physiological regulation and
disease pathogenesis. However, association analysis based on statistical modeling of microbiome data has
continued to be a challenge due to inherent noise, complexity of the data, and high cost of collecting large
number of samples. To address this challenge, we employed a deep learning framework to construct a data-
driven simulation of microbiome data using a conditional generative adversarial network. Conditional
generative adversarial networks train two models against each other while leveraging side information learn
from a given dataset to compute larger simulated datasets that are representative of the original dataset. In our
study, we used a cohorts of patients with inflammatory bowel disease to show that not only can the generative
adversarial network generate samples representative of the original data based on multiple diversity metrics,
but also that training machine learning models on the synthetic samples can improve disease prediction
through data augmentation. In addition, we also show that the synthetic samples generated by this cohort can
boost disease prediction of a different external cohort.
1 INTRODUCTION
The microbiome is a collection of microscopic
organisms cohabitating in a single environment.
These organisms have been shown to have a profound
impact on its environment. Of particular interest is the
human microbiome and how its composition can
affect the health and development of the host. In
particular, the microbiome of the human gut has been
linked to the pathogenesis of metabolic diseases such
as obesity, diabetes mellitus, and inflammatory bowel
disease (Barlow, Yu, & Mathur, 2015; Franzosa et al.,
2019; Tilg & Kaser, 2011). Additionally, the gut
microbiome has been shown to have an effect on the
development and modulation of the central nervous
system (Carabotti, Scirocco, Maselli, & Severi,
2015), stimulation of the immune system (Fung,
Olson, & Hsiao, 2017), and even impact the response
to cancer immunotherapy treatment (Gopalakrishnan,
Helmink, Spencer, Reuben, & Wargo, 2018).
Because of the profound effect that the microbiome
has on the human host, it is of increasing importance
a
https://orcid.org/0000-0002-7955-3980
b
https://orcid.org/0000-0002-7638-849X
to understand how the changes in its composition lead
to physiological changes in the host.
An important analysis in microbiome studies
involves uncovering underlying association between
microbes and the host’s health status. However,
statistical modelling of the underlying distribution of
microbiome data has been a long-standing challenge
due to the sparsity and over-dispersion found in
microbiome data. There have been many approaches
proposed over the past decade, however there is still
no consensus as to which models and underlying
assumptions are best suited for handling the
complexity of the data. (Kurilshikov, Wijmenga, Fu,
& Zhernakova, 2017; Xu, Paterson, Turpin, & Xu,
2015).
Recently, machine learning (ML) models have
been advocated for a data-driven approach for the
prediction of the host phenotype (Knights, Parfrey,
Zaneveld, Lozupone, & Knight, 2011; LaPierre, Ju,
Zhou, & Wang, 2019; Pasolli, Truong, Malik,
Waldron, & Segata, 2016). However, one persistent
challenge is the relatively small size of microbiome
datasets. It is often the case that datasets have a far
Reiman, D. and Dai, Y.
Using Conditional Generative Adversarial Networks to Boost the Performance of Machine Learning in Microbiome Datasets.
DOI: 10.5220/0009892601030110
In Proceedings of the 1st International Conference on Deep Learning Theory and Applications (DeLTA 2020), pages 103-110
ISBN: 978-989-758-441-1
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
103

greater number of features than the number of
samples, which can quickly lead to the overfitting of
models.
To address these challenges and limitations, we
construct a novel method for generating microbiome
data using a conditional generative adversarial
network (CGAN). We then construct synthetic
samples using the generative model in order to
augment the original training set. Data augmentation
is a technique often used in ML to improve task
performance and improve generalization (Bowles et
al., 2018; Mikołajczyk & Grochowski, 2018). By
generating a large number of synthetic microbiome
samples that resemble the original data, we show that
it is possible to improve the performance of ML
models trained on the generated synthetic samples.
Generative adversarial networks (GANs) involve
two neural networks competing against each other in
an adversarial fashion in order to learn a generative
model in a non-parametric data-driven approach
(Goodfellow et al., 2014). GAN models have shown
success in multiple domains including the generation
of medical images (Frid-Adar et al., 2018) and single
cell RNA-Seq gene expression profiles (Ghahramani,
Watt, & Luscombe, 2018). Additionally, synthetic
datasets generated using GAN models have shown to
be able to boost performance of prediction based
tasks through data augmentation (Che, Cheng, Zhai,
Sun, & Liu, 2017). A recent study has also explored
the behaviour of Wasserstein GAN models with
gradient penalty in microbiome data, showing success
in generating realistic data compared to other
simulation techniques (Rong et al., 2019). However,
the utility and benefits of using GANs to generate
microbial synthetic data has not been fully explored.
Specifically, we hypothesize that the synthetic data
generated using GAN models can boost the
performance of downstream analyses.
In our study, we use a variation of standard GAN
models called CGAN. CGANs incorporate side
information into the model to allow the generation of
samples from different distributions when certain
underlying conditions, such as disease status, are
given. CGAN has shown improvement from standard
GAN models (Mirza & Osindero, 2014). The
incorporation of side information also allows for the
training of a single generative model that can
incorporate different conditions.
The main contribution of this manuscript is the
utilization of the CGAN model in order to construct a
generator that can sample from different conditions to
provide synthetic data representative of the true data.
Additionally, we use the generator to synthesize
samples for data augmentation. We show that the
generated data not only are similar to the original data
with respect to diversity metrics, but also that the data
augmentation can lead to statistically significant
improvement in the performance of disease
prediction tasks in ML models.
2 MATERIALS AND METHODS
2.1 Datasets Used in Study
For our study, we use the data reported from two
different cohorts of patients with inflammatory bowel
disease (IBD). The Prospective Registry in IBD
Study at Massachusetts General Hospital (PRISM)
enrolled patients with a diagnosis of IBD based on
endoscopic, radiographic, and histological evidence
of either Crohn’s Disease or Ulcerative Colitis. The
second dataset is used specifically for external
validation and consists of two independent cohorts
from the Netherlands (Tigchelaar et al., 2015). The
first consists of 22 healthy subjects who participated
in the general population study LifeLines-DEEP in
the northern Netherlands. The second cohort consists
of subjects with with IBD from the Department of
Gastroenterology and Hepatology, University
Medical Center Groningen, Netherlands. This
will be used as the validation dataset.
Processing of the stool samples collected for both
datasets is described in the original study (Franzosa et
al., 2019). Briefly, metagenomic data generation and
processing were performed at the Broad Institute in
Cambridge, MA. Quality control for raw sequence
reads was performed and reads were taxonomically
profiled to the species level using MetaPhlAn2
(Segata et al., 2012). The relative abundance values
are publicly available and were obtained from the
original study (Franzosa et al., 2019). A summary
showing the number of IBD patients, healthy
subjects, and species level microbes for each dataset
is shown in Table 1.
Table 1: Datasets used in study.
# IBD # Healthy # Microbes
PRISM 121 34 195
Validation 43 33 115
2.2 CGAN Architecture
In order to generate synthetic microbial community
structures, we utilize a CGAN architecture. A
standard GAN is composed of two competing
networks: a generator and a discriminator. The task of
DeLTA 2020 - 1st International Conference on Deep Learning Theory and Applications
104

the generator is to learn to generate synthetic data
representative of real data while the discriminator
tries to determine if a given sample is synthetic or
real. The generator is trained to maximize the
probability of the discriminator in misclassifying
samples. At the same time, the discriminator is
trained to minimize this probability. A CGAN
expands on standard GAN models by feeding side
information, i.e., the disease status, to both the
generator and discriminator. This allows the
generator to generate synthetic samples conditioned
on the provided side information.
The generator, G, of the CGAN model requires
two sets of inputs: a set of priors and the conditional
side information. In our study, we sample our priors
from the uniform distribution ~U1,1. Both inputs
are fed through multiple fully connected hidden
layers of perceptrons and finally to an output layer.
The output of the generator represents a vector of
microbial abundance features.
The discriminator, 𝐷, takes a sample of microbial
abundance features as an input in addition to the side
information. The inputs are passed through multiple
fully connected layers and then to an output of a
single node using the sigmoid activation function.
The sigmoid function is used so that the output is a
value ranging from 0 and 1. The output of the
discriminator represents the prediction of the
probability that the given sample of data is real.
Both generator and discriminator networks are
trained in an iterative fashion such that in each epoch,
the discriminator is first trained on the generated and
real samples and the network weights are updated.
After the discriminator has been updated, the
generator is updated. The loss functions for the
discriminator and generator are shown below.
𝐿
1
𝑛
log 𝐷𝑥
,𝑠
log
1𝐷
𝐺
𝑧
,𝑠
,𝑠
(1)
𝐿
1
𝑛
log𝐷
𝐺
𝑧
,𝑠
,𝑠
(2)
Here 𝑛 represents the number of real samples, 𝑧
represents a vector of priors for the generator, 𝑥
is
the relative abundance vector of a real microbial
community sample, and 𝑠
is the side information that
the networks are conditioned on. 𝐷𝑥
,𝑠
is the
discriminator’s prediction if 𝑥
is real given the side
information 𝑠
. 𝐺𝑧
,𝑠
is the generator’s prediction
of a synthetic sample given the prior noise 𝑧
and side
information 𝑠
. A figure showing the architecture of
our CGAN is shown in Fig.1.
Figure 1: Visualization of the CGAN architecture. A set of
prior noise 𝑧
and side information 𝑠
corresponding to
sample 𝑥
are used to generate a synthetic sample. The
discriminator then uses the side information to predict if a
given sample is real or synthetic.
3 RESULTS
3.1 CGAN Training
CGAN models were trained only using the PRISM
dataset. Before training, microbial relative abundance
features present in less than 20% of samples or with a
mean abundance less than 0.1% across all samples of
both the PRISM and Validation sets were removed
from the analysis, resulting in a total of 93 microbial
features in the PRISM and Validation datasets.
In our analysis, we sample a vector of size 8 for
the input 𝑧
in the generator model. We add a vector
of size 2 representing the one-hot encoded value of
the disease state (IBD or healthy) as the input 𝑠
and
concatenate the two inputs together. The
concatenated input is then passed through two fully
connected layers of size 128. Batch normalization is
performed at each layer. The leaky ReLU activation
function with an alpha value of 0.1 is performed after
each batch normalization. Unlike the standard ReLU
activation function, leaky ReLU still allows a small
positive gradient for given negative values. The
output layer of the generator is a vector of size 93
representing the microbial features. The softmax
activation function in used in order to reconstruct the
relative abundance of the microbial community.
The discriminator network takes a vector of size
93 representing microbial relative abundance features
as an input in addition to vector of size 2 representing
the one-hot encoded disease state for that sample. The
two inputs are concatenated and fed through two fully
connected layers of size 128. The leaky ReLU
activation is again used for each fully connected
layer. The output of the discriminator is a single node
with a sigmoid activation to shrink the prediction
value to be between 0 and 1.
Using Conditional Generative Adversarial Networks to Boost the Performance of Machine Learning in Microbiome Datasets
105
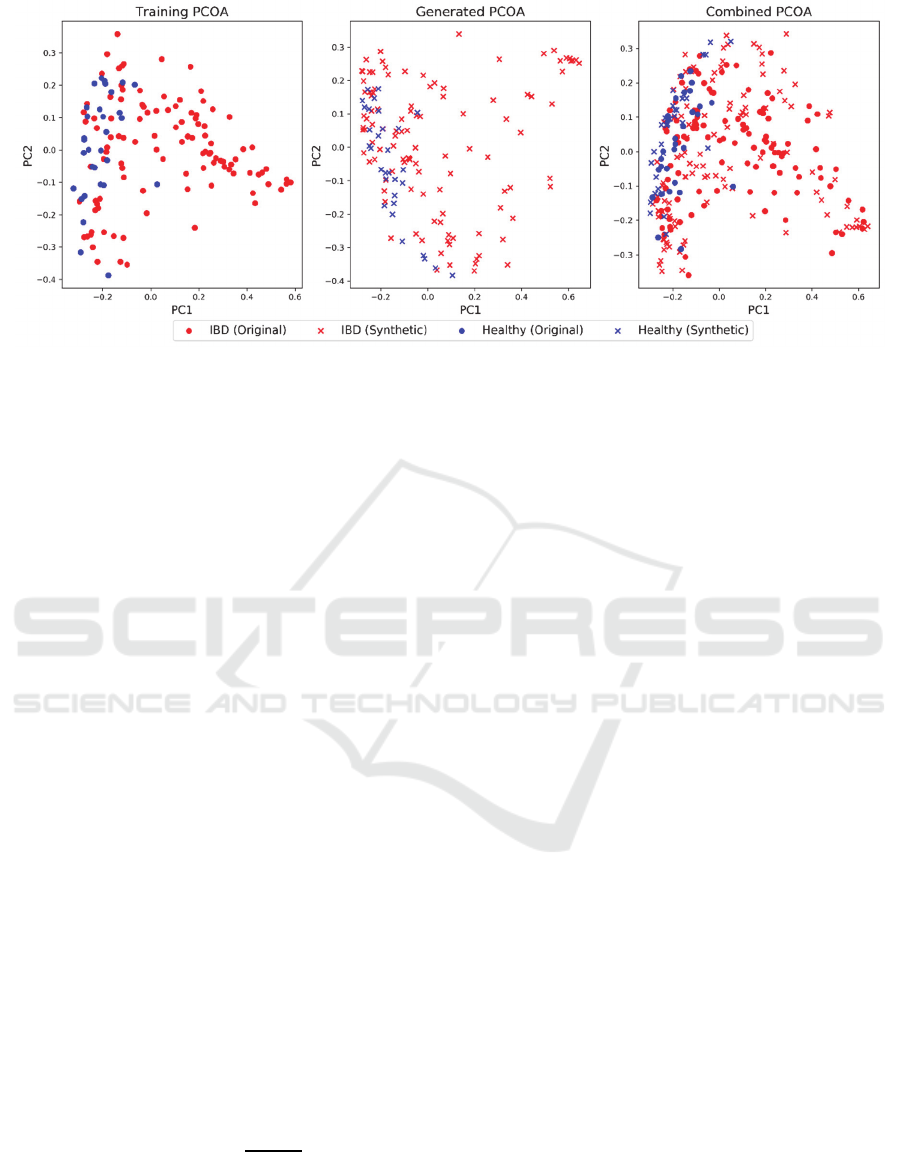
Figure 2: PCOA of the training (left), generated (middles), and combined (right) datasets using the Bray-Curtis dissimilarity.
Red points represent patients with IBD and blue points represent healthy subjects.
Models were trained using 10-fold cross-
validation. In each partition, 90% of the PRISM
dataset was used to train the CGAN model. CGAN
models were trained for 30,000 iterations in which 32
random samples were selected at each iteration as real
samples. A synthetic sample was generated for each
of the 32 real samples using the sample’s respective
disease state as the side information. The 32 real and
32 synthetic samples were then fed to the
discriminator for training and the discriminator was
updated based on Eq. 1. After updating the
discriminator, the discriminator is again used to
predict the synthetic samples and the generator is
updated based on Eq. 2. Both networks were trained
using the ADAM optimizer with a learning rate of
5x10
-5
(Kingma & Ba, 2014). For the implementation
and training of our CGAN models we used the
TensorFlow package in Python (Abadi et al., 2016).
During training, models were saved every 500
iterations. Additionally, the Principal Coordinate
Analysis (PCOA) (Wold, Esbensen, & Geladi, 1987)
of the training set, generated set, and the combination
of the two sets was visualized and stored. The Bray-
Curtis dissimilarity measure was used in calculating
the distance matrix for PCOA (Bray & Curtis, 1957).
The Bray-Curtis dissimilarity quantifies the microbial
compositional dissimilarity between two different
samples. Given two microbial samples, 𝑥
and 𝑥
,
the Bray-Curtis dissimilarity between the two
samples is calculated as
𝐵𝐶
𝑥
,𝑥
1
2𝐶
𝑆
𝑆
(3)
where 𝐶
is the sum of the lesser values for the
abundances of each species found in both 𝑥
and 𝑥
,
and 𝑆
and 𝑆
are the total number of species counted
in 𝑥
and 𝑥
respectively. Visual analysis of the
PCOA plots and the overlap of the original and
generated data was used to select the best model. An
example showing the PCOA of a selected model from
the cross-validated training is shown in Fig. 2.
3.2 Generated Data Improve
Prediction Performance
For each of the partitions in the 10-fold cross-
validation, we simulated 10,000 samples for both IBD
and healthy groups using the selected best model.
Relative abundance values were then log-transformed
and normalized to zero mean and unit variance. Next,
we trained logistic regression and multilayer
perceptron neural network (MLPNN) models to
predict disease status using microbial features. For
each partition of the cross-validation training, two
sets of MLPNN and logistic regression models were
trained. One set of models was trained using the
original samples in the partition of the training set.
The second set of models was trained using the
10,000 simulated samples generated by the CGAN
trained on the training set.
To train a logistic regression model on each 90%
used as training set, we performed internal 5-fold
cross-validation grid search over L1, L2, and Elastic
Net regularizations considering 10 penalty strengths
spaced evenly on a log scale ranging from 1 to 10,000.
Logistic regression models were trained using the
Python scikit-learn package (Pedregosa et al., 2011).
MLPNN models were trained using two fully
connected hidden layers with 256 nodes each and
dropout with a rate of 0.5 after each layer. Leaky
ReLU with an alpha of 0.1 was used as the activation
function. The output layer contained two nodes using
DeLTA 2020 - 1st International Conference on Deep Learning Theory and Applications
106
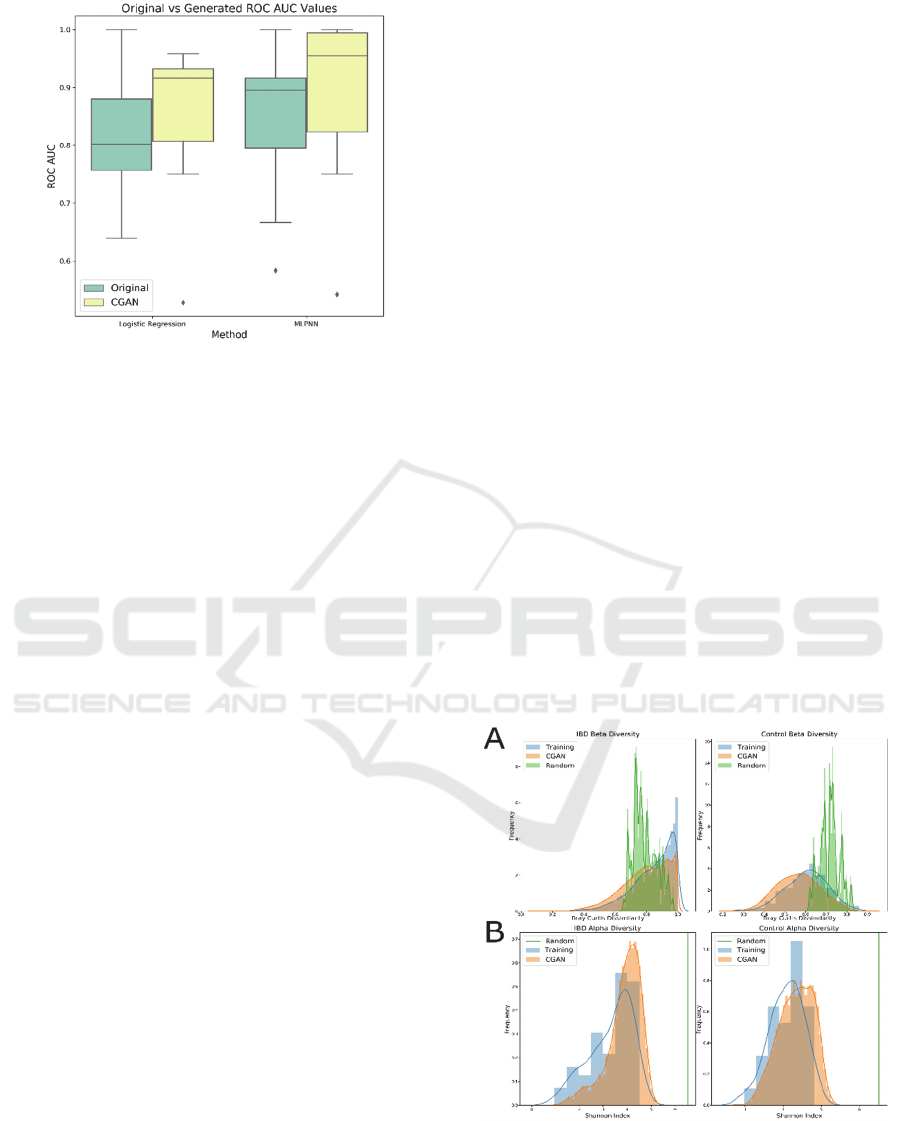
Figure 3: Boxplots for the ROC AUC values across 10-fold
cross-validation for logistic regression and MLPNN models
trained on original and synthetic data.
the softmax activation to predict the disease state.
Networks were trained using the ADAM optimizer
with a learning rate of 1x10
-4
. We set aside 20% of the
training set as a validation set and networks were
trained until the loss of the validation set had not
decreased for 100 epochs. The implementation and
training of the MLPNN models was again done using
the TensorFlow package in Python (Abadi et al., 2016).
Using the trained logistic regression and MLPNN
models generated from a fold’s training set as well as
the generated dataset, we calculated the area under the
receiver operating characteristic curve (ROC AUC)
using the fold’s 10% held out data of true observed
values. We observed that for logistic regression, the
models trained using the generated sets had an
average ROC AUC of 0.849 while the models trained
on the original data had an average ROC AUC of
0.778 across the 10 folds. Similarly, for MLPNN
models, the ROC AUC had a value of 0.889 when
training on the generated data and 0.847 when
training on the original data. Using a Wilcoxon
Signed-Rank test, the ROC AUC when using the
generated samples was significantly larger than that
of when using the original data with a p-value of
0.0249 for logistic regression models and a p-value of
0.0464 for MLPNN models. Boxplots of the ROC
AUC values when using original and generated
datasets is shown in Fig. 3. These results
demonstrated that the CGAN augmented datasets can
boost the predictive power of the ML models.
3.3 Diversity of Generated Data
Diversity metrics are often used to characterize
microbiome samples and datasets. In order to check
how well the generated samples represent the real
samples, we compare the distributions of the alpha
and beta diversities for IBD and healthy samples.
Alpha diversity is a local measure of species
diversity within a sample. It characterizes the
microbial richness of a community. For our analysis,
we use the Shannon Entropy metric to quantify the
alpha diversity of samples. Given a sample 𝑥 with 𝑚
relative abun-dance values, the Shannon Entropy is
calculated as
𝐻
𝑥
𝑥
log
𝑥
(4)
Beta diversity, on the other hand, allows us to
quantify how similar samples are to each other. In our
study, we use the Bray-Curtis dissimilarity as a
distance measure of beta diversity, calculated as
described in Eq. 3.
To demonstrate the behaviour of the CGAN
model, we visualize the diversity metrics for the
training set and for 10,000 generated samples using
the selected best model. In addition, we calculate the
diversity metrics of a set of 10,000 generated samples
using the random initialization of the CGAN before
any training to show the initial random distribution.
Before calculating the diversity metrics, we
clipped the generated samples in order to introduce
zero values. The softmax function used to generate
samples provides a vector entirely of positive values.
However, in reality microbiome data very sparse.
Figure 4: Distributions of (A) the beta diversity based on
the Bray-Curtis dissimilarity between the training set and
itself, the generated (CGAN), and random datasets, and (B)
the Shannon alpha diversity of training, generated, and
random samples for IBD (left) and healthy (right) samples.
Using Conditional Generative Adversarial Networks to Boost the Performance of Machine Learning in Microbiome Datasets
107
Therefore, to induce this sparsity into the
generated samples, we calculated the minimum value
across all species found in the training set. We used
this value as a threshold and set any generated value
less than the observed minimum to zero.
After clipping the generated sets, we calculated
the diversity metrics. When considering beta
diversity, we only considered the Bray-Curtis
dissimilarity from the training set to itself, the training
set to the best generated samples, and the training set
to the randomly generated samples. The distributions
of alpha and beta diversity for one of the cross-
validated partitions is shown in Fig. 4.
We observed that the data generated from the
selected best model followed very similar
distributions of the alpha and beta diversities of the
data used to train the CGAN. We did notice that the
beta diversity within the training set had a spike near
one, however upon post-analysis we discovered that
was caused by samples with only a few numbers of
microbial species present.
3.4 Generated Data Is Predictive of
External Dataset
To evaluate if the synthetic samples generated from
the CGAN model were generalizable to a dataset of a
similar study, we trained a CGAN model using the
entire PRISM dataset in the same manner as
described in Section 3.1. The CGAN is trained for
30,000 iterations and models as well as PCOA
visualization of the real and synthetic samples are
saved every 500 iterations. The best model is selected
based on the PCOA comparison between the training
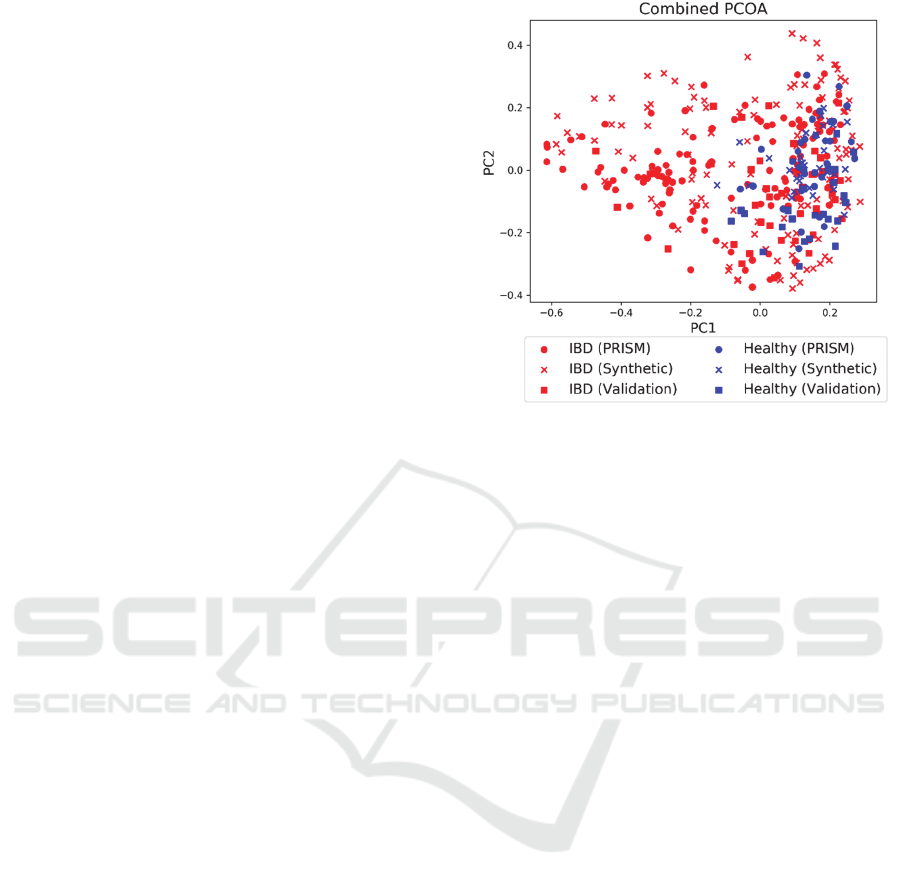
and generated sets. A PCOA visualization of the
PRISM dataset combined with the synthetic data
generated from the best model and the external
validation set is shown in Fig. 5.
Using the best model, we evaluate if the generated
samples can improve the task of predicting IBD
status. Logistic regression and MLPNN models are
trained in a similar fashion as outlined in Section 3.2.
The model was trained using 10,000 generated
samples from a CGAN model that was trained on the
entire PRISM dataset. We then evaluate the model
performance on the true observations of the external
validation IBD dataset. We observed an improvement
in ROC AUC from 0.734 to 0.832 in logistic
regression models and from 0.794 to 0.849 in
MLPNN models. This demonstrates that the synthetic
samples generated using one cohort can augment the
analysis of a different cohort.
Lastly, we analyse the distribution of alpha and
beta diversities of the original PRISM dataset, the
Figure 5: PCOA visualization of the combination of the
PRISM dataset, synthetic data generated by the best CGAN
model, and the external validation set. Red points represent
patients with IBD and blue points represent healthy
patients.
samples generated after training a CGAN on the
whole PRISM dataset, and the external validation
dataset. The alpha diversity is calculated for each
dataset using the Shannon Entropy metric. The beta
diversity within the PRISM dataset, from the PRISM
dataset to the generated samples, and from the
external validation dataset to the generated samples
was calculated. In addition, we compared the random
diversities from the randomly initialized CGAN
before training. The alpha and beta diversities are
shown in Fig. 6.
We observed that the beta diversity between the
PRISM dataset and the synthetic samples generated
from it displays similar distributions. Additionally,
the distribution of the beta diversity values between
the external validation set and the synthetic samples
follow a similar pattern, suggesting that the CGAN
model did not overfit the PRISM dataset and is robust
in generating synthetic samples. We also observed
that the alpha diversities within the PRISM, synthetic,
and external validation datasets showed similar
distributions. In particular, the alpha diversity within
the samples of IBD patients was very similar. The
distributions in the healthy samples were slightly
different in each of the datasets, however we suspect
this may be due to the fact that there were far fewer
cases of healthy samples in the original PRISM
dataset.
DeLTA 2020 - 1st International Conference on Deep Learning Theory and Applications
108
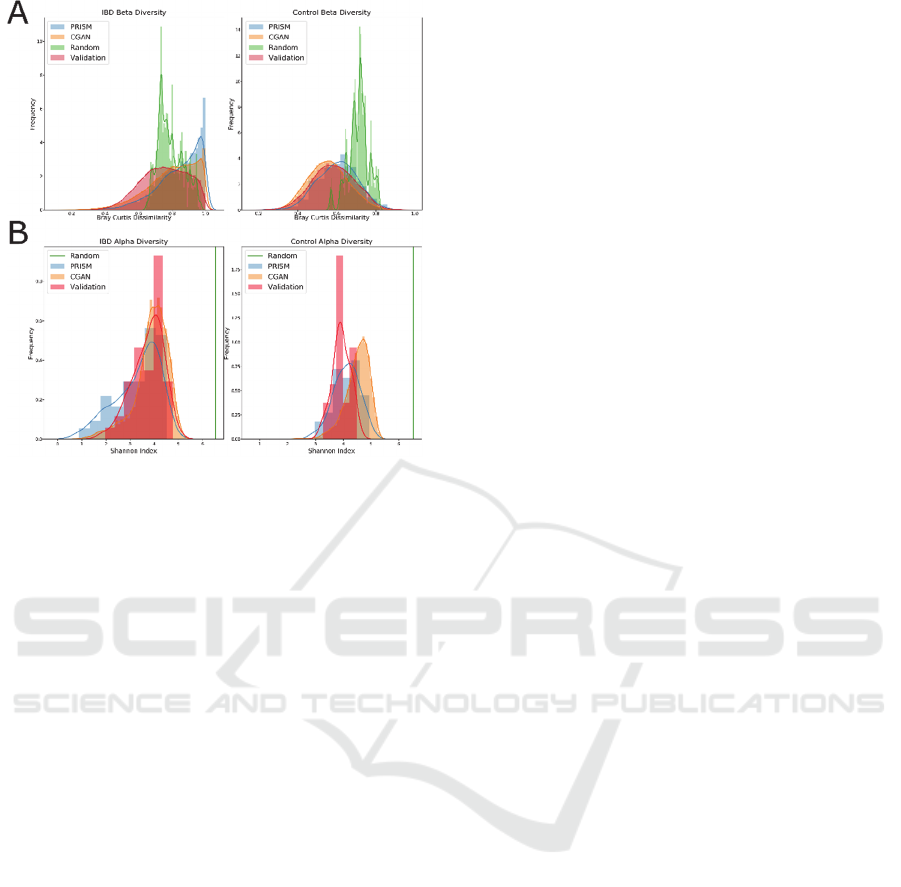
Figure 6: Distributions of (A) beta diversity based on the
Bray Curtis dissimilarity between the training set and itself,
the validation, the generated (CGAN), and random datasets
and (B) Shannon alpha diversity of training, validation,
generated, and random samples for IBD (left) and healthy
(right) samples.
4 CONCLUSIONS
In this study, we have developed a novel approach for
the generation of synthetic microbiome samples using
a CGAN architecture in order to augment ML
analyses. Using two different cohorts of subjects with
IBD, we have demonstrated that the synthetic
samples generated from the CGAN are similar to the
original data in both alpha and beta diversity metrics.
In addition, we have shown that augmenting the
training set by using a large number of synthetic
samples can improve the performance of logistic
regression and MLPNN in predicting host phenotype.
A current limitation to this approach involves
selecting the best CGAN model. Even though visual
inspection has been a common approach, it is a
subjective and may miss the optimal model. We plan
to further this study by investigating stopping criteria
using alpha and beta diversity metrics in order to
facilitate CGAN model selection. In addition, we plan
to evaluate other forms of side information such as
using time in longitudinal datasets.
REFERENCES
Abadi, M., Barham, P., Chen, J., Chen, Z., Davis, A., Dean,
J., Isard, M. (2016). Tensorflow: A system for large-
scale machine learning. Paper presented at the 12th
{USENIX} Symposium on Operating Systems Design
and Implementation ({OSDI} 16).
Barlow, G. M., Yu, A., & Mathur, R. (2015). Role of the
gut microbiome in obesity and diabetes mellitus.
Nutrition in clinical practice, 30(6), 787-797.
Bowles, C., Chen, L., Guerrero, R., Bentley, P., Gunn, R.,
Hammers, A., Rueckert, D. (2018). GAN
Augmentation: Augmenting Training Data using
Generative Adversarial Networks.
Bray, J. R., & Curtis, J. T. (1957). An Ordination of the
Upland Forest Communities of Southern Wisconsin.
Ecological Monographs, 27(4), 326-349.
doi:10.2307/1942268
Carabotti, M., Scirocco, A., Maselli, M. A., & Severi, C.
(2015). The gut-brain axis: interactions between enteric
microbiota, central and enteric nervous systems. Annals
of gastroenterology, 28(2), 203-209.
Che, Z., Cheng, Y., Zhai, S., Sun, Z., & Liu, Y. (2017, 18-
21 Nov. 2017). Boosting Deep Learning Risk
Prediction with Generative Adversarial Networks for
Electronic Health Records. Paper presented at the 2017
IEEE International Conference on Data Mining
(ICDM).
Franzosa, E. A., Sirota-Madi, A., Avila-Pacheco, J.,
Fornelos, N., Haiser, H. J., Reinker, S., Xavier, R. J.
(2019). Gut microbiome structure and metabolic
activity in inflammatory bowel disease. Nature
microbiology, 4(2), 293-305. doi:10.1038/s41564-018-
0306-4
Frid-Adar, M., Diamant, I., Klang, E., Amitai, M.,
Goldberger, J., & Greenspan, H. (2018). GAN-based
synthetic medical image augmentation for increased
CNN performance in liver lesion classification.
Neurocomputing, 321, 321-331.
Fung, T. C., Olson, C. A., & Hsiao, E. Y. (2017).
Interactions between the microbiota, immune and
nervous systems in health and disease. Nature
neuroscience, 20(2), 145.
Ghahramani, A., Watt, F. M., & Luscombe, N. M. (2018).
Generative adversarial networks simulate gene
expression and predict perturbations in single cells.
BioRxiv, 262501.
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B.,
Warde-Farley, D., Ozair, S., Bengio, Y. (2014).
Generative adversarial nets. Paper presented at the
Advances in neural information processing systems.
Gopalakrishnan, V., Helmink, B. A., Spencer, C. N.,
Reuben, A., & Wargo, J. A. (2018). The influence of
the gut microbiome on cancer, immunity, and cancer
immunotherapy. Cancer cell, 33(4), 570-580.
Kingma, D., & Ba, J. (2014). Adam: A Method for
Stochastic Optimization. International Conference on
Learning Representations.
Knights, D., Parfrey, L. W., Zaneveld, J., Lozupone, C., &
Knight, R. (2011). Human-associated microbial
Using Conditional Generative Adversarial Networks to Boost the Performance of Machine Learning in Microbiome Datasets
109

signatures: examining their predictive value. Cell host
& microbe, 10(4), 292-296. doi:10.1016/j.chom.
2011.09.003
Kurilshikov, A., Wijmenga, C., Fu, J., & Zhernakova, A.
(2017). Host Genetics and Gut Microbiome: Challenges
and Perspectives. Trends in Immunology, 38(9), 633-
647. doi:https://doi.org/10.1016/j.it.2017.06.003
LaPierre, N., Ju, C. J. T., Zhou, G., & Wang, W. (2019).
MetaPheno: A critical evaluation of deep learning and
machine learning in metagenome-based disease
prediction. Methods. doi: https://doi.org/10.1016/
j.ymeth.2019.03.003
Mikołajczyk, A., & Grochowski, M. (2018, 9-12 May
2018). Data augmentation for improving deep learning
in image classification problem. Paper presented at the
2018 International Interdisciplinary PhD Workshop
(IIPhDW).
Mirza, M., & Osindero, S. (2014). Conditional generative
adversarial nets. arXiv preprint arXiv:1411.1784.
Pasolli, E., Truong, D. T., Malik, F., Waldron, L., & Segata,
N. (2016). Machine learning meta-analysis of large
metagenomic datasets: tools and biological insights.
PLOS Computational Biology, 12(7).
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V.,
Thirion, B., Grisel, O., Duchesnay, É. (2011). Scikit-
learn: Machine Learning in Python. J. Mach. Learn.
Res., 12(null), 2825–2830.
Rong, R., Jiang, S., Xu, L., Xiao, G., Xie, Y., Liu, D. J.,
Zhan, X. (2019). MB-GAN: Microbiome Simulation
via Generative Adversarial Network. BioRxiv, 863977.
doi:10.1101/863977
Segata, N., Waldron, L., Ballarini, A., Narasimhan, V.,
Jousson, O., & Huttenhower, C. (2012). Metagenomic
microbial community profiling using unique clade-
specific marker genes. Nature Methods, 9(8), 811-814.
doi:10.1038/nmeth.2066
Tigchelaar, E. F., Zhernakova, A., Dekens, J. A. M.,
Hermes, G., Baranska, A., Mujagic, Z., Feskens, E. J.
M. (2015). Cohort profile: LifeLines DEEP, a
prospective, general population cohort study in the
northern Netherlands: study design and baseline
characteristics. BMJ open, 5(8), e006772-e006772.
doi:10.1136/bmjopen-2014-006772.
Tilg, H., & Kaser, A. (2011). Gut microbiome, obesity, and
metabolic dysfunction. The Journal of Clinical
Investigation, 121(6), 2126-2132.
Wold, S., Esbensen, K., & Geladi, P. (1987). Principal
component analysis. Chemometrics and intelligent
laboratory systems, 2(1-3), 37-52.
Xu, L., Paterson, A. D., Turpin, W., & Xu, W. (2015).
Assessment and Selection of Competing Models for
Zero-Inflated Microbiome Data. PloS one, 10(7),
e0129606-.doi:10.1371/journal.pone.0129606.
DeLTA 2020 - 1st International Conference on Deep Learning Theory and Applications
110
