
Multi-channel ConvNet Approach to Predict the Risk of in-Hospital
Mortality for ICU Patients
Fabien Viton, Mahmoud Elbattah, Jean-Luc Guérin and Gilles Dequen
Laboratoire MIS, Université de Picardie Jules Verne, Amiens, France
Keywords: ConvNet, Deep Learning, Multivariate Time Series, in-Hospital Mortality.
Abstract: The healthcare arena has been undergoing impressive transformations thanks to advances in the capacity to
capture, store, process, and learn from data. This paper re-visits the problem of predicting the risk of in-
hospital mortality based on Time Series (TS) records emanating from ICU monitoring devices. The problem
basically represents an application of multi-variate TS classification. Our approach is based on utilizing
multiple channels of Convolutional Neural Networks (ConvNets) in parallel. The key idea is to disaggregate
multi-variate TS into separate channels, where a ConvNet is used to extract features from each univariate TS
individually. Subsequently, the features extracted are concatenated altogether into a single vector that can be
fed into a standard MLP classification module. The approach was experimented using a dataset extracted from
the MIMIC-III database, which included about 13K ICU-related records. Our experimental results show a
promising accuracy of classification that is competitive to the state-of-the-art.
1 INTRODUCTION
Healthcare services are delivered in data-rich
environments where a wealth of data is created at
multiple levels of operational and medical records. In
view of that, data analytics is increasingly becoming
a key enabling thrust to leverage of such massive data
amounts. An important part of the analytics
capabilities is based on Time Series (TS) data.
Applications of TS analytics are essential in a wide
diversity of domains, especially healthcare where the
use of temporal data is ubiquitous.
However, the multi-dimensionality of TS data
brings up further challenges regarding the extraction
and selection of features. In this respect, Deep
Learning (LeCun, Bengio, and Hinton 2015) could
present as an appropriate approach. Deep Learning
allows for learning hierarchical feature
representations from raw data automatically. Deep
architectures of Convolutional Neural Networks
(ConvNets) (LeCun et al. 1989; LeCun et al. 1998)
have been successfully implemented in complex
tasks. Examples include Computer Vision and
Machine Translation (e.g. Krizhevsky, Sutskever,
and Hinton 2012; Gehring et al. 2017).
Correspondingly, Deep Learning has also been
considered as an attractive path for tackling tough TS
problems. Particularly, in the case of multiple
variates, complex relationships, and large amounts of
data. There has been a growing interest over the past
few years in this regard (e.g. Karim et al. 2019), as an
alternative approach to avoid the need for developing
conventional hand-crafted features.
In this context, this study approaches a multi-
variate TS problem. The task is to predict the risk of
in-hospital mortality among ICU patients. The
problem under consideration represents a typical
application of multi-variate TS classification. Using a
multi-channel architecture, our approach utilizes
multiple ConvNets to extract features from each
univariate TS individually. The experiments used a
dataset extracted from the MIMIC-III database,
which provides a freely accessible repository of ICU
records (Johnson et al. 2016).
The study attempts to make contributions in two
aspects. On one hand, the study is conceived to
contribute to the ongoing efforts towards availing of
Deep Learning methods for multi-variate TS
problems. While from a practical standpoint, the
performance of channel-wise ConvNet architectures
is explored with respect to the problem of predicting
in-hospital mortality.
98
Viton, F., Elbattah, M., Guérin, J. and Dequen, G.
Multi-channel ConvNet Approach to Predict the Risk of in-Hospital Mortality for ICU Patients.
DOI: 10.5220/0009891900980102
In Proceedings of the 1st International Conference on Deep Learning Theory and Applications (DeLTA 2020), pages 98-102
ISBN: 978-989-758-441-1
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All r ights reserved

2 RELATED WORK
The problem of TS classification was notably
identified as one of the key challenges in Data Mining
research (Yang and Wu, 2006). Distance-based
methods such as Dynamic Time Warping (DTW)
have been long recognized as the most performing
technique in this respect (Berndt and Clifford, 1994).
The development of feature-based similarity
measures was also explored (Fulcher, 2018).
However, the intensive process of pre-processing and
feature extraction was generally considered as a
limiting factor. While the analysis and forecasting of
TS have been dominated by regression-based
modelling methods such as Auto Regressive
Integrated Moving Average (ARIMA).
With recent advances, Machine Learning (ML)
has increasingly come into prominence, especially for
complex multivariate TS problems. Various
implementations of ConvNet and Recurrent Neural
Network (RNN) architectures were successfully
applied for TS classification tasks. For instance, a
ConvNet-based framework was proposed by (Cui,
Chen, and Chen, 2016). ConvNets were exploited to
automatically extract features through a sequence of
convolution and pooling operations. The extracted
features could represent the internal structure of the
input TS. Furthermore, (Wang, Yan, and Oates, 2017)
demonstrated that ConvNet models could provide
better performance over traditional DTW methods.
Further efforts concentrated on employing Deep
Learning potentials for complex TS problems that
involve large-scale datasets and multiple variables.
For example, a ConvNet-based feature extractor was
developed for multivariate TS classification (Zheng
et al. 2016). (Purushotham et al. 2017) provided an
exhaustive evaluation of Deep Learning against other
ML models based on the MIMIC dataset. They
demonstrated that Deep learning consistently
outperformed other approaches, especially in the case
of large multi-variate TS data.
Other studies experimented Long Short-Term
Memory (LSTM) models. For example, (Siami-
Namini, Tavakoli, and Namin, 2018) reported that
LSTM outperformed traditional algorithms including
ARIMA. Another study explored the use of bi-
directional LSTMs, which provided a better
performance as well (Siami-Namini, Tavakoli, and
Namin, 2019). A detailed presentation of such efforts
would go beyond the scope of this study, but (Fawaz
et al. 2019) provides a comprehensive review of the
state-of-the-art Deep Learning implementations for
TS classification.
3 DATA DESCRIPTION
The study used a dataset extracted from the MIMIC-
III database (Johnson et al. 2016). The MIMIC
database provides a rich repository of ICU admissions
to the Beth Israel Deaconess Medical Center in
Boston between 2001 and 2012. It is considered to be
one of the largest databases of its kind publicly
available. It has been utilized in plentiful studies (e.g.
Desautels et al. 2016; Komorowski et al. 2018).
The dataset comprised more than 13K patient
records related to a variety of ICU admissions
including cardiac, medical, surgical, trauma, and
others. The TS variables described the patient status
over the 48-hour timespan after admission (e.g. heart
rate, blood pressure, temperature, etc.). Specifically,
the dataset included 17 temporal measurements,
which represented the typical readings used during
ICU monitoring (Silva et al. 2012). As such, a
(17x48) matrix could describe the development of
each ICU patient. The dataset variables are listed in
Table1 below.
For each case, a binary label corresponded to the
outcome (i.e. in-hospital mortality). The mortality
rate among patients was about 9%. To establish a
benchmark for comparison, the dataset was prepared
following the set of procedures provided by
(Harutyunyan et al. 2019). Under normal conditions,
some variables could suffer from missing values (e.g.
blood or urine samples). To fill in missing values, we
applied values from the previous time point. All
variables were normalized with zero mean and unit
standard deviation.
Table 1: Dataset variables.
Variables
Heart Rate
Respiratory Rate
Capillary Refill Rate
Systolic Blood Pressure
Diastolic Blood Pressure
Mean Blood Pressure
Fraction Inspired Oxygen (FiO
2
)
Oxygen Saturation (SaO
2
)
Temperature
Glucose
pH
Glascow Coma Scale Eye Opening
Glascow Coma Scale Motor Response
Glascow Coma Scale Verbal Response
Glascow Coma Scale Total
Height
Weight
Multi-channel ConvNet Approach to Predict the Risk of in-Hospital Mortality for ICU Patients
99
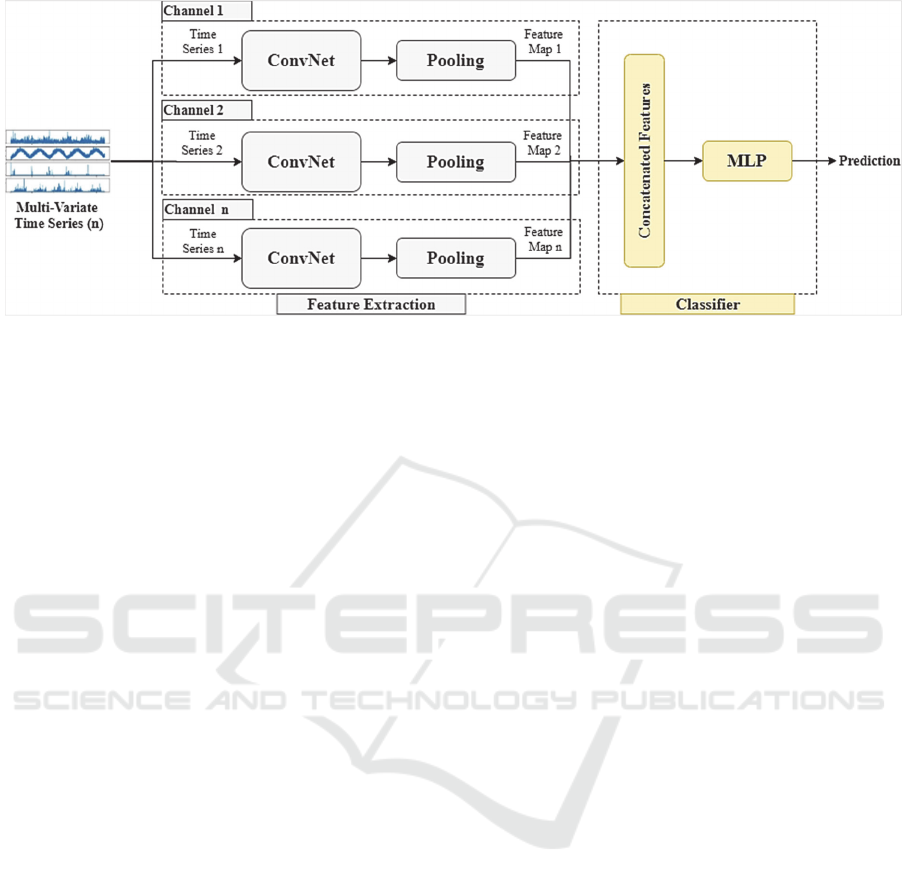
Figure 1: Approach overview.
4 APPROACH OVERVIEW
The approach is based on the multi-channel ConvNet
architecture proposed by (Zheng et al. 2014). The
architecture includes a combination of unsupervised
and supervised learning over two stages as follows.
4.1 Feature Extraction
Learning representations is a fundamental question
for ML. In this respect, ConvNets introduced a potent
mechanism to automatically learn complex patterns
from raw data. ConvNets were cleverly designed to
deal with the spatial or temporal variability
underlying data. Likewise, ConvNets were utilized in
our case to learn features from the TS data, and hence
eliminating the need to develop hand-crafted features.
Initially, the multi-variate TS is separated into
univariate channels. For each channel, hierarchical
features are extracted through operations of
convolution and pooling. The convolutional layers
extract temporal patterns by applying 1-D filters over
TS sequences. The convolutions are followed by a
ReLU activation layer, which introduces the non-
linearity into the learning process. Subsequently, a
global average pooling is applied, which computes
the mean value of filters across the time dimension.
The operations conducted on each TS channel are
described in the equations below.
y
i
= Wi ⁎ TS
i
+ b (1)
h
i
= ReLU(y
i
) (2)
feat
i
= GlobalAveragePooling(h
i
) (3)
Where y
i
is the output filter and ⁎ is the convolution
operation, and feat
i
is the output feature map.
4.2 MLP Classifier
The output of each ConvNet channel is a feature map
that can be regarded as a compressed representation
of the input TS. The feature maps are subsequently
concatenated to jointly form a single feature map.
Eventually, the aggregated feature map is fed to a
conventional MLP classifier, which would be trained
to perform the classification task.
Supervised training is basically performed at this
stage. The filter coefficients output from ConvNet
channels are updated simultaneously during the MLP
learning process. The classifier may include a single
layer or multiple hidden layers, which would perform
further non-linear transformation of the feature map.
Figure 1 sketches the approach architecture.
5 EMPIRICAL EXPERIMENTS
As alluded earlier, the goal was to predict the risk of
in-hospital mortality based on the initial 48h interval
of ICU monitoring. The MIMIC dataset was
randomly divided into 75% train and 25% test
portions. The hyperparameters (e.g. filter size) of
ConvNet channels were decided empirically.
Specifically, we could achieve the highest accuracy
with filter size= 8, and number of filters=8. Given 17
TS variables, the model included 136 convolutional
filters (i.e. 17*8), and the output layer was composed
of 136 weights. The model was trained using Adam
optimizer (Kingma and Ba, 2014).
DeLTA 2020 - 1st International Conference on Deep Learning Theory and Applications
100
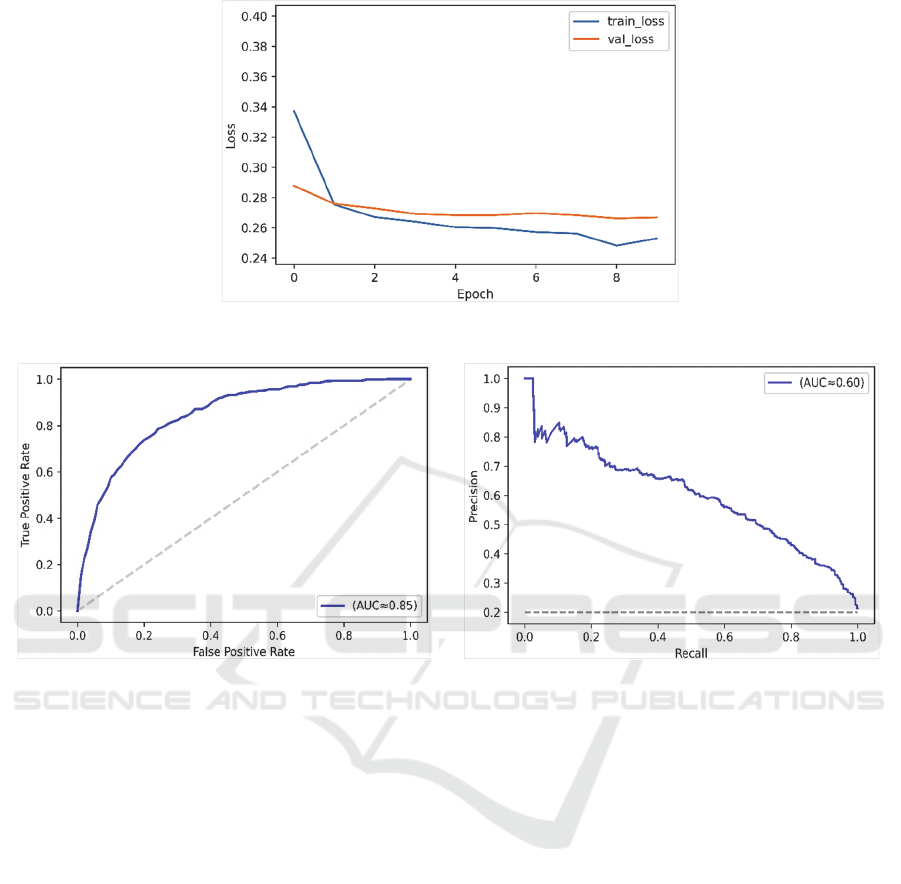
Figure 2: Model loss in training and validation sets.
Figure 3: ROC curve.
Figure 4: Precision-Recall curve.
Various structures of MLP were experimented for
training the model. It turned out that the best
performance could be achieved using 3 fully
connected layers. Specifically, the hidden layers
consisted of 64, 32 and 16 neurons, respectively.
Further, the dropout technique was applied to help
reduce the model over-fitting (Srivastava et al, 2014).
Figure 2 plots the model loss in training and
validation over 10 epochs with 20% of the dataset
used for validation.
Figure 3 examines the classification accuracy
based on the Receiver Operating Characteristics
(ROC) curve. The ROC curve plots the relationship
between the true positive rate and the false positive
rate across a full range of possible thresholds. The
model could achieve a very good accuracy (AUC-
ROC≈0.85). Figure 4 plots the Precision-Recall
curve, which is particularly important in the case of
imbalanced datasets (AUC-PR≈0.60).
Overall, the model could largely provide
comparable performance to the literature.
Furthermore, we could achieve a relatively higher
AUC-PR compared to the work conducted by
(Harutyunyan et al. 2019), which did not include the
multi-channel ConvNet approach. The experiments
were implemented using the Keras library (Chollet,
2015) with the TensorFlow (Abadi et al. 2016)
backend. The model implementation is shared on the
GitHub repository (Elbattah, 2020).
6 CONCLUSIONS
The multi-channel ConvNet approach could yield
promising results applied to the problem of predicting
in-hospital mortality. Despite using a relatively
simple ConvNet architecture, the accuracy achieved
is competitive to the state-of-the-art. It is conceived
that further improvements could be realized by
applying more sophisticated architectures.
Our future work aims to explore further
interesting prospects. On one hand, we are concerned
with the explainability of predictions. Analyzing and
visualizing the output of ConvNet channels may be
employed to bring insights into the most influential
Multi-channel ConvNet Approach to Predict the Risk of in-Hospital Mortality for ICU Patients
101

variables on the predicted outcome. On the other
hand, with the mounting successful applications of
Transfer Learning, we endeavor to explore that path
as well. Transfer Learning methods might be a key to
improve the performance by fine-tuning pre-trained
models rather than training from scratch.
REFERENCES
Abadi, M., Barham, P., Chen, J., Chen, Z., Davis, A., Dean,
J., ... & Kudlur, M. (2016). Tensorflow: A system for
large-scale machine learning. In Proceedings of the
12th USENIX Symposium on Operating Systems Design
and Implementation (pp. 265-283).
Berndt, D. J., & Clifford, J. (1994). Using dynamic time
warping to find patterns in time series. In Proceedings
of KDD workshop (Vol. 10, No. 16, pp. 359-370).
Chollet, F., (2015). Keras. https://github.com/fchollet/keras
Cui, Z., Chen, W., & Chen, Y. (2016). Multi-scale
convolutional neural networks for time series
classification. arXiv preprint arXiv:1603.06995.
Desautels, T., Calvert, J., Hoffman, J., Jay, M., Kerem, Y.,
Shieh, L., ... & Wales, D. J. (2016). Prediction of sepsis
in the intensive care unit with minimal electronic health
record data: a machine learning approach. JMIR
Medical Informatics, 4(3), e28.
Elbattah, M., 2020. https://github.com/Mahmoud-
Elbattah/DeLTA2020.
Fawaz, H. I., Forestier, G., Weber, J., Idoumghar, L., &
Muller, P. A. (2019). Deep learning for time series
classification: a review. Data Mining and Knowledge
Discovery, 33(4), 917-963.
Fulcher, B. D. (2018). Feature-based time-series analysis.
In Feature Engineering for Machine Learning and
Data Analytics (pp. 87-116). CRC Press.
Gehring, J., Auli, M., Grangier, D., Yarats, D., & Dauphin,
Y. N. (2017). Convolutional sequence to sequence
learning. In Proceedings of the 34th International
Conference on Machine Learning (ICML)-Volume 70
(pp. 1243-1252).
Harutyunyan, H., Khachatrian, H., Kale, D. C., Ver Steeg,
G., & Galstyan, A. (2019). Multitask learning and
benchmarking with clinical time series data. Scientific
Data, 6(1), 1-18.
Karim, F., Majumdar, S., Darabi, H., & Harford, S. (2019).
Multivariate LSTM-FCNs for time series classification.
Neural Networks, 116, 237-245.
Kingma, D. P., and Ba, J. (2014). Adam: A method for
stochastic optimization. arXiv preprint arXiv:
1412.6980.
Komorowski, M., Celi, L. A., Badawi, O., Gordon, A. C.,
& Faisal, A. A. (2018). The artificial intelligence
clinician learns optimal treatment strategies for sepsis
in intensive care. Nature Medicine, 24(11), 1716-1720.
Krizhevsky, A., Sutskever, I., & Hinton, G. E. (2012).
ImageNet classification with deep convolutional neural
networks. In Proceedings of Advances in Neural
Information Processing Systems (pp. 1097-1105).
Kumar, M., & Singh, S. (2009). Engineering Mathematics-
III. KRISHNA Prakashan Media, Meerut, India.
LeCun, Y., Boser, B. E., Denker, J. S., Henderson, D.,
Howard, R. E., Hubbard, W. E., and Jackel, L. D.
(1989). Handwritten digit recognition with a back-
propagation network. In Proceedings of Advances in
Neural Information Processing Systems (NIPS) (pp.
396-404).
LeCun, Y., Bottou, L., Bengio, Y., & Haffner, P. (1998).
Gradient-based learning applied to document
recognition. In Proceedings of the IEEE, 86(11), 2278-
2324.
LeCun, Y.; Bengio, Y.; and Hinton, G. (2015). Deep
learning. Nature 521(7553):436–444.
Purushotham, S., Meng, C., Che, Z., & Liu, Y. (2017).
Benchmark of deep learning models on large healthcare
MIMIC datasets. arXiv preprint arXiv:1710.08531.
Johnson, A. E., Pollard, T. J., Shen, L., Li-wei, H. L., Feng,
M., Ghassemi, M., ... & Mark, R. G. (2016). MIMIC-
III, a freely accessible critical care database. Scientific
Data, 3, 160035.
Siami-Namini, S., Tavakoli, N., & Namin, A. S. (2018,
December). A comparison of ARIMA and LSTM in
forecasting time series. In 2018 17th IEEE International
Conference on Machine Learning and Applications
(ICMLA) (pp. 1394-1401). IEEE.
Siami-Namini, S., Tavakoli, N., & Namin, A. S. (2019). A
Comparative Analysis of Forecasting Financial Time
Series Using ARIMA, LSTM, and BiLSTM. arXiv
preprint arXiv:1911.09512.
Silva, I., Moody, G., Scott, D. J., Celi, L. A., & Mark, R. G.
(2012). Predicting in-hospital mortality of ICU patients:
The PhysioNet/Computing in Cardiology Challenge
2012. In Proceedings of the 2012 Computing in
Cardiology (pp. 245-248). IEEE.
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I., &
Salakhutdinov, R. (2014). Dropout: a simple way to
prevent neural networks from overfitting. The Journal
of Machine Learning Research, 15(1), 1929-1958.
Wang, Z., Yan, W., & Oates, T. (2017). Time series
classification from scratch with deep neural networks:
A strong baseline. In Proceedings of the 2017
International Joint Conference on Neural Networks
(IJCNN) (pp. 1578-1585). IEEE.
Yang, Q., & Wu, X. (2006). 10 Challenging problems in
data mining research. International Journal of
Information Technology & Decision Making, 5(04),
597-604.
Zheng, Y., Liu, Q., Chen, E., Ge, Y., & Zhao, J. L. (2014,
June). Time series classification using multi-channels
deep convolutional neural networks. In International
Conference on Web-Age Information Management (pp.
298-310). Springer, Cham.
Zheng, Y., Liu, Q., Chen, E., Ge, Y., & Zhao, J. L. (2016).
Exploiting multi-channels deep convolutional neural
networks for multivariate time series classification.
Frontiers of Computer Science, 10(1), 96-112.
DeLTA 2020 - 1st International Conference on Deep Learning Theory and Applications
102
