
Semantic Overlapping in Translational Bioinformatics Applied to the
Matching between Clinical Trial Eligibility Criteria and Patient
Needs
Radmila Juric
1
, Eton Williams
1
and Inhwa Kim
2
1
University of South Eastern Norway, Kongsberg, Norway
2
Samsung SDS Europe Ltd, U.K.
Keywords: Clinical Trials, Eligibility Criteria, OWL/SWR.
Abstract: Software technologies play an important role in defining clinical trials, their eligibility criteria and recruitment
process, in which patient enrol to a trial if they satisfy eligibility criteria. In this research we address the
problem of semantic overlapping between eligibility criteria and patient needs through a software architectural
model which houses a specific computational model based on reasoning upon the overlapping semantics. The
architectural model is deployed using semantic technologies in order to explore the meaning of the
relationships between trials, eligibility criteria and patient needs. The novelty is in the reusability and thus
converting of the existing conceptual models on deriving eligibility criteria, available in literature, into the
proposed OWL model, which can serve any clinical trial and requirements patients may have. This paper is
written by computer scientists interested in manipulating semantics of data through computational models
using modern software technologies. It serves as an invitation to researchers from the biomedical and
translational informatics to debate the future of software support in managing clinical trials.
1 INTRODUCTION
The complex problem of designing Clinical Trials
(CT) and systemizing patient eligibility, using
software technologies, has been in the focus of
research interest for more than a decade (Kopcke and
Prokosch, 2014), (Ross et al., 2010) (Cimino et al.,
2007) (Shankar et al., 2006). The work in this field is
vast and has resulted in numerous solutions, which
address the complexity in defining the purpose of CT
and patents clinical and personal needs.
Unfortunately, in the third decade of the 21
st
century,
we still do not have a powerful software solution,
which could bring us closer to resolving the problem
and creating an universal environment for
pharmaceuticals, medical professionals and patients,
to address the problem in its entirety. The reasons are
numerous, but we would like to draw the reader’s
attention to the following three facts.
First, creating CT and matching their eligibility
criteria (ET) to patient needs, through software
technologies, is a transdisciplinary work. It would
require a high level of collaboration across many
disciplines. However, if we expect that software
technologies, which constantly offer innovations in
the way we collect and process data, can help in
resolving this problem, then we should put computer
scientists in charge of a new computational model for
one important reason. We should avoid using old-
fashioned software solutions, which proved to have
success in the past and think that they will bring
progress in future and in this particular problem
domain. Computer science, software engineering and
computational modelling are fast moving disciplines,
which require constant engagement if we wish to use
them properly and successfully in any problem
domain. We believe that processing the data
generated form bioscience research needs new
computational models created by computer scientists
and thus it is worthwhile to investigate if this could
help to create an universal solution for matching the
semantic between CT and their ET and patent needs.
Second, the research on creating and running CT,
using software technologies, is dispersed and
scattered. After 10 years, we may say that it is
confined to the existence of numerous repositories,
including ontologies and vocabulary of terms,
generated through natural language processing (NLP)
(Elkin et al., 2016), for the purpose of storing
knowledge and manipulating it, mostly through
Juric, R., Williams, E. and Kim, I.
Semantic Overlapping in Translational Bioinformatics Applied to the Matching between Clinical Trial Eligibility Criteria and Patient Needs.
DOI: 10.5220/0009406503150322
In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2020) - Volume 3: BIOINFORMATICS, pages 315-322
ISBN: 978-989-758-398-8; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
315

queries (Baader t al., 2018). This is far from
expectations we have in software engineering, where
software application manage relevant data and
computations, based on a specifically designed
computational model for this problem domain. None
of them exist in published work.
Third, the vast knowledge generated from
biomedical and pharmacological sciences is
expanding fast and it is almost impossible to bridge
the gap between these fields and clinical practices. In
spite of talking about translational informatics, since
the late 2000s (Payne et al., 2015), (Butte, 2008),
(Tsafnat et al., 2013) and pushing software
technologies into this field to help to bridge the gap
between biomedicine and clinical practices, we have
not even started thinking on how these new advances
in biomedical science may have an impact on CT. We
can not resolve this problem by keeping on creating
new knowledge-bases, new ontologies, new database
or similar repositories, and performing queries upon
them. This will not take us forward. What we need
are new computational models, which would assist in
collecting and managing the semantic of relevant and
shared data across these complex research fields
(Almami et al., 2016), (Juric et al., 2018), (Juric,
2019). New computational models can perform a
miracle in collecting and managing the semantic of
data and their matching in order to answer any
question we may have across this complex, but
semantically rich research field.
In order to understand our contribution, it is
important to note that we do NOT wish to
• propose a new “software” for dealing with the
problem of creating CT and finding eligible
patients,
• address a fraction or a slice of this problem by
going into details on how we would implement it
using software technologies and
• create a new repository of knowledge (called a
knowledge-base in the past), controlled
vocabularies often associated with ontologies, and
add them to the existing pool of sources available
in this problem domain.
What we wish to promote in this paper is a generic
computational model, based on the reasoning upon
the semantic of requirements in CT and patient’s
eligibility criteria. The ultimate goal would be to
exploit the semantic overlapping between the two and
define, through the reasoning process, either a CT, or
ET or patient’s best possible match with the two in
particular circumstances.
Software engineering solutions, which would
support data sharing across disciplines of
pharmaceuticals, biomedical sciences and clinical
practices, including patient clinical data, in order to
create and manage CT, would require a generic
software architectural model (Tarabi and Juric, 2018)
((Juric, 2020). 13,14). It is essential in specifying
sources of shared data and computational models for
identifying the best matching between a CT and
patient needs.
For proving the concept, we illustrate our proposal
by using Semantic Web Technology (SWT) and its
languages OWL/SWRL for defining the reasoning
process in which data is shared from biomedical
research published in the literature.
The paper is organized as follows. In section 2 we
specify why and how the SWT and its languages can
be used in this problem domain. In section 3 we
highlight similar work which influenced this research.
The proposal of the software architectural model is in
section 4 and section 5 illustrates an example of an
ad-hoc creation of an OWL model using existing
knowledge from conceptual modelling of Ct and ET
available in the literature. In section 6 we outline the
deployment of the proposal and in the last section we
debate results of this research and comment on future
steps in the last section.
2 WHY SWT
SWT and its layered cake has widely been used, since
its standardisation in 2004, for interpreting the
meaning of data available on the Internet. In
biomedical science OWL has been used for building
common ontologies and controlled vocabularies
across domains, enriched with reasoning rules in
SWRL for bringing inference and more semantics to
biomedical repositories. Knowledge presentation
with SWRL enabled OWL ontologies is extremely
powerful. It is description logic which allows
definition OWL classes and their constraints, in the
form of object and data properties, which enable the
definition of all concept and relationships between
them. This leads to numerous possibilities of
(i) using and exploiting SWRL enabled OWL
ontologies in many problem domains, and outside
the web (Juric, 2016)
(ii) creating OWL ontologies which are not controlled
vocabularies. They may still represent relevant
knowledge, but they will never become
knowledge-bases
(iii) reasoning upon the content of oWL ontologies
using SWRL rule for either strengthening its
knowledge or adding inference to it.
C2C 2020 - Workshop on COMP2CLINIC: Biomedical Researchers Clinicians Closing The Gap Between Translational Research And
Healthcare Practice
316

If we wished to avoid building knowledge bases and
still use SWRL enabled OWL ontologies for
manipulating the semantic of data we process, then
computational models, which house SWRL enabled
reasoning upon OWL concepts can bring inference
without having complex knowledge systems in the
background or using AI algorithms for creating and
manipulating inference. In this paper we talk about
software engineering applications of the SWT
technology which is OUTSIDE formal ontologies and
knowledge-bases and as such, might be a
promisingfstart for addressing the problem of CT /ET
and paitent needs.
We would like to use SWRL enabled OWL
ontologies in order to define
a) the semantic specific for defining and
manipulating EC for CT,
b) the way of converting the semantic of existing
solutions, which use different
method/technologies for defining EC into an
OWL model and
c) semantic overlapping between CT and their EC
and patient needs.
This semantic overlapping would create a cradle
for reasoning upon OWL concepts, which gives a
semantically rich pool of all possible combinations of
ET and patient needs. The power of reasoning,
secured through semantics overlapping between
relevant OWL concepts, infers either new individuals
or constraints in OWL through SWRL rules.
For readers interested in exploring the ways SWT
helps in the creation and manipulation of semantic
overlapping in biomedicine, we suggest reading our
previous publications (Almami et al., 2017), (Juric et
al., 2018), (Juric, 2019), (Juric 2016).
3 RELATED WORK
The section illustrates how scattered the research on
CT is and how it is impossible to find a thread
between the publications.
(Shankar et al., 2006) propose a knowledge-based
framework, named Epoch, and tailored it to the
Immune Tolerance Network research consortium in
order to cover a spectrum of clinical trials
management activities, by tracking study participants
and biological specimens processed in trial
laboratories. The role of developed ontologies in
their software architectural model is to conceptualize
knowledge in the relevant CT domain. In (Mucke et
al., 2009) a semantic model for representing items in
CT is proposed. Its purpose is to move away from
known database technologies and model the semantic
of the problem domain differently. However, their
semantic model does not feed any software
application and reasoning is not introduced for
decision making relevant for CT. In (Besana et al.,
2010) the SWT is used for CT recruitment and their
ontology contains data from patient heath records in
order to verify eligibility of patients for CT. A
consumer centric tool from (Pate et al., 2015), named
TrailX, which matches patients to CT uses numerous
sources of data, such as patient health records, Google
health and Microsoft Health Vault. However, the
matching of patient information and CT is done using
Columbus Matching technology, which relies on
NLP with the assistance of the Unified Medical
Language System. In (Damen et al., 2013) we can
read about the PASTEL platform which assists in CT
recruitment, by using the semantic generated through
topic maps and in (Dameron, 2013) the authors show
an OWL model which systemizes the ET with
partially known information. The authors of (Lee et
al., 2010) introduce the MindTrial system which
facilitates specific matches between clinical trial
criteria and patient volunteers, using a set of
ontologies and semantic queries. In (Elkin et al.,
2016) we can read how local clinical trials can be
enhanced with ontologies and Internet of Things, with
assistance of technologies such as natural language
processing.
It is obvious from the paragraph above that
ontologies are used for a variety of purposes and no
ontological model, generated in one study/project has
been used in another. Furthermore, the power of logic
reasoning with SWRL, suitable for ontology
matching, has not been exploited at all, and therefore
if there were a need for matching the semantic of CT
to patient eligibilities, some other technologies are
used. Finally, emantic overlapping, which is essential
in logic inference, enabled with SWT, is also not
used. This shows that the SWT has not been
completely utilized in this problem domain, except
for creating or retrieving knowledge-bases.
The inference, secured by the semantic
overlapping between OWL concepts, are not to be
confused with the term “reuse and overlapping” in
biomedical science, as described in (Maulik et al.,
2017). In the SWT world, the semantic overlapping
is a computational mechanism which secures
reasoning and inference as in (Almami et al., 2016),
(Juric et al., 2018), (Juric, 2019), and therefore its use
might be associated to semantic mapping evolution
known in biomedical ontologies, as described in (Dos
Res et al., 2014).
Semantic Overlapping in Translational Bioinformatics Applied to the Matching between Clinical Trial Eligibility Criteria and Patient Needs
317
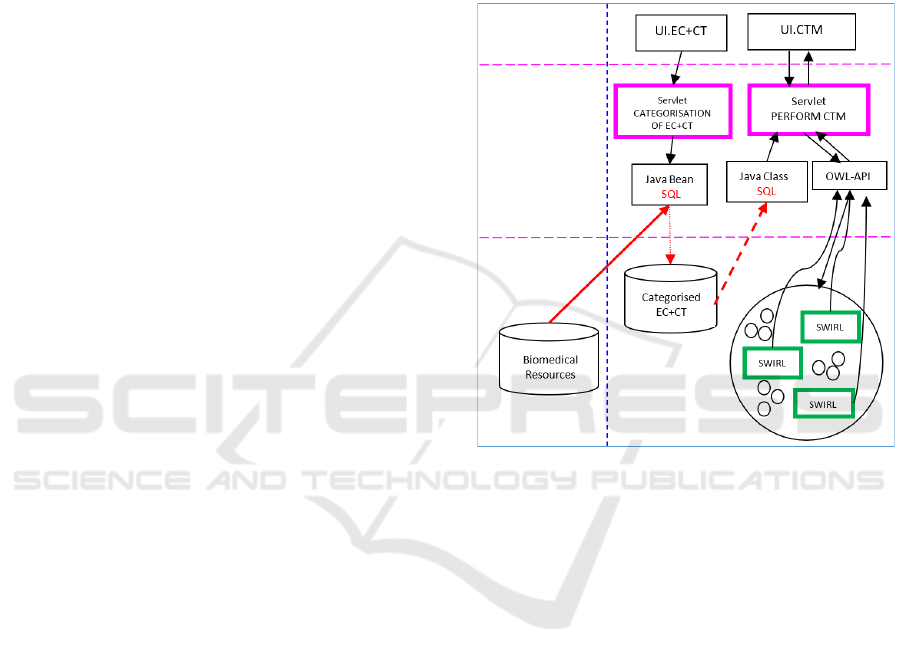
4 SOFTWARE ARCHITECTURE
If we wish to propose a solution which would address
problems with CT as described in the introduction,
and use the SWT which would infer the matching
between CT/EC and patient needs, Figure 1 shows the
essential software architectural (SA) model.
Software components in Figure 1 are technology
specific, i.e. we can use Java Servlet and Enterprise
Java beans technology in order to create a
computational model from the proposed architecture.
Therefore the applications generated from the SA
in Figure 1 would have a computational model
consisting of two types of computations:
a) typical transactional processing with SQL
databases in the background (left part in Fig. 1)
b) reasoning with OWL concepts through SWRL in
order to perform matching of CL/EC and patients
needs (right part in Fig. 1).
These types of computations are not very
common, but they are feasible and they do have
applications across many problem domains (Juric,
2016). For readers interested in software engineering
aspects of the implementations of applications from
the software architecture in Figure 1, we suggest
sources similar to (Patadia et al., 2011), (Shojanoori,
2013) (Tarabi and Juric, 2018).
SA In Figure 1 is component based and layered,
and allows a synergy between computations with
SQL in Java environments and reasoning with SWRL
in the OWL environment though OWL-API.
Obviously, UI.EC+CT interface (left part of the SA
model) would lead us towards the categorization of
EC and CT, which could be converted into
ontological concepts to secure reasoning with SWRL
for the matching with patient needs. (right part of the
SA model) Semantic Overlapping between CT/EC
and patient needs.
There is one important aspect of the proposed SA.
It will generate a software application suitable for
patients. The CATEGORISATION EC+CT
computations (servlet, left side of Figure 1)) would
collect relevant information from Biomedical
Sources and perform the categorisation of EC for
each CT in order to create an environment for
ontological matching. However, patient requests and
needs have already been categorised through the
PERFORM CTM computations (servlet, right side of
Figure 1) and asserted in the application though
UI.CTM interface (Kataria and Juric, 2014).
However, we could have turned the SA around and
start with categorisation of PATIENTS requests (left
side of Figure 1) and create an environment for
matching with EC for all CT, which have already
been categorised and enter into the PERFORM CTM
computations (right part of Figure 1.) The SA and
computations remain the same, only data and entry
mechanisms with UI change. To summarise, the SA
form Figure 1 creates software application, for
pharmaceutics/ clinicians/ patients, in order to secure
the best possible definition of eligibility criteria and
patient needs.
Figure 1: Software Architecture for enabling ontological
matching (CTM stands for Clinical Trial Matching).
4.1 Semantics of Biomedical Resources
Repository named Biomedical Resources from
Figure 1 is an abstract repository illustrated in Figure
2. There exists a vast biomedical knowledge in
various formats, available through a variety of
software solutions such as
(i) Public databases which exist across Biomedical
Informatics, including potential repositories
where various CT and their EC are advertised.
We should also look at sources such as UMLS
(UMLS) and SemDB (Kilicoglu, 2012) which
describe biomedical semantics and co-relation
between therapeutic drugs and diseases.
(ii) Numerous data sets which are generated from
biomedical experiments, and some of them might
be publically available. They are often associated
with the computational analytics and could be
used with popular leaning and predictive
technologies;
(iii) Biomedical data which available on the web /
social media / dedicated web applications, for the
C2C 2020 - Workshop on COMP2CLINIC: Biomedical Researchers Clinicians Closing The Gap Between Translational Research And
Healthcare Practice
318
purpose of disseminating advances in biomedical
research through either publications or social
media means (blogs, twitter, Facebook).
(i)-(iii) are collated in the red box in Figure 2. It is
important to note that Extraction of Biom. DATA
does not mean pure data retrieval. It should include
both: extraction of relevant data and their semantics
at the same time (Kataria and Juric, 2010), (Saaidi et
al., 2010).
Figure 2: Resources of Biomedical Data.
Figure 2 can also be implemented as a software
application using java technologies (Kataria and
Juric, 2010), but the creation of Biomedical
Resources repository from Figure 1, outlined in
Figure 2, is outside the scope of this work. It would
depend on each particular CT, and would vary
between suitable sources. The options on reusing
biomedical knowledge across a variety of repositories
must be open for future work.
In the study we show an example of using public
databases focusing on CT, but we do not exclude data
collected from publications in which categorisation of
EC for a particular CT is debated.
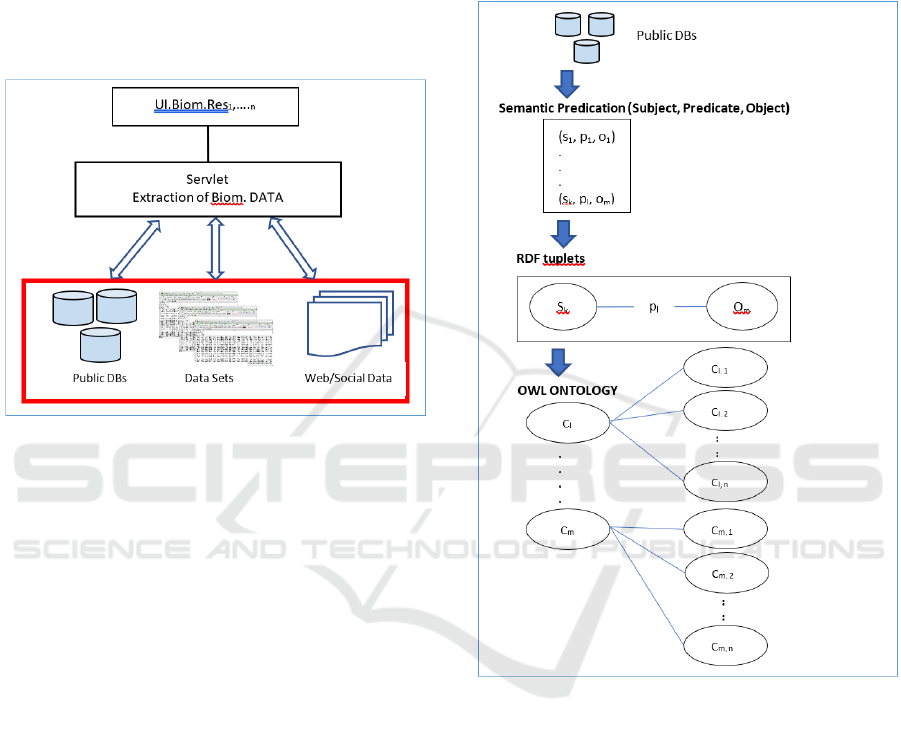
4.2 OWL Model for CT/ET
Figure 3 shows a potential benefit of using the
semantic from UMLS in order to categorise
biomedical knowledge relevant to CT and convert it
into OWL concepts. Sematic Predications (Ahlers et
al., 20107), (Zhang, 2014), (Machado, 2015) have
already been exploited in biomedical science and
could be an excellent starting point for categorizing
semantic of CT/EC and patient needs.
In Figure 3 we show the pathway from the
predications in the form of triplets:
(subject, predication, object)
where we can define the relationships between
subjects and objects through predications. It is
important to note, that a set of triplets: (s,p,o) could
be directly converted into RDF triplets and
consequently create an ontological model according
to the SWT stack.
Figure 3: From Triplets to OWL Model.
Set {C
1
,…C
n
} from Figure 3 are OWL classes,
converted from subjects and objects of semantic
predications. Predicates are converted into OWL
constraints, such as object properties. Ontological
hierarchies, shown at the bottom part of Figure 3 are
the main OWL concepts. Potential object properties
might be of either the “is-a” or “has” format or any
other type of relationship which may have existed
between subject and object in definition of CT. In this
particular example, where we need to define a CT
through its EC, “has” object property is more suitable
for explaining the semantic of a CT through EC
In Figure 3 C
i
denotes a CT and a set of {O
1,1
,…
O
1,n
} denote a categorised EC for that C
i
.
If we wish to perform semantic matching between
CT/ET and patient needs then ontological structures
Semantic Overlapping in Translational Bioinformatics Applied to the Matching between Clinical Trial Eligibility Criteria and Patient Needs
319
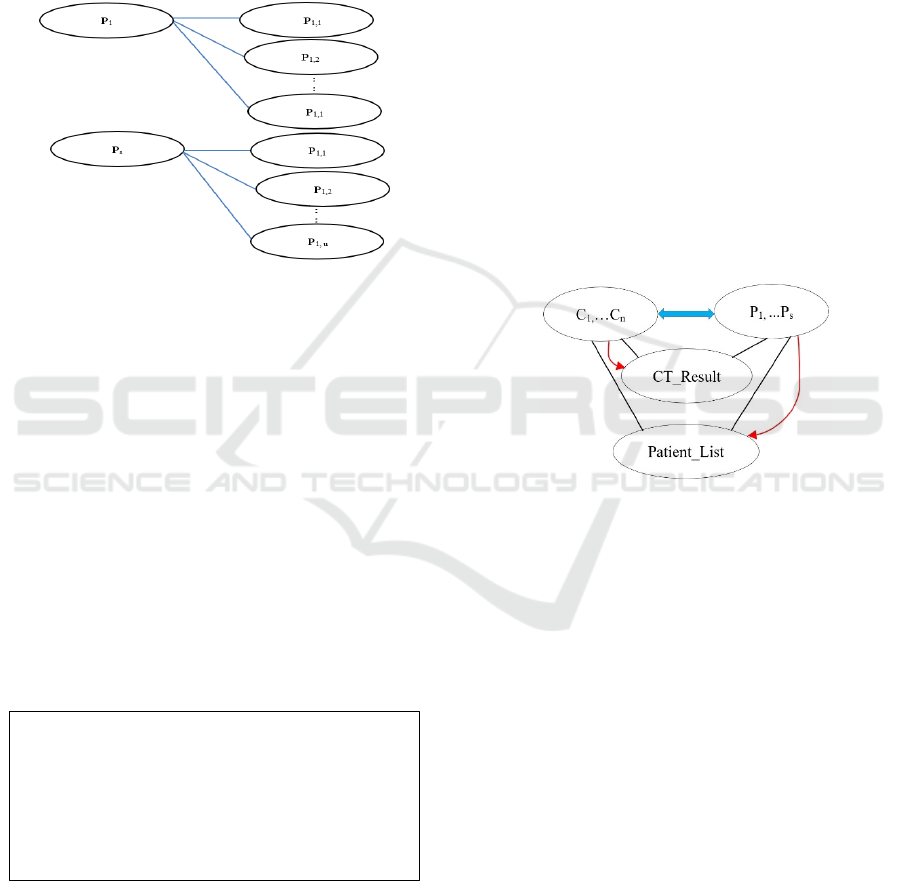
based on triplets (s,p,o) should be available for
describing patient requirements for CT. Therefore
Fig. 4 mirrors Figure 3: P
1
denotes a patient and a set
of {R
1,1
,… R
1,n
} denote his/her requirements for a CT.
The similarity between Fig. 3 and 4 means that if
we wish to match patient requests with CT/EC then
we should use similar categorisation for both: C
i
classes are described though {O
1,1
,… O
1,n
} and P
j
classes though {R
1,1
,… R
1,n
}.
Figure 4: OWL Model for Patient Requirements.
5 REASONING PROCESS
We illustrate the way of identifying semantic triplets
for categorizing the complexity of EC in CL as
described in (Ross et al., 2010). They have a specific
categorization of criteria (EC), affected by complex
relationships between disease diagnosis, clinical
phenotypes, which in turn are refined by their
severity, associated complications or response to a
specific treatment. We use a data sample from (Ross
et al., 2010) in order to illustrate that existing
knowledge (publication) is reusable, i.e. their
categorisation of EC could b entered into our OWL
model as individuals and properties, as long as we can
find semantic triplets (s,p,o).
Figure 5: A selection of triplets for OWL model.
A set of triplets could relate symptoms and
disease, treatment/intervention for the disease,
behaviour of a patient, clinical content related to the
disease, temporal criteria and similar. Predications
are available and range from “caused-by”,
“described-with” and “diagnosed-by”, to “without”,
“at least n times per week”, “at hospital discharge”
and “contains-normal-values”.
Figure 5 shows a selection of triplets, where p1 is
an object/subject representing a patient. Predicates
are defined as object properties between individuals
of classes. Individuals of these classes are
subject/objects defined in these triplets.
If we connect all individuals of these OWL classes
(subject/objects) with appropriate object properties
derived from predicates, we could run SWRL
reasoning upon such classes and create an answer if a
particular patient would be eligible for a CL. The
decision will depend on the reasoning process in
which individual(s) of the PATIENT class will be
eligible for a CT only if the reasoning with SWRL
confirms that the particular patient(s) have the same
object properties (predicates) defined between
him/her (them) and eligibility of a clinical trial
(subjects and objects in triplets).
Figure 6: The Reasoning Process.
The reasoning process from Fig 6 is self
explanatory. Blue arrow specifies definition of object
properties between C
i
and P
j
classes, black lines
indicate classes involved in reasoning and red one-
directional arrows show inference: only suitable CT
are moved into the CT_Result class for a patient and
only patients which satisfy the EC would be moved
to Patient_List class. The reasoning process is
programed through the following SWRL rules.
CLINICAL_TRIAL (?, D) and has_EC1(?D, EC1)
and has_ECn (?D,ECn) - > CT_Result (?D).
PATIENT (?, XXX) and has_Req1(?XXX, Req1)
and has_Rwqn (?XXX,Reqn) - > PATIENT_LIST
(?XXX).
where EC
1
, …. ECn and Req
1
, …. Req
s
are predicates
(object properties) identified in triplets from the
above and D/XXX are variable for patient/CT.
The message from the OWL model and reasoning
with SWRL, based on semantic overlapping between
ET for a CT and patient requirements, is that the
semantic overlapping secures an almost instant
(p1, diagnosed, disease1)
(p1,exludes, disease2)
(lab-tests, contains-normal-values, p1)
(disease2, treated-by, antibiotics)
(treatment2, unsuccessful-for, p1)
(visits-repeated, required-for, tretament1)
(p
1, refuses, re
p
eated visits
)
C2C 2020 - Workshop on COMP2CLINIC: Biomedical Researchers Clinicians Closing The Gap Between Translational Research And
Healthcare Practice
320

answers to the questions we may have. We filtered all
the patients (?D) which satisfy ET for a CT for SWRL
rule 1. SWRL rule 2 answers this question: Which CT
are available for a particular patient? The
computational model remains the same, and SWRL
rule uses the same object properties (predicates). The
only difference would be in the format of the rule:
6 CONCLUSIONS
This paper is written from the computer science and
software engineering perspectives and therefore should
show concepts upon which we can build an application
for finding semantic overlapping between CT and
patient needs. Data sharing and computations upon
biomedical and clinical data will reside within one
dedicated light-weight software application. It should
be, suitable for running in mobile and wireless
environments, where updates and constant changes are
welcome and not seen as obstacles. The application
will NOT build an excessive new knowledge base, but
will reuse existing biomedical knowledge, which has
been growing rapidly on a daily basis and add value to
the way we conduct CT.
This study is one of many attempts of using the
SWT and ontological modelling for the purpose of
creating semantic overlapping for matching of CT and
their EC with patient needs. The OWL model and its
constraints, as introduced in (Juric, 2019) can fit the SA
model from Figure 1 and individuals needed for the
OWL model, which would secure reasoning, could be
taken from any of the available sources which
document information on CT. If we could represent
this knowledge in the form of semantic predications
(triplets), than the implementation of the application,
defined by Figures 1,2 and 3 would be straight forward.
In cases when we do not have structures of triplets, the
categorization of data for CT and EC and patient needs
should be performed with OWL principles in mind.
SWRL enabled OWL ontologies are very powerful and
they can really make a difference in this problem
domain, considering that the computational model and
the SA allow software solutions which could run on
modern environments, including Android. Therefore,
from the computational science and software
engineering perspectives, there should be no obstacles
in commercializing the proposal.
However, this study may send message to
researchers from the biomedical field. Repositories
which may contribute towards Biomedical Resources
in Figure 1 should be available, accessible and shared,
but never integrated or changed due to their role in
creating semantic matching. We should leave their
manipulation to computer scientists who can make this
proposal operational and commercial. This implies
that variations of possible implementations of the SA
from Figure 1 would keep up with advances in
software technologies, as long as we can interpret the
semantic overlapping between CT and ET with patient
needs.
We are currently looking at the possibilities of
using biomedical data sets for running predictive
analytics with learning technologies in order to predict
possible predications between various ranges of
subjects and objects, applicable to clinical trials. The
proposed computational model remains the same.
REFERENCES
Ahlers, M. Fiszman, D. Demner-Fushman, F. M. Lang, T. C.
Rindflesch, 2007. Extracting Semantic Predications from
Medline, Pacific Symposium on Biocomputing 12:209-
220(2007).
Almami, E. Almami, I., Juric, R, 2016. Knowledge
Dissemination In Biomedical Science: Using
Ontological Reasoning For The Analyses Of Activated
Enzymes In Various Diseases, In Proceedings of the 21
st
SDPS 2016 conference, December 2016
Almami, E. Almami, I., Juric, R, 2016. Knowledge
Dissemination In Biomedical Science: Using
Ontological Reasoning For The Analyses Of Activated
Enzymes In Various Diseases, In Proceedings of the 21
st
SDPS 2016 conference, December 2016
Baader, F., Stefan Borgwardt, Walter Forkel, 2018. Patient
Selection for Clinical Trials Using Temporalized
Ontology-Mediated Query Answering WWW ’18
Companion, April 23–27, 2018, Lyon, France,
Besana P, Cuggia M, Zekri O, Bourde A, Burgun A. 2010.
Using semantic web technologies for clinical trial
recruitment. The Semantic Web-ISWC 2010: 9th
International Semantic Web Conference, ISWC 2010;
November 7-11, 2010; Shanghai, China. Springer; 2010.
p. 7748.
Butte AJ, 2008. Translational bioinformatics: coming of age.
J Am Med Inform Assoc 15(6):709–714.
Damen, D., Kim Luyckx, Geert Hellebaut, Tim Van Den
Bulcke. Åastel 2013. A Sematic Platform for Assisted
Clinical Trial Patient Recruitment, In Proceedings of the
2013 IEEE Intern. Conference on Healthcare
Informatics, 9-11 September 2013, Philadelphia, PA,
USA.
Dameron, O., Besana, P., Zekri, O., Bourde, A., Burgun, A.,
Cuggia, M. (2013) OWL Model of clinical trial eligibility
criteria compatible with partially-known information,
Journal of Biomedical Semantics, BioMed Cnetral, 2013,
4(1), pp 17.
Dos Reis et al. 2014. Understanding Semantic mapping
evolution by observing changes in Biomedical
Ontologies, inJournal of Biomedical Semantics,
47(2014) pp 71-82.
Semantic Overlapping in Translational Bioinformatics Applied to the Matching between Clinical Trial Eligibility Criteria and Patient Needs
321

Elkin, P.L., SCHLEGEL, D.R. ANAND, E. 2016. Recruiting
Participants to Local Clinical Trials using Ontology and
the IoT, in Studies in Health Technology and Informatics,
January 2016
Juric R. (2019) Semantic matching between Eligible Paitents
and clinical Trials using SWRL enabled OWL
Ontologies, in Proceedings of the 24
th
–SDPS 2019
Conferece, Taiwan, July.
Juric, R. 2016. Could Semantic Technologies Create a new
Computational Model outside Semantic Web, in
Proceedings of the 21
st
SDPS Conference, December,
FL, US.
Juric, R., Kim, I. 2017. Software Architectures for Smart
Applications Which Merge Ontological Reasoning With
Big Data Analytics, In Proceedings of the 22
nd
SDPS
2017 conference November 2017
Juric, R., Almami, E. Almami, I. 2018. Semantic
Overlapping of OWL Models in Biomedicine, In
Proceedings of the SDPS 2018 Workshop on
Accountability of AI, December 2018, Bologna, Italy
Kataria, P., Juric, R. 2010. Creating Semantics From User
Inputs Through Ontological Reasoning, in Proceedings
of the 15
th
International Conference SDPS 2010,
University of Texas in Dallas, US, June 6-11.
Kataria, P., Juric, R. 2014 Transferring Ontological
Individuals In OWL/SWRL Enabled Ontologies, In
Proceedings of 18th International Conference SDPS
2013, Campinas, Sao Paulo, Brazil, October 27-31.
Kilicoglu H, Shin D, Fiszman M, Rosemblat G, Rindflesch
TC. 2012. SemMedDB: a PubMed-scale repository of
biomedical semantic predications. Bioinformatics
2012;28:3158–60.
Köpcke,F., Hans-Ulrich Prokosch, 2014. Employing
Computers for the Recruitment into Clinical Trials: A
Comprehensive Systematic Review, J Med Internet Res.
2014 Jul; 16(7): e161.
Lee Y, Dinakarpandian D, Katakam N, Owens D. (2010)
MindTrial: An Intelligent System for Clinical Trials,
AMIA Annu Symp Proc. 2010;2010:442–6.
http://europepmc.org/abstract/MED/21347017.
Machado, C. M.; Rebholz-Schuhmann, D.; Freitas, A. T.;
Couto, F. M. 2015. The semantic web in translational
medicine: current applications and future directions.
Briefings Bioinf. 2015, 16, 89.
Maulik R. Kamdar, Tudorache, T., Musen, Daniel R., 2017.
A Systematic Analysis of Term Reuse and Term overlap
across Biomedical Ontologies, in Semantic Web 2017;
8(6): 853–871.
Moore, R., Lopes, J., 1999. Paper templates. In
TEMPLATE’06, 1st International Conference on
Template Production. SCITEPRESS.
Mucke, R., Mathias Lobe, Magnus Knuth, Frank Loebe,
2009. Semantic model for Representing Items in Clinical
Trials. Proceedings of the 2009 22nd IEEE International
Symposium On Computer-Based Medical Systems, 2-5
August 2009; Albuquerque, NM, USA.
Ontology Web Language https://www.w3.org/TR/owl-
features/ and Semantic Web Rule Language, available
atT https://www.w3.org/Submission/SWRL/
Patadia, R.; Kataria, P., Juric, R., Kim, I. 2011. Conceptual
Design Of Semantic Software Applications, in
Proceedings of the 16th International Conference on
System Design and Processing Science, SDPS 2011, Jeju
Island, South Korea, June 12-16.
Patel C, Cimino JJ, Dolby J, Fokoue a, Kalyanpur a,
Kershenbaum a, Ma L, Schonberg E, Srinivas K. 2007.
Matching patient records to clinical trials using
ontologies. Proceedings of the 6th International
Semantic Web Conference, 2nd Asian Semantic Web
Conference, ISWC 2007 and ASWC 2007; Nov. 11-15,
Busan, Korea. Springer; 2007. p. 6297
Payne, P., Embi, R.O. Peter, J. (Eds.) (2015) Translational
Informatics: Realizing the Promise of Knowledge-
Driven Healthcare. Springer, London 2015.
Ross J, Tu S, Carini S, Sim I. 2010. Analysis of eligibility
criteria complexity in clinical trials. AMIA Summits
Transl Sci Proc AMIA Summit Transl
Sci. 2010;2010:46–50.
Saaidi, R., Kataria, P., Juric R. 2011. Semantic Management
of Submissions of Applications for Marketing
Authorisation of Medicines”, book chapter, in: Suh, S.C.,
Tanik, M., Gurupur, V. P. (Eds.) Biomedical
Engineering: Healthcare Systems, Technology and
Techniques (,Springer; 1st Edition (20 Aug 2011).
Shankar, R.D. S.B. Martins, M.J. O’Connor, D.B. Parish,
A.K. Das, 2006. Towards Semantic Interoåerability in a
Clinical Trials Management Systems in Proc of ISWC,
LNCS 4273, pp 901-912, 2006, Springer Verlag, Berlin
2006
SWT Semantic Web Technology SWT Road Map, available
at W3C website https://www.w3.org/2001/10/03-sww-
1/slide7-0.html
Shojanoori, R. (2013) “Towards Formalization of Situation-
Specific Computations In Pervasive Computing
Environments”, PhD Thesis, University of Westminster,
London, UK, 2013
Shankar, R.D., S.B. Martins, M.J. O’Connor, D.B. Parish,
A.K. Das, 2006, EPOCH An Ontological Framework to
Support Clinical Trials Management, in ACM
Proceedings of HIKM 06, November, VA, US
Tsafnat G., Lin F., Choong M.K. (2013) Translational
Biomedical Informatics. In: Dubitzky W., Wolkenhauer
O., Cho KH., Yokota H. (eds) Encyclopaedia of Systems
Biology. Springer, New York, NY
UMLS Reference Manual, National library of Medicine (last
update September 2009) available at
https://www.ncbi.nlm.nih.gov/books/NBK9676/pdf/Bo
okshelf_NBK9676.pdf)
Tarabi, M., Juric, R. 2018. Software Architectures For Smart
Applications In The Management Of Chronic Diseases:
A Case Study Of Reversibility Of Diabetes 2, In
Proceedings of the 51
st
HICSS Conference January 2018
R. Zhang, M. J. Cairelli, M. Fiszman, G. Rosemblat, H.
Kilicoglu, T. C., Rindflesch, S. V. Pakhomova, G. B.
Melton, 2013. Using semantic predications to uncover
drug–drug interactions in clinical data. Journal of
Biomedical Informatics 49 (2014) 134–147
C2C 2020 - Workshop on COMP2CLINIC: Biomedical Researchers Clinicians Closing The Gap Between Translational Research And
Healthcare Practice
322
