
FLOPTICS: A Novel Automated Gating Technique for Flow
Cytometry Data
Wiwat Sriphum, Gary Wills and Nicolas G. Green
School of Electronics and Computer Science, University of Southampton, Southampton, U.K.
Keywords: Flow Cytometry, Automated Gating, Density-based Clustering, Optics Clustering.
Abstract: Flow cytometry (FCM) involves the use of optical and fluorescence measurements of the characteristics of
individual biological cells, typically in blood samples. It is a widely used standard method of analysing blood
samples for the purpose of identifying and quantifying the different types of cells in the sample, the result of
which are used in medical diagnoses. The multidimensional dataset obtained from FCM is large and complex,
so it is difficult and time-consuming to analyse manually. The main process of differentiation and therefore
labelling of the populations in the data which represent types of cells is referred to as Gating: gating is the
first step of FCM data analysis and highly subjective. Significant amounts of research have focussed on
reducing this subjectivity, however a faster standard gating technique is still needed. Existing automated
gating techniques are time-consuming or need many user-defined parameters which affect the differentiation
to different clustering results. This paper presents and discusses FLOPTICS: a novel automated gating
technique that is a combination of density-based and grid-based clustering algorithms. FLOPTICS has an
ability to classify cells on FCM data faster and with fewer user-defined parameters than many state-of-the-art
techniques, such as FlowGrid, FlowPeaks, and FLOCK.
1 INTRODUCTION
Flow cytometry (FCM) is a high-throughput
technology that is used to identify characteristics of
cells by using the concept of cell-scatter measurement
and light emission after receiving a laser beam
stimulation (Bio-Rad, 2018). The technique provides
a set of chemical and physical characteristics for each
individual cell in fluid samples such as blood (Lo et
al., 2008) and can process large numbers, giving a
detailed information on size and distribution of the
different cell populations. It is a standard diagnostic
tool in general healthcare and has been widely applied
in medical research, especially in haematology and
immunology, and broadly adopted in clinical
environments to diagnose and monitor treatments,
such as: leukaemia, chemical healing responsiveness,
and stem cell transplantation monitoring (Jahan-Tigh
et al., 2012). The process provides multi-dimensional
data, including relative size, relative granularity, and
relative fluorescence intensity (BD-Biociences,
2002). The data is highly complicated and difficult to
analyse as a result (Bashashati and Brinkman, 2009).
Flow cytometry measures individual cells by
compressing them into a narrow stream of fluid
passing through at least one laser beam, with
detectors to measure transmission, reflection, scatter
and fluorescence emission. Cell properties that can be
measured include
relative size, relative granularity,
and relative fluorescence. This
technique was
developed over 40 years ago but was limited initially
because the cytometer was too large, difficult to
maintain and an expensive instrument. As with many
modern pieces of equipment (Robinson et al., 2012),
FCM is now more accurate, cheaper, and more
convenient to use, hence its wide application in
clinical research, particularly haematology and
immunology.
Flow cytometry is able to detect cells from 0.2
microns to 150 microns in diameter, but actual
capability depends on the equipment used (Rowley,
2019). In FCM, the fluid containing the cells is driven
through a narrow nozzle, with the resulting ejected
stream or droplets thin enough to have only one cell
at a time passing through the laser beams. Typically,
scattered light and fluorescent emissions from each
cell are measured by a detector; light scattered by less
than 5 degrees is called forward scatter (FSC) and is
used to identify the size of cells, while larger
deflections are called side scatter (SSC) and are used
96
Sriphum, W., Wills, G. and Green, N.
FLOPTICS: A Novel Automated Gating Technique for Flow Cytometry Data.
DOI: 10.5220/0009426300960102
In Proceedings of the 5th International Conference on Complexity, Future Information Systems and Risk (COMPLEXIS 2020), pages 96-102
ISBN: 978-989-758-427-5
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
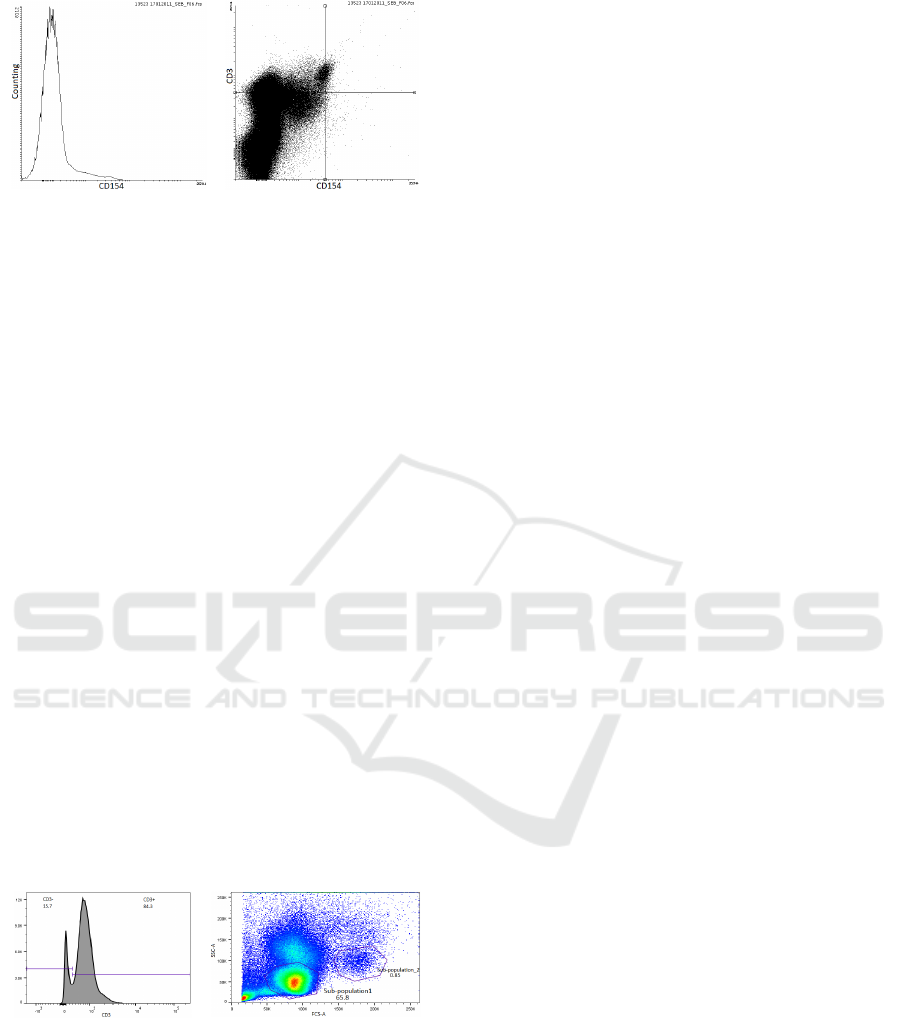
a. Histogram b. Scatter plot
Figure 1: Flow cytometry data examples. (a) a histogram of
the number of cells measured at different fluorescent
intensity values for the CD154 marker. (b) a scatter plot of
the fluorescent intensity values for the CD3 versus the
CD154 marker, used for identifying smaller populations,
with the quadrant markers demonstrating that most cells
have a low response to both CD154 and CD3.
to measure granularity and membrane roughness
(World Health Organization, 2009). Different
fluorescence molecules or “markers” are used to label
particular types of cells to improve the identification
and quantification of different populations and sub-
populations. The standard identification system for
markers is referred to as Cluster of Differentiation
(CD).
Flow cytometry data is a multidimensional dataset
and the data is generally displayed in one or two
parameters (Moloney and Shreffler, 2008). For one
parameter, it can be displayed as a histogram with the
parameter value on the x-axis and the frequency
(number) of cells on the y-axis (Figure 1a). For two
parameters, the data is displayed as a scatter plot, with
points representing the cell as an (x,y) pair of the
values of the two parameters (Figure 1b). Up to 50
cell parameters can be determined (Lee et al., 2017),
with the number of features dependent on the flow
cytometer and experimental design. Viewing the
entire dataset is involved and complex.
a. 1D histogram b. 2D scatter plot
Figure 2: Manual gating examples using either drawn lines
in 1 dimension (histogram) or polylines in 2 dimensions
(scatter plot), to visually identify populations.
After obtaining the data, an expert operator
identifies the populations - known Gating. Gating is
the process of identifying cells by drawing shapes
around populations (Bashashati and Brinkman,
2009), as shown in Figure 2. The expert needs to
know about the characteristics of the cells of interest,
and the populations and sub-populations of cells
before starting the analysis.
Manual Gating is, therefore, highly subjective and
time consuming - with machine learning being
proposed to support this process (Lo et al., 2008).
FCM data is so large and complex that it is
difficult to analyse without computational tools.
There are three main problems in FCM analysis;
firstly, manual gating (identify cells of interest) is
highly subjective (Lo et al., 2008); secondly,
sometimes the number of key events is very low
(Groeneveld-Krentz et al., 2016), which makes them
harder to detect and may result in false positives;
thirdly, manual gating is a time consuming process
(Rahim et al., 2018), especially when the number of
parameters and cells are large. Although some
applications have been developed to help clinical
experts, flow cytometry data analysis application still
have limitations, as mentioned before. The paper
presents the application of machine learning
techniques to implement a novel automated gating
method which can provide appropriate clustering of
cells in blood samples.
2 METHOD
Ye and Ho, 2018 proposed a state-of-the-art
automated gating technique, FlowGrid, and claimed
higher accuracy and better time efficiency compared
with flowPeaks (Ge and Sealfon, 2012), FlowSOM
(Van Gassen et al., 2015), and FLOCK (Qian et al.,
2010). However, FlowGrid still has the problem with
requirement of too many user-defined parameters.
The method proposed here has the aim of
improving the performance of FlowGrid, by reducing
both process time and user-defined parameters. This
improved method, the FLOPTICS algorithm, begins
by partitioning data into equal-sized grids for each
dimension (‘bins’) – with then only non-empty bins
being processed as data points. An example of
partitioning 2-dimensional data in this way is shown
in Figure 3. Partitioning data is not appropriate for
low density datasets, but FCM data is always high
density (as can be seen in Figure 1 and 2), so the
accuracy results of gating are acceptable and the run
time is faster than many state-of-the-art techniques.
2.1 DBSCAN
Density-Based Spatial Clustering and Application
with Noise (DBSCAN) was proposed by Ester et al.
(1996).
DBSCAN is a density-based algorithm for
FLOPTICS: A Novel Automated Gating Technique for Flow Cytometry Data
97
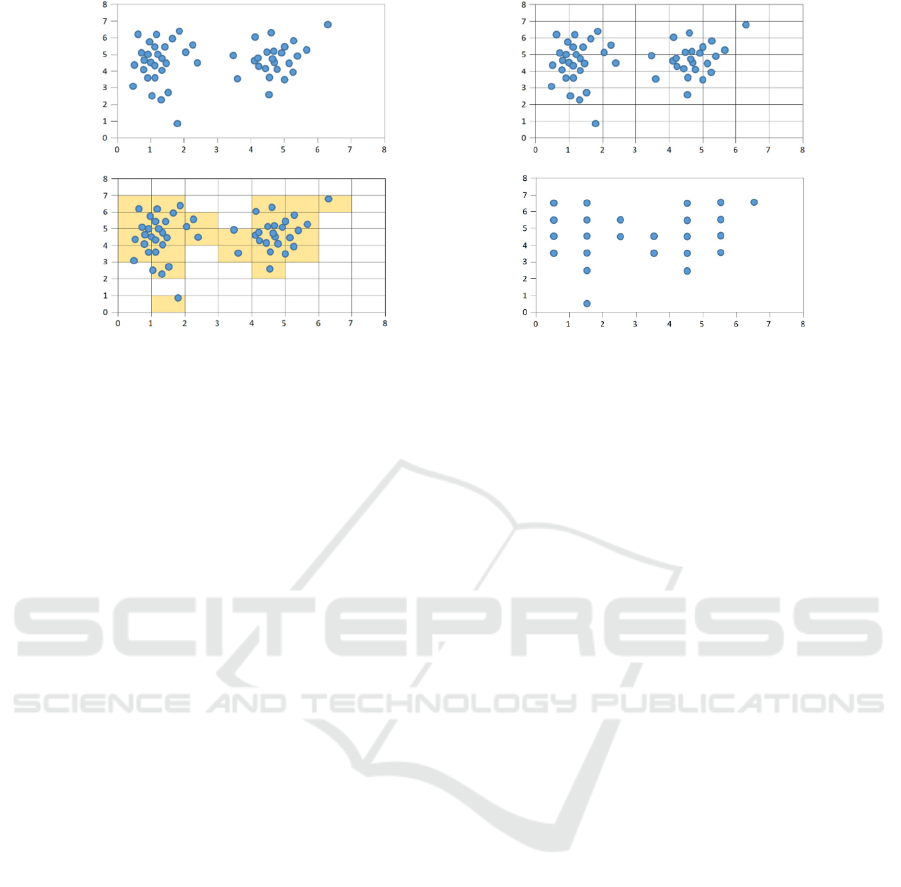
a. original data b. drawing grids on original data
c. selecting non-empty bins d. transformed data
Figure 3: Partitioning 2-dimensional data into equal-sized bin for each dimensions.
clusters, so it can identify non-convex shapes.
Methods based on density must have some
parameters defined in advance, and for DBSCAN,
there are two such parameters (defined by the user):
Eps (
ɛ
)
and
MinPts
.
A point that is considered as a
member of a cluster needs to have at least one
neighbour (another data point) where the distance
between the pair is closer than
ɛ.
In other words, the
data point
p
is a neighbour of point
q
when the
distance between
p
and
q
is less than or equal to
ɛ.
MinPts
is the minimum number of neighbours for a
data point to be a member of a cluster. The algorithm
for DBSCAN clustering can be summarised as:
Step 1: Label all data points as core points, border points,
and noise points.
Step 2: Treat a core point as the centre of a group.
Step 3: Merge each group together if they have at least
one overlapping neighbour.
A core point is a point that has at least MinPts
neighbours. A border point is a point that has less
than MinPts neighbours, but is a neighbour of at least
one core point. A noise point is a point that is not a
neighbour of any core point.
Although DBSCAN is able to identify convex
shapes, the number of clusters does not need to be
defined in advance and, has the ability to identify
noise - which leads to more robustness than partition-
based
clustering. However, it only works properly for
datasets with
uniform densities and parameters need
to be defined before clustering is performed.
2.2 FlowGrid
This framework is a combination of DBSCAN and a
grid-based clustering algorithm that provides high
accuracy. DBSCAN can detect outliers and identify
arbitrarily-shaped clusters. FlowGrid combined the
benefits of DBSCAN and reduced computational
time by using equal-sized grids, similar to the
FLOCK algorithm (Qian et al., 2010). Each
dimension is partitioned into an equal-sized bin, so
the total number of bins for d-dimensional data is
, where
is the number of bins for each
dimension. All data points in the same bin are treated
as a single point by using a representative, which acts
as an index or label for the bin; moreover, only non-
empty bins are considered, which is the reason why
this framework is faster than previous ones.
Every
is labelled with a row of d positive
numbers. For example, if
5,2,3 is a coordinate
of
, it means that the dataset has three
dimensions, and the corresponding data points are
located in the fifth bin of dimension one, the second
bin of dimension two and the third bin of dimension
three. Although FlowGrid is faster than many
automated gating algorithms, provides high accuracy,
and can deal with noise, there are still some user-
defined parameters that can significantly affect the
clustering result. Moreover, FlowGrid is based on
DBCSCAN, meaning it is not suitable for datasets
with different density distributions.
2.3 OPTICS
Ordering Points To Identify the Clustering Structure
(OPTICS) was proposed by Ankerst et al. (1999). The
algorithm was derived from DBSCAN in order to
deal with the need for two parameters, which could
provide different clustering results for different
density thresholds. However, OPTICS does not
produce an explicit clustering; instead, it generates an
ordering
density clustering. The main idea behind
COMPLEXIS 2020 - 5th International Conference on Complexity, Future Information Systems and Risk
98

Figure 4: Higher-density clusters A1 and A2 are completely
within lower-density cluster A.
these algorithms is that higher-density clusters are
completely contained in a lower-density one, as
shown in Figure 4. Local higher-density clusters,
therefore, should be processed first. Key terms
involved in OPTICS algorithm are defined as follows:
Core-distance: Assuming p is an object in the
dataset, ɛ is a value of the distance between two
objects, N
ɛ
(p) is a set of neighbours of object p, and
MinPts the minimum number of neighbours, then
core-distance
ɛ,MinPts(p)
is equal to:
Infinity or undefined, if the cardinality of N
ɛ
(p) is
less than MinPts.
Otherwise, the minimum distance from p to its
neighbour that can cover at least MinPts members.
Reachability-distance: If p and o are objects in
the dataset, ɛ is the distance between two objects,
N
ɛ
(p) is a set of neighbours of object p, and MinPts
the minimum number of neighbours, then
reachability-distance
ɛ,MinPts(o,p)
is equal to:
Infinity or undefined, if the cardinality of N
ɛ
(p) is
less than MinPts.
Otherwise, Max (core-distance(p), distance(o,p)).
Figure 5: The difference between core-distance and
reachability-distance, given ɛ and MinPts = 5.
Therefore, in accordance with these definitions,
reachability-distance must be equal to or greater than
core-distance, as shown in Figure 5. Then,
reachability-distance(p, q) is equal to reachability-
distance(p, r) and equal to core-distance(p), while,
reachability-distance(p, o) is greater than core-
distance(p).
The algorithm for OPTICS clustering can be
summarised as follows:
Step 1: Read an unprocessed object (p) from the dataset
Step 2: If p is a core-object, update core-distance of p
For each q ϵ N
ɛ
(p)
Update reachability of object q
Update the OrderSeeds list, which contains the
objects ordered by reachability-distance
(from smallest-to-largest)
Mark p as processed
Step 3: Read an unprocessed object p from the OrderSeeds
list if the list is not empty; otherwise, read the next
unprocessed object from the dataset
Step 4: Repeat Step 2 - Step 3 until the end of the dataset
Most density-based methods, such as DBSCAN
and OPTICS, can detect non-convex cluster shapes,
identify noise and automatically identify the number
of clusters. However, the density thresholds and other
parameters need to be carefully defined, because
different identification of the parameters in this
method could lead to different clustering results
.
2.4 FLOPTICS
For the algorithm presented here, termed FLOPTICS,
data is partitioned into equal-sized bins, and the data is
clustered using the OPTICS algorithm (Ankerst et al.,
1999). The radius distance (ɛ) has to be defined by the
user, as in the DBSCAN (Ester et al., 1996) algorithm,
but OPTICS can provide the optimal value of ɛ by
showing the structure of data. Therefore, the number of
user-defined parameters for FLOPTICS is fewer than
FlowGrid, which is based on DBSCAN. The
FLOPTICS algorithm can be summarized as follow:
Step1: All data points are partitioned into equal sized bins
for each dimension
Step2: Only non-empty bins are processed
Step3: Read an unprocessed bin (b) from the dataset
obtained from Step 2
Step4: If b is a core-bin, update core-distance of b
For each a ϵ Nɛ(b),
update reachability of object a
Update the OrderSeeds list, which contains the
objects ordered by reachability-distance (from
smallest-to-largest)
Mark b as processed
FLOPTICS: A Novel Automated Gating Technique for Flow Cytometry Data
99
Step5: Read an unprocessed bin b from the OrderSeeds list
if the list is not empty; otherwise, read the next
unprocessed bin from the dataset
Step6: Repeat Step 3 - Step 5 until the end of the dataset
The key terms involved in the algorithm are defined:
N
ɛ
(b) is a set of neighbours of bin b for radius
distance value ɛ, identified by the user
Core-bin is the bin that its number of neighbour
(regarding the radius ɛ) more than or equals to
MinPts, which is identified by a user
Core-distance(b) is the minimum distance that lead
the number of neighbour of bin b reach MinPts
Directly connected: Bin a is directly connected to
Bin b if Distance(a,b)≤ ɛ
Reachability-distance: If b and o are bins in the
grid space, ɛ is the distance between two bins,
Nɛ(b) is a set of neighbours of bin b, and MinPts is
the minimum number of neighbours, then
reachability-distanceɛ,MinPts(o,b) is equal to:
o Infinity or undefined, if the number of
members in Nɛ(p) is less than MinPts
o Otherwise the maximum of core-distance(b) or
distance(o, b)
The OrderSeeds list is the list (queue) of bins in the
grid space ordered by reachability-distance
3 RESULTS AND DISCUSSION
DBSCAN, OPTICS, FlowGrid and FLOPTICS were
applied to a synthetic dataset, is generated to mimic a
real FCM dataset with control over data features. The
experiments were conducted on a computer with
specification as follows: Intel(R) Core(TM) i7-6700
CPU @ 3.40GHz; RAM 16.0 GB; Operating System
- Windows 10 Enterprise, 64-bit.
3.1 Reference Dataset
Patterns or clusters in real sample datasets obtained
from different donors will be similar but different,
even they are obtained from the same flow cytometry
experimental setup. The cell populations in any
sample generally have a normal distribution for the
measurements of any given marker or optical
characteristic. Therefore, cluster shapes formed from
two normally distributed value sets are usually found
but can be symmetric or asymmetric depending on the
donors and markers used. The clusters might be, for
example, circle-shaped with different radiuses, or
cigar-shaped with different widths, heights and angles
and can be different from donor to donor. In order to
provide some clear comparative analysis as well as to
explore the limitations of the methods, imitative
datasets were generated based on model blood
sample, rather than randomly choosing a donor blood
sample. The parameters of the data could then easily
be modified to test the performance of each method.
3.2 Generation of Imitative Datasets
The imitative datasets used in the experiment were 2-
dimensional datasets generated by the function
rmvnorm (n, mean, sigma) in RStudio 3.5.2; this
function randomly generates data from a multivariate
normal distribution, which is often found in FCM
data. For this function, three arguments are required:
the number of data points (
n), an average of the data
(mean), and a covariance matrix (sigma). The structure
of the imitative datasets consisted of three clusters for
each dataset. The number of data points in Clusters 1,
2 and 3 were 5000, 2500 and 2500 respectively. They
were generated with four different argument sets,
which mean four different overlapping levels (shown
in Table 1) and generated three times for each set of
arguments; in total, 12 datasets were used in
experiments. Examples of these imitative datasets are
shown in Figure 6.
Table 1: Parameter values for the generation of the imitative
datasets used in this work.
Cluster
Sigma
(Covariance
matrix)
N
Means (Centres)
Level 1 Level 2 Level 3 Level 4
x y x y x y x y
1
[(6,15), (15,120)]
5000
5 35 3 30 1 25 - 1 20
2
[(2,0.3), (0.3,5)]
2500
-5
-10
-5
-10
-5
-10
-5
-10
3
[(3,2), (2,10)]
2500
16 1 1 2 1 8 1 4 1
3.3 Results
The datasets were clustered using DBSCAN,
OPTICS, FlowGrid, and FLOPTICS, with user-
defined parameters shown in Table 2. The values of ɛ
for DBSCAN and FlowGrid which provided the best
average accuracy results were selected (0.8 and 6.0
respectively).
Table 2: The parameter values for each technique.
DBSCAN OPTICS FlowGrid FLOPTICS
ɛ = 0.8
MinPts = 10
ɛ = optimal
MinPts = 10
ɛ = 6
MinDenB = 3
MinDenC = 40
Bin_size = 100
ɛ = optimal
MinPts = 10
Bin_size = 100
All techniques were implemented and run on RStudio
3.5.2, and the result are presented in Table 3.
COMPLEXIS 2020 - 5th International Conference on Complexity, Future Information Systems and Risk
100
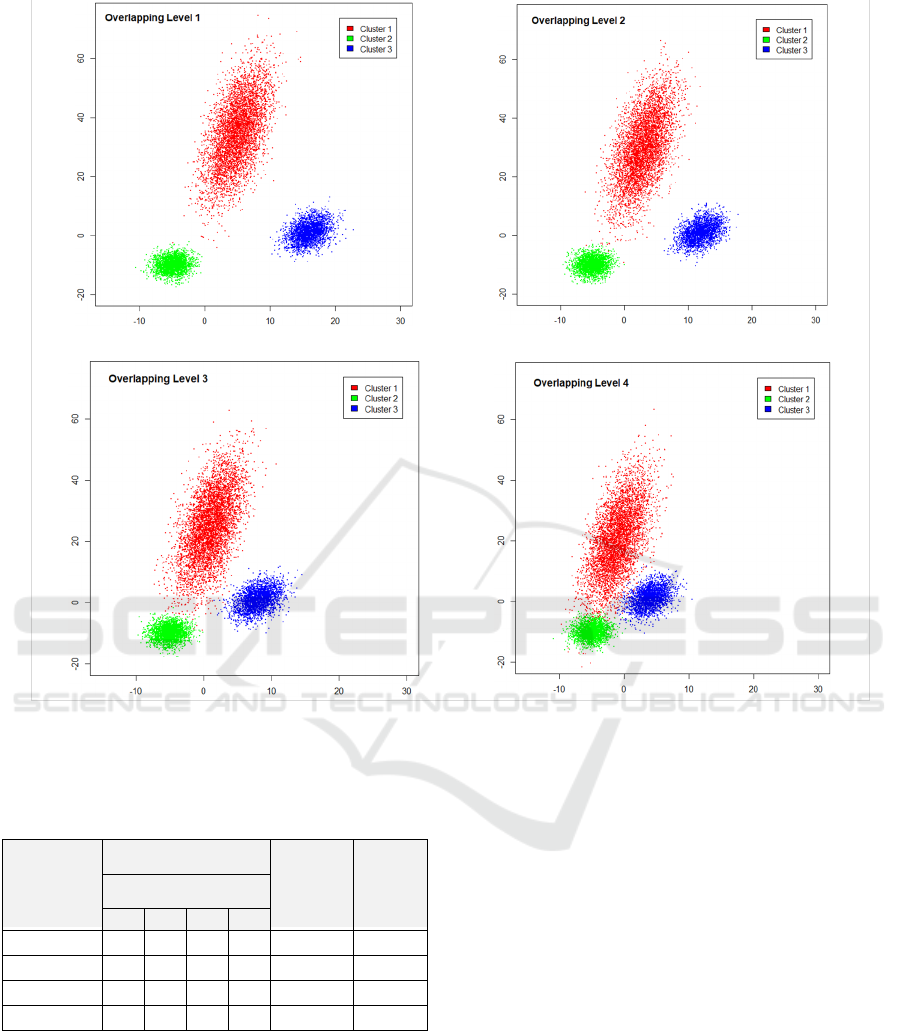
a Imitative dataset with overlapping level 1 b. Imitative dataset with overlapping level 2
c. Imitative dataset with overlapping level 3
d. Imitative dataset with overlapping level 4
Figure 6: Scatter plot of imitative datasets.
Table 3: The results of applying the analysis methods
techniques to the datasets.
Techniques
Average accuracy
(%)
Overall
average
accuracy
(%)
Average
Runtime
(milli-
second)
Overlapping dataset
Level
1 2 3 4
DBSCAN
94.99 94.70 94.80 62.70 86.80 6,728.60
OPTICS
99.93 99.75 98.60 92.07 97.59 1,961.12
FlowGrid
96.00 95.75 95.01 93.59 95.09 723.80
FLOPTICS
99.87 99.49 97.60 90.45 96.85 265.88
4 CONCLUSIONS
According to the results, OPTICS provided the best
average accuracy of 97.59%, though FLOPTICS gave
a higher accuracy result than DBSCAN and
FlowGrid. Although OPTICS gave the highest
accuracy, it was approximately 7.4 times slower than
the FLOPTICS technique. FLOPTICS was the fastest
technique applied to the imitative datasets, compared
with DBSCAN, OPTICS, and FlowGrid. In terms of
the number of user-defined parameters, FLOPTICS
requires two parameters, which are MinPts and
bin_size, while FlowGrid requires four parameters,
which are ɛ, bin_size, MinDenB, and MinDenC. In
conclusion, FLOPTICS has better performance than
comparative state-of-the-art automated gating
techniques.
5 FUTURE WORK
Although the FLOPTICS algorithm provides better
accuracy and a fast run time, its performance can be
further improved. In the process of partitioning data
into equal-sized bins, only non-empty bins are
processed, but both high-density bins and low-density
bins are treated equally; moreover, core points are
FLOPTICS: A Novel Automated Gating Technique for Flow Cytometry Data
101

identified by consideration of the number of
neighbours. An improvement would be to identify
core points not only by the number of neighbours, but
also the density of individual bins. Moreover, the
proposed technique is tested on a single specialised
machine. The next stage will be to revise the
algorithm to be machine-independent.
REFERENCES
Ankerst, M. et al. (1999) ‘OPTICS: Ordering Points To
Identify the Clustering Structure’, in Proc. ACM
SIGMOD’99 Int. Conf. on Management of Data.
Philadelphia.
Bashashati, A. and Brinkman, R. R. (2009) ‘A Survey of
Flow Cytometry Data Analysis Methods’, Advances in
Bioinformatics, 2009, pp. 1–19. doi:
10.1155/2009/584603.
BD-Biociences (2002) Introduction to Flow Cytometry :
Ester, M. et al. (1996) ‘A Density-Based Algotithm for
Discovering Clusters in Large Spatial Database with
Noise’, Comprehensive Chemometrics, 2, pp. 635–654.
doi: 10.1016/B978-044452701-1.00067-3.
Van Gassen, S. et al. (2015) ‘FlowSOM: Using self-
organizing maps for visualization and interpretation of
cytometry data’, Cytometry Part A, 87(7), pp. 636–645.
doi: 10.1002/cyto.a.22625.
Ge, Y. and Sealfon, S. C. (2012) ‘Flowpeaks: A fast
unsupervised clustering for flow cytometry data via K-
means and density peak finding’, Bioinformatics,
28(15), pp. 2052–2058. doi: 10.1093/bioinformatics/
bts300.
Groeneveld-Krentz, S. et al. (2016) ‘The Role of Machine
Learning in Medical Data Analysis. A Case Study:
Flow Cytometry’, (January 2016), pp. 303–310. doi:
10.5220/0005675903030310.
Jahan-Tigh, R. R. et al. (2012) ‘Flow Cytometry’, J. Invest.
Dermatol. Nature Publishing Group, 132(10), p. e1.
doi: 10.1038/jid.2012.282.
Lee, H. C. et al. (2017) ‘Automated cell type discovery and
classification through knowledge transfer’,
Bioinformatics, 33(11), pp. 1689–1695. doi:
10.1093/bioinformatics/btx054.
Lo, K., Brinkman, R. R. and Gottardo, R. (2008)
‘Automated gating of flow cytometry data via robust
model-based clustering’, Cytometry Part A, 73(4), pp.
321–332. doi: 10.1002/cyto.a.20531.
Moloney, M. and Shreffler, W. G. (2008) ‘Special Series :
Basic Science for the Practicing Clinician Basic science
for the practicing physician : flow cytometry and cell
sorting’, p. 2008.
Qian, Y. et al. (2010) ‘Elucidation of seventeen human
peripheral blood B-cell subsets and quantification of the
tetanus response using a density-based method for the
automated identification of cell populations in
multidimensional flow cytometry data’, Cytometry Part
B: Clinical Cytometry, 78B(S1), pp. S69–S82. doi:
10.1002/cyto.b.20554.
Rahim, A. et al. (2018) ‘High throughput automated
analysis of big flow cytometry data’, 135, pp. 164–176.
doi: 10.1016/j.ymeth.2017.12.015.
Robinson, J. P. et al. (2012) ‘Computational analysis of
high- throughput flow cytometry data’, (June). doi:
10.1517/17460441.2012.693475.
Rowley, T. (2019) Flow Cytometry - A Survey and the
Basics. doi: //dx.doi.org/10.13070/mm.en.2.125.
World Health Organization, R. O. for S.-E. A. (2009)
‘Laboratory guidelines for enumerating CD4 T
lymphocytes in the context of HIV/AIDS’, WHO
Regional Office for South-East Asia, pp. 1–86.
Ye, X. and Ho, J. W. K. (2018) ‘Ultrafast clustering of
single-cell flow cytometry data using FlowGrid’, BMC
Systems Biology, 13. doi: 10.1186/s12918-019-0690-2.
COMPLEXIS 2020 - 5th International Conference on Complexity, Future Information Systems and Risk
102
