
A Case Study on Performance Optimization Techniques in Java
Programming
Ciprian Khlud
a
and Cristian Fr
˘
asinaru
b
Alexandru Ioan Cuza University, Ias¸i, Romania
Keywords:
Java, Runtime Performance, Memory Usage, Garbage Collection, Sequence Analysis, SAM/BAM Files.
Abstract:
Choosing the right programming platform for processor or memory intensive applications is a subject that is
debated in all types of contexts. In this paper we investigate how a state-of-the art implementation, part of a
multi-threaded framework for sequence analysis (elPrep) could benefit from various optimization techniques
dedicated to improving the runtime performance of Java applications. We show that, without changing the
semantics of the algorithm, by using appropriate programming techniques we are able to significantly improve
the behavior of the Java implementation to a point that may even alter the conclusions of the original study.
We also show that, by changing the manner in which data is represented, to better fit the particulars of the Java
memory management, we are able to improve the original scoring (based on computing time and memory
consumption) to around one order of magnitude better on the most expensive component (read/write).
1 INTRODUCTION
In the field of bioinformatics, DNA sequence anal-
ysis (D
¨
oring et al., 2008) generally consists of pro-
cessing large amounts of data and performing various
operations on it, such as sequence alignment, variant
detection, searches against biological databases, etc.
A large variety of software tools exist for these opera-
tions, most of them having specific uses cases but with
a common denominator regarding the fact they need
to perform processor and memory intensive tasks: I/O
operations on large file, compression/decompression,
text processing, etc.
Choosing a programming platform that offers all
the required instruments to handle the specific chal-
lenges in bioinformatics is important, as pointed out
in a recent study dedicated to migrating an existing
Common Lisp application, called elPrep, to another
platform with better support for memory manage-
ment and concurrency (Costanza et al., 2019). El-
Prep (Herzeel et al., 2019) is a multi-threaded tool for
preparing sequence alignment/map files (SAM/BAM)
for variant calling in DNA sequencing pipelines. A
key feature of elPrep is the ability to avoid the stan-
dard practice of creating a pipeline consisting of
multiple command line tools invocations, by exe-
a
https://orcid.org/0000-0001-6211-3199
b
https://orcid.org/0000-0002-5246-7396
cuting a single pass through a SAM/BAM file and
keeping data as much as possible in main memory.
In (Costanza et al., 2019) the authors investigated Go,
Java and C++ programming platforms, as an alterna-
tive to Common Lisp. The result of their study con-
cluded that the Go implementation performed best,
using a metric that involved both the RAM usage
and the runtime performance. The benchmarks of the
study showed that Java had a faster runtime, but a sig-
nificantly higher memory usage, while Go offered a
better balance between the two.
As the Java source code for elPrep is available at
https://github.com/exascience/elprep-bench, we
have analyzed key aspects regarding the memory
management and thread synchronization, and propose
a series of improvements that could increase signifi-
cantly the performance of the Java implementation.
2 BACKGROUND
2.1 Garbage Collection
In order to analyze the behavior of memory in-
tensive applications, it is important to understand
how garbage collection works and especially how
Java (Java Platform, Standard Edition, 2019) imple-
ments its garbage collectors.
84
Khlud, C. and Fr
˘
asinaru, C.
A Case Study on Performance Optimization Techniques in Java Programming.
DOI: 10.5220/0009591200840091
In Proceedings of the 15th International Conference on Software Technologies (ICSOFT 2020), pages 84-91
ISBN: 978-989-758-443-5
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

The Java Virtual Machine (JVM) (Lindholm et al.,
2014) offers an automatic storage management sys-
tem, called garbage collector (GC) which reclaims
heap storage occupied by objects which are no longer
used. The garbage collection process (Oracle GC,
2019) works typically by splitting the heap into two
regions: a young generation region and an old gener-
ation. All new objects are allocated in the young re-
gion, in a very fast manner, using typically a ”bump-
pointer” strategy. When this region becomes full, a
minor garbage collection occurs and all dead objects
are deleted very quickly. The objects which are still
referenced survive, and then they are moved to the old
generation. This minor collection is always a ”stop
the world” event, meaning that all of the application
threads will be paused until the GC is finished. In
the old generation, objects are expected to live longer
and they are collected more seldom but with a more
expensive algorithm, called major garbage collection.
We will analyze the impact of some simple tweaks
meant to reduce the impact of GC over the application
performance, such as reducing the unnecessary small
allocations in young region, controlling the scope in
which objects are referenced in order to minimize the
number of times when expensive collection of old re-
gion is triggered, simplifying the object graph and
controlling the amount of memory JVM is allowed
to use.
2.2 Memory Usage
The Java Virtual Machine allocates memory either on
stack or on heap (Lindholm et al., 2014), (Gosling
et al., 2014). An object allocated on the heap has a
header which contains information used for locking,
garbage collection or the identity of that object. The
size of the header depends on the operating system,
and it may be 8 bytes on 32 bit architectures or 16
bytes on 64 bit architectures. Also, for performance
reasons and in order to conform with most of the hard-
ware architectures, JVM will align data. It means
that if we have an object that wraps just one byte, it
will not use 8(object header) + 1(content) = 9 bytes
of memory on the heap, but it will use 16 bytes as it
needs to be aligned to the next 8 byte boundary.
In Java, strings are objects and they are allocated
on the heap. Inspecting the source code of the String
class, one can observe the following instance fields:
byte[] value, byte coder and int hash. As ex-
pected, a String object keeps a reference to an in-
ternal byte array. However, the other two fields will
make the size of the object equal to 8 (header) +
4 (value reference) + 1 (coder value) + 4 (hash
value) = 17 bytes. Being aligned to 8 bytes, it will
actually use 24 bytes. When creating many String
instances (like millions of them, as in our case study),
the extra information included in this class will add
up, consuming memory and triggering the garbage
collector more often than necessary.
We will show that replacing the String usage to
the underlying value byte array will improve the per-
formance of the application, and this approach should
be implemented in every scenario that involves pro-
cessing large amounts of text data.
2.3 Memory Compaction
Another important part of working with large data sets
that have to be accessible in memory regards the for-
mat in which they are represented. Choosing the right
format will not only reduce the amount of consumed
memory but it will also reduce the GC cost to copy the
objects between regions and the cost of visiting and
marking them (Schatzl et al., 2011), (Eimouri et al.,
2017).
The most common approach of representing infor-
mation is in row based form, where a row is a record
of some kind and a column is a certain property of
that row. This type of representation is used in most
relational databases management systems, where sets
of rows of the same type form tables.
A column store model (Abadi et al., 2013) ”re-
verses” the orientation of the tables. It stores data by
columns and uses row identifiers in order to access
a specific cell of the table. By storing each column
separately, query performance is increased in certain
contexts as they are able to read only the required at-
tributes, rather than having to read entire rows from
disk and discard unneeded attributes once they are in
memory. Another benefit is that column stores are
very efficient at data compression, since representing
information of the same type inside of a column helps
the data alignment process that we have previously
mentioned.
Let’s consider a simple example, using the class
Point, defined as a pair of two integer fields x and y.
The basic idea is that instead of having a row-based
model consisting of an array Point[] of instances
(each Point object is a row and its members x and
y are the columns), to use two arrays of integers x[]
and y[], representing the two columns. This way we
can store the same data, minus the object headers cor-
responding to all the Point instances. Not only the
memory consumption will be lower (so the GC will
be triggered less often), but the structure will also take
shorter time to visit, since there are only two objects
now (the two arrays).
Though memory compaction is a very good so-
A Case Study on Performance Optimization Techniques in Java Programming
85

lution for size reduction, it has the downside of re-
quiring more computational effort in order to work
with multiple properties of the same object. However,
when saving memory is the major concern, and espe-
cially when it comes to hundreds of GB per instance,
the execution slowdown becomes far less important
if we can achieve significant reductions in consumed
memory.
3 DATA REPRESENTATION
3.1 The Row-based Model
The data structure used in the original elPrep algo-
rithm is represented by the class SamAlignment, an
object of this type storing one row of a SAM file. The
class contains the following declarations of instance
variables:
Slice QNAME, RNAME, CIGAR, RNEXT, SEQ, QUAL;
char FLAG; byte MAPQ; int POS, PNEXT, TLEN;
List<Field> TAGS, temps;
For a small BAM file of 144 MB there will be cre-
ated around 2.1 million SamAlignment instances and
for a 1.27 GB BAM file there will be created around
17.6 million objects.
For simplicity, let’s disregard TAGS and temps
fields (which can have different lengths) as it makes
the calculation simpler and analyze the memory con-
sumption in both cases. We suppose also that the JVM
uses 32 bits for representing an object header.
One SamAlignment object contains 8 bytes (ob-
ject header), 6 instances of Slice objects (QNAME,
RNAME, CIGAR, RNEXT, SEQ, QUAL) of 4 bytes each,
3 integer fields (POS, PNEXT, TLEN) of 4 bytes each,
1 character (FLAG) of 2 bytes, and an additional byte
(MAPQ). So, the total size of the object is 8+ 6 ∗ 4 + 3∗
4 + 1 ∗ 2 + 1 ∗ 1 = 47 bytes, and as it is rounded up to
a multiple of 8, the result is 48 bytes.
In order to save memory, the string representing a
row scanned from the original file is shared between
multiple objects. All 6 Slice instances contain a ref-
erence to the underlying string and two integers point-
ing to the start index and length. So, a Slice instance
uses 8 (object header) +4 (reference to the string)
+4 + 4 = 20 bytes, being rounded to 24.
As Slice instances point to a String object, the
String itself adds another 24 bytes, as we have al-
ready seen, and the byte array object referenced from
the String adds another 24 bytes (not counting its
content size). Adding all these numbers up, we con-
clude that for representing a SamAlignment object
the JVM needs 48 (the object itself) +6 ∗ 24 (Slice)
+24 + 24 = 240 bytes.
For a 144 MB file there are 2.1 million entries,
so the memory requirement for storing the graph
of objects and the integer fields is approximately
504, 000, 000 bytes, which equals to more than 480
MB (not counting the 144 MB actual content of byte
array). For the 1.27 GB BAM file, the numbers are
much larger as there are a 17.6 million rows. The total
is 4, 224, 000, 000 bytes, representing almost 4 GB.
3.2 The Column-based Model
Let us analyze how much memory can be saved by
switching to a column-based approach. We have de-
fined the following data structures: StringSequence
for representing in a compact manner a col-
lection of strings, DeduplicatedDictionary for
eliminating duplicate copies of repeating strings,
DnaEncodingSequence for storing A,C,G,T,N se-
quences using an encoding of 21 letters per long
and TagSequence for representing tags encoded in
an array of short values. We have also used
the classes CharArrayList, IntArrayList and
ByteArrayList from FastUtil library (Vigna, 2019),
which offers implementations with a small memory
footprint and fast access and insertion.
The new definition of the data store is described
by the class SamBatch, containing the following:
StringSequence QNAME, QUAL;
DeduplicatedDictionary RNAME, CIGAR, RNEXT;
IntArrayList POS, PNEXT,TLEN;
CharArrayList FLAG;
ByteArrayList MAPQ;
DnaEncodingSequence SeqPacked;
So, instead of having a large number of
SamAlignment instances, we will have a single ob-
ject of type SamBatch which contains references to
the ”columns”, i.e. our data structures holding all the
information of a specific type.
Regardless of VM bitness, the memory consump-
tion for representing one row of the input file is
2 ∗ 4 + 3 ∗ 4 + 3 ∗ 4 + 2 + 1 + 4 = 39 bytes. Consid-
ering that no rounding up is necessary, for 2.1 million
rows this sums up to 81, 900, 000 bytes, equivalent to
78 MB. The header sizes of the column objects (11 ∗8
bytes) become negligible in this context.
The basic idea is that instead of storing an array
of String objects, for example: String items[]
= {"abc", "def"}, each consuming memory due
to their headers, we can use a single object of type
String, storing all the characters, and an additional
array for their lengths.
String dataPool = "abcdef";
int[] endLengths = {3, 6};
ICSOFT 2020 - 15th International Conference on Software Technologies
86

For such a small array, the save is minor, but for
a large number of items (millions), the memory re-
duction becomes significant. Even more important,
the GC work is also reduced, since no matter how
many items are in the dataPool and endLengths
fields, there are only two objects to visit. The tech-
nique described above was implemented in the class
StringSequence.
If the strings that are to be stored are repeated fre-
quently, we can apply another optimization: instead
of keeping them joined, we will use an indexed col-
lection containing all the distinct strings and an ar-
ray holding one index for each string. For example, {
”abc”, ”def”, ”abc”, ”xyz”, ”abc”} becomes:
table : {abc=>0, def=>1, xyz=>2}
items : [0, 1, 0, 2, 0]
This data deduplication technique (He et al.,
2010), (Manogar and Abirami, 2014) was imple-
mented in the class DeduplicatedDictionary.
When storing strings containing characters from
a restricted alphabet, one optimization that can be
performed is using an array of primitive values,
for example a long[], and encoding each character
into a block of bits. The number of bits required
for a character depends on the size of the alphabet.
DNA sequences use four letters A,C,G,T, but it is
possible for a machine to read incorrectly a symbol
and to return N. In order to represent 5 possible
characters we need at least 3 bits, which as a result
long can store in its 64 bits 21 DNA letters. For
example, encoding the 21 letters string ”AAAACC-
CCGGGGTTTTNNNNA” would produce a single
long value, containing the bits (from right to left, 000
represents A, 001 represents C, and so on):
00001001001001000110110110110100
10010010001001001001000000000000
In the sample files, one DNA sequence is typically
around 100 letters, so the memory needed in order to
represent it would be 1 int (encoding length) and 5
longs (the content), that is 44 bytes. This reduces the
memory consumption by a factor of two.
Running the smaller input file (144 MB), we have
estimated that the original elPrep algorithm would use
around three times more memory than the size of the
input SAM file. However, when trying to process the
larger input file (1.2 GB), on a 32 GB machine, we
have obtained an OutOfMemoryError, meaning that
the penalty of using too many objects in order to rep-
resent the information was preventing us in loading
the entire data set into memory.
On top of this, there is the cost of tags. In order
to address temps and TAGS we have implemented the
TagSequence class, which is a combination between
StringSequence and DeduplicatedDictionary.
Without detailing all the calculations, in the original
implementation, for the smallest file (144 MB), con-
taining 2.1 million rows, the JVM will use almost one
gigabyte (1, 001, 700, 000 bytes) just for object book-
keeping. Our model will use much less additional
memory, a 10× saving for object bookkeeping. For
2.1 million entries, the tags memory consumption is
now 50, 400, 000 bytes and the combined value for the
whole model is 132, 300, 000 bytes (about 126 MB).
4 OPTIMIZING I/O OPERATIONS
4.1 Buffering and Synchronization
The elPrep algorithm starts by reading the input file.
Based on the read information, it creates a large data
set in memory and, in the end, it writes the pro-
cessed data into another file. In the writing step, the
algorithm creates parallel tasks, which in turn take
all SamAlignment instances and serialize them into
the string format of SAM files. Especially when the
full data set is loaded into memory, these tasks cre-
ate many small objects that have a negative impact in
terms of memory management. Using a simple tech-
nique of pre-allocating the buffer sizes based on the
specific context of the problem, we can prevent the
creation of many of these intermediate objects.
The code sequence that captures the writing pro-
cess is presented below:
var outputStream = alnStream
.parallel()
.map((aln) -> {
var sw = new StringWriter();
try (var swout = new PrintWriter(sw))
{ aln.format(swout); }
return sw.toString();
});
An important technique that could improve per-
formance when working with data is buffering
(Oaks, 2014). If we take a closer look at the
StringWriter class we notice that it uses an inter-
nal StringBuffer object for storing its data, which
in turn has an internal primitve buffer which has 16
characters as default. The average length of a row in
the input file is 325 − 330 characters. As 350 is about
10% larger than the average line, based on the regular
statistical distribution, thus most lines would require
no extra resizes of their corresponding buffers. This
prevents the creation of extra garbage, which in turn
reduces the number of times when GC is executed. In
the following sections, we will denote the algorithm
that employs this technique as PresizedBuffers .
A Case Study on Performance Optimization Techniques in Java Programming
87

We will further analyze the usage of the
StringWriter and its StringBuffer helper. To
quote from its documentation, a StringBuffer is a
thread-safe, mutable sequence of characters. Mean-
ing that most of its methods are declared as synchro-
nized in order to control the access to the buffer of
characters in a multi-threaded environment. How-
ever, in our case, each writing thread spawned by the
parallel stream implementation gets its own copy of
a StringWriter so there is no resource contention
that would require synchronization. Instead of using
a StringWriter, it would be better to simply use a
common buffer, with no synchronization. Instead of
creating and storing individual string representations
of all the SamAlignment objects, and writing them
one by one to disk, we add progressively informa-
tion into the buffer. Only when the buffer becomes
full, we write its content on disk. Obviously, all these
steps will be performed in a single thread, but the bot-
tleneck will be less obvious. The class implementing
the byte array is called StreamByteWriter.
4.2 Chunking-batching and Extracting
Parallelism
In the elPrep original algorithm, a SamAlignment ob-
ject is created for each row in the input file. In order to
extract parallelism, there is an explicit .parallel()
call, a construct that would create a task for each row
that must be processed. These tasks are queued and
executed in a concurrent fashion using threads created
transparently by the Java Stream API.
var alnStream = inputStream.parallel()
.map((s) -> new SamAlignment(s));
In the column-based model, described in sec-
tion 3.2, there is a single SamBatch instance storing
all the data. Because we perform data compaction, the
read operation must take into account the dependen-
cies between rows. Just like in the case of removing
duplicates, in order to process a new row we have to
inspect the values of the previously read rows. There-
fore the reading process cannot be fully parallel per-
row. In order to obtain a better performance than read-
ing using a single thread, we propose a technique that
splits the SamBatch structure in several ”chunks”. In-
stead of having a single large object, we will represent
the data using an array of smaller SamBatch objects.
A sketch of our Compact algorithm is described by
the following pseudo code:
int chunkSize = 20000;
int nbOfThreads=cpuCoreCount * 4;
var samBatches=new ArrayList<SamBatch>();
while (!endOfFile) {
var rowsRead=parallelReadRows(
batchSize, nbOfThreads);
parallelTransformRowsIntoBatches(
samBatches, rowsRead);
}
The readRows method reads chunkSize ∗
nbO f T hreads rows out of the SAM file for the next
processing step. The actual reading is done using an
appropriate number of threads (based on the CPU
configuration), each thread reading sequentially a
fixed number of rows, calculated taking into account
the nature of the information being encoded. Having
the data split into chunks, we can process in parallel
the mapping between the associated text and the
SamBatch data structure, where we perform data
compaction and deduplication. The techinque of
grouping similar tasks requiring the same resources
in order to streamline their completion is called
batching.
In our case, the advantages of the chunking-
batching approach are multiple. Since a
DeduplicatedDictionary will now have less
than 20000 unique strings, the values needed to
encode the strings could be represented on 2 bytes,
instead of 4. Similarly, we can reduce the tag
size from 4 to 2 bytes. After the algorithm is
fully executed, we will have instead of 2.1 million
SamAlignment objects, about 105 SamBatch in-
stances, each of them having around 4 orders of
magnitude less objects overall. This translates into 2
orders of magnitude less objects. When creating the
output file, the SamBatch array could be processed in
parallel, with no blocking except the actual operation
of writing to disk.
We have seen that the column-based model saves
memory at the expense of the running time. This opti-
mization, however, reduces the overall execution time
of the read operation, which is now on par with the
original implementation.
When it comes to writing, compared to our
StreamByteWriter algorithm, which is anyway much
faster than the original implementation, the execution
time is drastically reduced from 28 seconds (for the
12 GB BAM file) to 12 seconds. Preparing the strings
that are to be written in the file can be done in almost
perfect parallelism, using the available cores.
In order to make sure there are no dead times when
using the external device, we have also implemented
the async/await pattern. This allows the program to
perform in advance reading operations, using a ded-
icated thread, while waiting for the data processing
threads to complete their execution. This new algo-
rithm, called Compact/Par, offers a small improve-
ment in the running time, as we will see in the next
ICSOFT 2020 - 15th International Conference on Software Technologies
88
section, but with the disadvantage of a significant in-
crease in code complexity.
5 EXPERIMENTAL RESULTS
5.1 Overview
We have created five implementations that address the
most expensive parts of the elPrep algorithm, which
are reading, storing all data in the memory and writ-
ing. Except for the original version, which was taken
from elPrep public repository, all other algorithm im-
plementations contain various types of optimization
that are meant to improve runtime performance and
to lower the memory usage, especially on large files
where GC becomes a limiting factor.
The computer we have used in order to perform
the experiments is a Ryzen 9-3900X, having 12 cores
and using 48 GB of RAM. Since we didn’t have
access to the hardware necessary to run all the tests
in memory, as the original paper, we have used the
smaller SAM files and ran the same processing re-
peatedly in order to obtain an accurate result of the
running time. For example, running 10 times the al-
gorithm on the smallest input SAM file, which is ap-
proximately 700 MB, will produce a total running
time of around 70 seconds. Running the algorithm
repeatedly will trigger the garbage collector and this
will be the cause of variations in the collected results,
ranging from around 2 to 3 seconds, when reading the
smallest file, and 3 to 4 seconds for writing.
The original elPrep benchmarks have been per-
formed on a Supermicro SuperServer 1029U-TR4T
node with two Intel Xeon Gold 6126 processors con-
sisting of 12 processor cores each, clocked at 2.6
GHz, with 384 GB RAM (Costanza et al., 2019). The
authors claim to do the processing of the 8 GB BAM
file in 6 min:54sec to 7 min:31sec and memory usage
is 330 − 340 GB.
As we didn’t have access to such a performing ma-
chine, we did most of testing with the smallest file, the
144 MB BAM file (673.3 MB SAM file). For the 8
GB BAM file (27.18 GB SAM) our results will show
only the Compact algorithm but we will make some
inferences over the scaling of the algorithms across
file sizes and cores.
5.2 Runtime Performance
The following table shows a brief comparison of the
running times obtained by our algorithms, in three
configurations: 144 MB file using 4 cores and 12
cores, and 1.2 GB file using 12 cores.
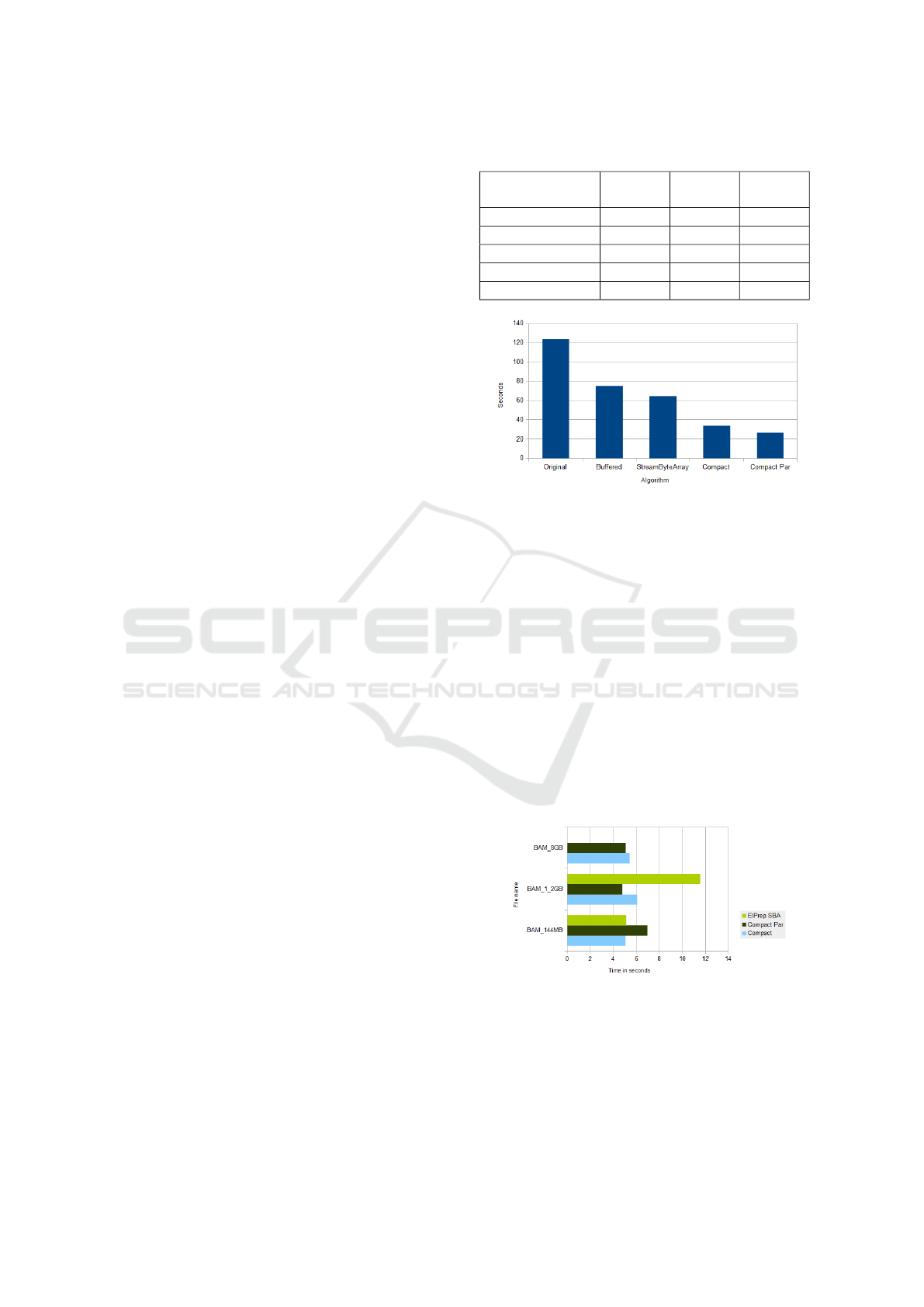
Table 1: Running time per algorithm in seconds.
144M
(4c)
144M
(12c)
1.2G
(12c)
Original 9.13 3.938 123.91
PresizedBuffers 8.98 4.19 75.1
StreamByteArray 8.09 3.43 64.62
Compact 5.64 3.4 34.5
Compact/Par 5.42 4.68 26.7
Figure 1: Runtime Performance in seconds (1.2 GB BAM).
Comparing 4 cores to 12 cores, we notice that the
Original algorithm scales with a factor of 2.3, Pre-
sizedBuffers by a factor of 2.14, StreamByteArray
scales by 2.36 and Compact would scale by 1.69. So,
at least for the small file, it seems that using larger
machines will offer a better performance. It is im-
portant to notice that, in its current implementation,
the Compact algorithm has an explicit sequential part
that reduces its scalability. Some potential fixes are
described in section 6, where we describe some tech-
niques aimed at improving the single threaded part.
A more useful way to present the algorithm is to
show how it scales based on input size, and at least
on the 12 core machine, we can see that both compact
algorithms remain roughly in the same speed, so GC’s
runtime cost doesn’t become in impediment:
Figure 2: Seconds per GB by algorithm and file size.
5.3 Memory Usage
To measure the live data set, we have used Java Visu-
alVM (VisualVM, 2019) which provides a visual in-
terface for profiling a running application. Using Vi-
A Case Study on Performance Optimization Techniques in Java Programming
89
sualVM, we have analyzed the memory consumption
in each scenario and we have estimated the minimum
amount of memory that JVM requires in order to load
a specific data set.
Unlike the original paper, which measures pro-
cess memory size, we have measured live data size.
This is possible due to VisualVM which offers very
precise information regarding the objects consuming
memory.
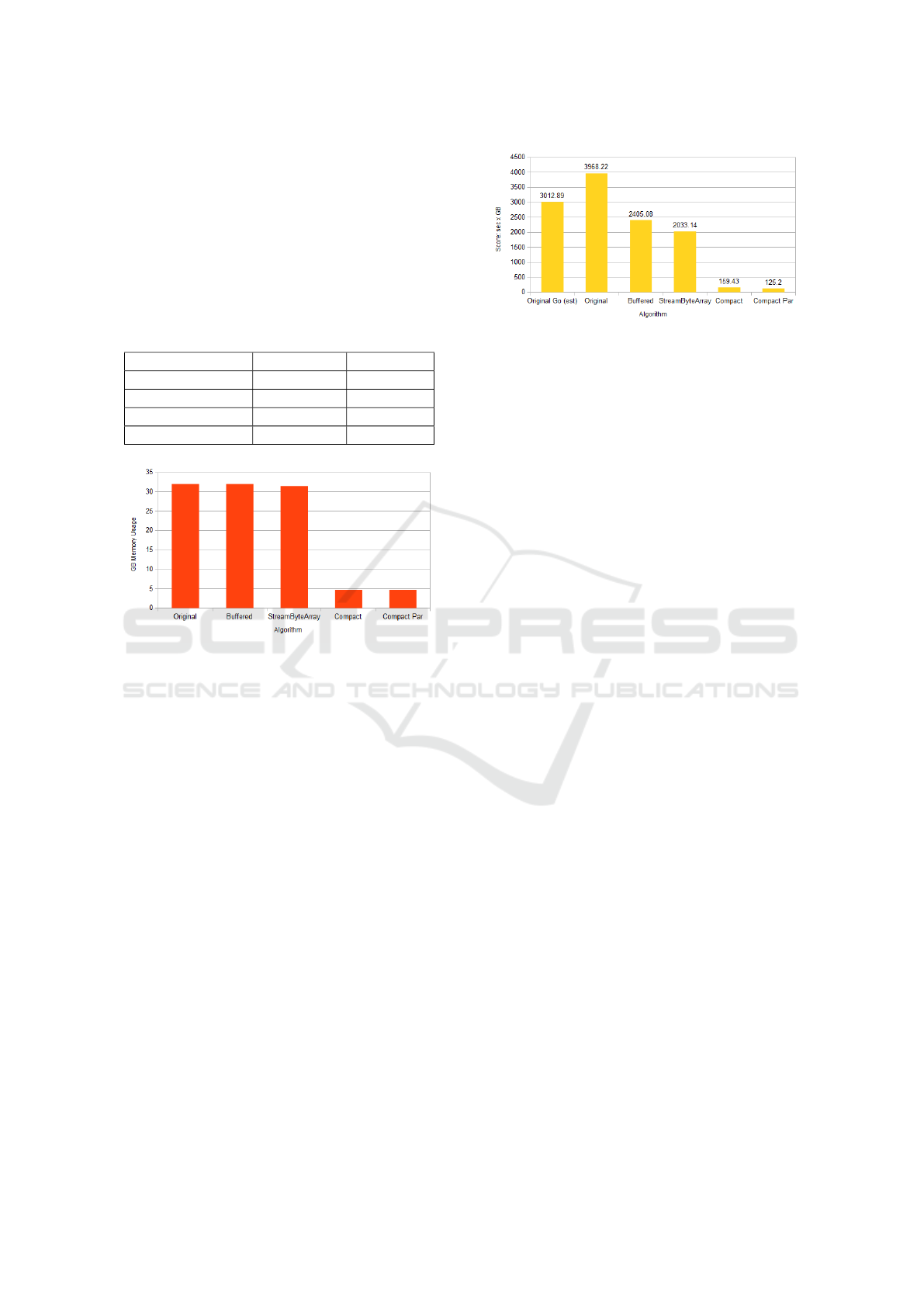
Table 2: Memory usage per algorithm in MB.
144 MB file 1.2 GB file
Original 2326 MB 32025 MB
PreSizedBuffers 2326 MB 32025 MB
StreamByteArray 2275 MB 31463 MB
Compact/Par 606 MB 4689 MB
Figure 3: Live Memory Usage (in GB).
The peak usage was measured by suspending the
program at the moment when the whole file was read.
We notice that for the 1.2 GB BAM file, the Original
and Presized Buffers algorithms are using around 32
GB of memory. In order to offer this amount of mem-
ory to JVM we used a machine with 48 GB of RAM.
To further reduce the overhead of GC, 64 GB would
certainly have been better.
5.4 Calculating Performance
The goal of elPrep was to keep both the running time
and the the memory consumption low. The evalua-
tion function was defined as the multiplication of the
average elapsed wall-clock time (in hours) with the
average maximum memory use (in GB), with lower
values (in GBh) being better (Costanza et al., 2019).
We have used the same approach, changing only the
measurement units to MB and seconds.
On the medium file (1.2 GB BAM) the values are
conclusive from the point of view of scaling the re-
sults. The improvements resulting from our optimiza-
tion techniques are now clearly visible.
Tests performed up-to the 8 GB file showed that
algorithms scale as expected, GC does not become
Figure 4: Memory x Times Score (Seconds x GB).
a large impediment for the Compact implementa-
tions and the overall performance seems limited only
by other hardware components, such as disk drive
read/write speeds. Both Compact algorithms keep a
steady pace of processing at around 180 MB/second
(under 6 seconds to process every 1 GB of BAM).
StreamByteArray has a sharp loss of speed as GC is
triggered more often, processing data at half speed
when increasing the BAM file size from 0.144 GB
to 1.2 GB.
6 FUTURE WORK
Batch Reader Scalability.
As we have described in section 4.2, our batch reader
works in two steps: initially, on the main thread, it
extracts from the original file the rows for the number
of the expected batches, and then it executes in paral-
lel, using all cores, the data compaction step. Before
chunking and batching is done, we split the full byte
array read from file into distinct List<byte[]> in-
stances. This separation may not be necessary, an al-
ternate approach being to store inside a large byte[]
structure all the information and to use an additional
array of indices in order to retrieve the actual lines of
text. This would reduce the number of allocations and
eventually speed up the execution of the main thread.
Value Types.
When Java specifications were elaborated, more than
25 years ago, the cost of a retrieving an object from
the memory and executing an arithmetic operation
was approximately the same. On modern hardware
however, the memory fetch operations are up to 1000
times more expensive than arithmetic ones. This is
why, the Project Valhalla (Valhalla, 2019), that is ex-
pected to be integrated in modern JDK releases, in-
troduces new data structures and language constructs
that improve various aspects regarding data manipu-
lation. For example, Value Types provide the nec-
essary infrastructure for working with immutable and
reference-free objects. In our context, this would al-
ICSOFT 2020 - 15th International Conference on Software Technologies
90

low us to further reduce the memory used by the Com-
pact algorithm by using an efficient by-value compu-
tation with non-primitive types.
7 CONCLUSIONS
This paper addresses the situation when one has to
manipulate a large textual data set by reading it from
a file, transforming it into objects, processing it and
then writing it back to a file, and all these operations
must be performed in a single in-memory session.
We have analyzed a modern implementation of an al-
gorithm for processing SAM and BAM files, elPrep
(Herzeel et al., 2015), (Herzeel et al., 2019), which
must handle input files up to 100 GB. The conclu-
sion of the elPrep authors was that a Java implemen-
tation for this specific problem suffers from the mem-
ory management offered by JVM (Costanza et al.,
2019). However, when using an object-oriented pro-
gramming platform, one has to take into considera-
tion all aspects regarding memory allocation offered
by that specific platform and to adapt its model and
programming techniques.
We have showed that major improvements can
be obtained by using techniques that are aimed at
reducing the number of created objects. This will
not only save memory but it will also improve run-
time performance by decreasing the overhead of the
Garbage Collector. Using a column-based represen-
tation we have compacted the data set in a man-
ner that boosted the overall score calculated as the
multiplication between used memory and running
time. The penalty incurred by the more elaborate
data model was compensated by a multi-threaded ap-
proach, called chunking-batching, that actually allows
the algorithm to use all available machine cores when
processing the input file.
Given the hardware differences between the ma-
chine used by the elPrep authors and ours, there are
limits on the testing that could be done with the tech-
niques used by this paper. However, using input files
ranging in size from 144 MB to 12 GB, we have
proved that our algorithms are scalable and could per-
form as expected for files of any size, provided the
machine has sufficient memory.
REFERENCES
Abadi, D., Boncz, P., and Harizopoulos, S. (2013). The De-
sign and Implementation of Modern Column-Oriented
Database Systems. Now Publishers Inc., Hanover,
MA, USA.
Costanza, P., Herzeel, C., and Verachtert, W. (2019). Com-
paring ease of programming in C++, Go, and Java for
implementing a next-generation sequencing tool. Evo-
lutionary Bioinformatics, 15:1176934319869015.
D
¨
oring, A., Weese, D., Rausch, T., and Knut, R. (2008).
Seqan an efficient, generic c++ library for sequence
analysis. BMC bioinformatics, 9:11.
Eimouri, T., Kent, K. B., and Micic, A. (2017). Optimiz-
ing the JVM Object Model Using Object Splitting. In
Proceedings of the 27th Annual International Confer-
ence on Computer Science and Software Engineering,
CASCON 17, page 170179, USA. IBM Corp.
Gosling, J., Joy, B., Steele, G. L., Bracha, G., and Buckley,
A. (2014). The Java Language Specification, Java SE
8 Edition. Addison-Wesley Professional, 1st edition.
He, Q., Li, Z., and Zhang, X. (2010). Data deduplication
techniques. volume 21, pages 430 – 433.
Herzeel, C., Costanza, P., Decap, D., Fostier, J., and
Reumers, J. (2015). elprep: High-performance prepa-
ration of sequence alignment/map files for variant
calling. PloS one, 10:e0132868.
Herzeel, C., Costanza, P., Decap, D., Fostier, J., and Ver-
achtert, W. (2019). elprep 4: A multithreaded frame-
work for sequence analysis. PLOS ONE, 14(2):1–16.
Java Platform, Standard Edition (2019). Java De-
velopment Kit Version 11 API Specification.
https://docs.oracle.com/en/java/javase/11/docs/api.
Accessed: 2019-06-01.
Lindholm, T., Yellin, F., Bracha, G., and Buckley, A.
(2014). The Java Virtual Machine Specification, Java
SE 8 Edition. Addison-Wesley Professional, 1st edi-
tion.
Manogar, E. and Abirami, S. (2014). A study on data
deduplication techniques for optimized storage. pages
161–166.
Oaks, S. (2014). Java Performance: The Definitive Guide.
O’Reilly Media, Inc., 1st edition.
Oracle GC (2019). Java Garbage Collection Basics.
https://www.oracle.com/webfolder/technetwork/ tuto-
rials/obe/java/gc01/index.html. Accessed: 2019-06-
01.
Schatzl, T., Dayns, L., and Mssenbck, H. (2011). Optimized
memory management for class metadata in a JVM.
Valhalla, P. (2019). OpenJDK Project Valhalla.
https://openjdk.java.net/projects/valhalla/. Accessed:
2019-06-01.
Vigna, S. (2019). Fastutil 8.1.0. http://fastutil.di.unimi.it/.
Accessed: 2019-06-01.
VisualVM, O. (2019). Java VisualVM.
https://docs.oracle.com/javase/8/docs/technotes/
guides/visualvm/index.html. Accessed: 2019-06-01.
A Case Study on Performance Optimization Techniques in Java Programming
91
