
An Effective Sparse Autoencoders based Deep Learning Framework
for fMRI Scans Classification
Abeer M. Mahmoud
1,2 a
, Hanen Karamti
2,3 b
and Fadwa Alrowais
2c
1
Faculty of Computer and Information Sciences, Ain Shams University, Cairo, Egypt
2
Computer Sciences Department, College of Computer and Information Sciences, Princess Nourah bint Abdulrahman
University, PO Box 84428, Riyadh, Saudi Arabia
3
MIRACL Laboratory, ISIMS, University of Sfax, B.P. 242, 3021 Sakiet Ezzit, Sfax, Tunisia
Keywords: Sparse Autoencoder, Autism, Medical Image Classification.
Abstract: Deep Learning (DL) identifies features of medical scans automatically in a way very near to expert doctors
and sometimes over beats in treatment procedures. In fact, it increases model generalization as it doesn’t
focus on low level features and reduces difficulties (eg: overfitting) of training high dimensional data.
Therefore, DL becomes a prioritized choice in building most recent Computer-Aided Diagnosis (CAD)
systems. From other prospective, Autism Spectrum Disorder (ASD) is a brain disorder characterized by
social miscommunication and confusing repetitive behaviours. The accurate diagnosis of ASD through
analysing brain scans of patients is considered a research challenge. Some appreciated efforts has been
reported in literature, however the problem still needs enhancement and examination of different models. A
multi-phase learning algorithm combining supervised and unsupervised approaches is proposed in this paper
to classify brain scans of individuals as ASD or controlled patients (TC). First, unsupervised learning is
adopted using two sparse autoencoders for feature extraction and refinement of optimal network weights
using back-propagation error minimization. Then, third autoencoder act as a supervised classifier. The
Autism Brain fMRI (ABIDE-I) dataset is used for evaluation and cross-validation is performed. The
proposed model recorded effective and promising results compared to literatures.
a
https://orcid.org/0000-0002-0362-0059
b
https://orcid.org/0000-0001-5162-2692
c
https://orcid.org/0000-0002-8447-198X
1 INTRODUCTION
Deep Learning is an artificial intelligence approach
that offers automatic learning features similar to
experts in many fields but especially in computer
vision and imaging domains. The potential satisfying
feedback of applying deep learning methods in
medical imaging encouraged many researchers to
prioritize the approach while solving their research
challenges and faced problems (Krizhevsky et al.,
2012, Najafabadi et al., 2015;Litjens et al., 2017,
Ravì D., et al., 2017). Many neural network models
of varies number of layers to transform input images
to outputs through accurate extraction of most
discriminative features were proposed. However, the
convolutional neural network (CNN) (Bengio et al.,
2013) is highly a recommended selection. CNN
contains layers that transform the input with
convolution filters of a small extend. It is preferred
as it doesn’t waste time in learning separate
detectors for identical objects placed differently in
an image, and it reduce the number of network
training parameters as weight have no direct relation
with image size.
For many years, autism disorder has received
more attention as an important disease. It is a
significant crisis for many families in Arabic society
because unknown reasons for causing and poor
background of the characteristic of the disease.
Autism appears since birth and it recorded a variant
and multiple symptoms of illness. Many researches
tried to diagnose the disease from different data
types using machine learning techniques (SE.
Schipul et al., 2012; M. Plitt et al,2015; A. Abraham,
540
Mahmoud, A., Karamti, H. and Alrowais, F.
An Effective Sparse Autoencoders based Deep Learning Framework for fMRI Scans Classification.
DOI: 10.5220/0009397605400547
In Proceedings of the 22nd International Conference on Enterprise Information Systems (ICEIS 2020) - Volume 1, pages 540-547
ISBN: 978-989-758-423-7
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

et al.,2017; ). ABIDE-I is a large autism brain scans
images dataset. Knowledge discovery in this datasets
is a challenge for many researchers. Therefore
several machine learning approaches (G. Chanel, et
al., 2016; XA. Bi,2018; G. Chanel, et al.,2019 ) and
deep learning (Xi. Li, et al., 2018; H. Li, et al., 2018)
have been reported on ABIDE dataset. The data
contains 1035 individuals between autistic (ASD)
and controlled patients (TCs). These models may
suffer from lack of generalization of low reported
classification accuracy. Also, some researchers used
the full set of data while others used it partially due
to availability of resources and/or characteristics of
the used technique. However, the domain still opens
and encourages competition with different novel
hybridization methods and/or enrichment of
knowledge extraction or mining results from
medical data such ABIDE.
In this paper, an attempt to build a generalized
model for dealing with large medical imaging data is
proposed. It is a new Deep Learning framework
based on sparse autoencoder where it allows
machines to learn very complex data representation
that can subsequently be used to perform accurate
data classification in a fully treatment for ABIDE
datasets. Two conventional sparse autoencoders
(SAE) are unsupervised learning used in training to
extract discriminative features. The third one is a
supervised learning used to classify the extracted
features and to diagnose the ASD. These three stages
were jointed to form a stacked framework. The rest
of the paper is as follow: Section 2 presents related
work on ABIDE. Section 3 shows our contributions
framework construction, features extraction and
images classification. Section4 discusses computa-
tional results and section 5 concludes the paper.
2 RELATED WORK
Autism is neurological developmental disorder that
the disorder ,affect many families and recently
spread due to many unclarifed reasons. More
attention focuses on rehabilitation of families to
handle their cases however an intensive research
currently focusing on the treatment and/or
knowledge discovery in autism medical imaging.
fMRI scans of brain are a kind of medical imaging.
Each fMRI scan is actually a group of tiny cubic
elements called voxels(X,Y, Z or 3D) and if time is
added during data gathering , it become 4D. A time
series is extracted from each voxel to save its
activity change over time. One of the famous brain
scans for autism is ABIDE. In the following, some
related work on ABIDE with different techniques.
(H. Chen, et al., 2016), investigated the effect of
different frequency bands for constructing brain
functional network, and obtained 79.17% accuracy
using SVM technique applied to 112 ASD and 128
healthy control subjects. (Brown et al., 2018),
proposed framework based on an element-wise layer
for deep neural networks. Then they incorporate the
data-driven structural priors. They select 1013 of
539 healthy control and 474 with ASD and reported
68.7%. (XA. Bi, et al., 2018), selected the support
vector machines (SVM) as a classifier but used
multiply SVM architecture to enhance the results as
single SVM gives poor results. Their selected
samples included 46 TC and 61 ASD and recorded
96.15%. (Bi Xia-an, et al, 2018), proposed genetic-
evolutionary SVM and validated by data of 157
participants (86 AS and 71 TC). The classification
accuracy reached to 97.5%. (XA. Bi, et al, 2018),
presented multiple Random Neural network (NNs)
based model on ABIDE. They focused on 50 ASD
and 42 TCs samples. A random 5 NN clusters were
built using 5 different NNs. The highest accuracy
cluster is selected as the best base classifier. Then,
valuable features were used to retrieve abnormal
brain regions.
In addition, several deep learning models have
been proposed recently. (Xi. Li, et al, 2018), used
3D CNN to detect features based on the spatial
characteristics, then they voted the results through
visualization and interpretation to choose the most
competent for ASD or TCs output. They
implemented their proposed on subset of 82
diagnosed with autism child and 48 controlled. They
obtained higher accuracy. (X. Guo, et al., 2017),
proposed a DNN with a novel method for extraction
of features for high dimensional rs-fMRI. They used
multiple trained sparse auto-encoders for feature
extraction, and then used DNN for high-quality
representations of the whole-brain function
connectivity patterns. They considered 110 samples
(55 ASD and 55 TCs) and recorded 86.36%. (M.
Khosla, et al., 2018), proposed 3D deep learning
model and used subset of data. They classified
healthy controld individuals by 76 67% classifcation
accuracy of using subset of 178 samples for training.
However generalization of proposed models is
affected by small samples.
A two phase’s method was proposed in (Xi. Li,
et al., 2018). First, a deep neural network classifier
was trained with original scans. Then a corruption
on the regions of interest (ROIs) of the brain scans
An Effective Sparse Autoencoders based Deep Learning Framework for fMRI Scans Classification
541
were feed to the trained network to enhance
perdition. Their approach was tested on 82 subset
and reached 85.3% accuracy. (Heinsfeld et al.,
2018), proposed the usage of respective neural
patterns of functional connectivity in rs-fMRI as a
main discriminative features guide in classifying
healthy versus autism patients. They implemented
unsupervised phase, where they used 2 stacked
denoising autoencoders. Then they used multi-layer
perceptron as a supervised classifier. They applied
the proposed method on the whole data set and
reported 70%, it is considered an improvement with
almost full data.
(H. Li,, et al., 2018), proposed a deep transfer
learning NN framework. They trained a stacked
sparse autoencoder offline extract the functional
connectivity patterns. They selected a subset of 310
samples, and reported a range between 62.3% and
70.4%. (Eslami, T., et al., 2019), proposed a new
joint learning procedure combining autoencoder and
a single layer perceptron. Then continued with
results of first phase and applied a data
augmentation strategy, based on linear interpolation
to produce synthetic training datasets. They used
only 13 sites from 17 and reported 80%.
The literature showed that a few studies have
considered sparse autoencoder in the classification
of individuals with autism based on fMRI. Also,
only one reference implemented their proposed
system on the dataset fully. Hence, the motivation of
our proposed method where a full treatment of
ABIDE database is evaluated.
3 PROPOSED SPARSE AES
BASED DL FRAMEWORK
3.1 Sparse Autoencoder
Autoencoder consists of basically three-layers, input,
hidden, and output, respectively. The hidden layer is
fully connected to the input and output layers
through weighted connections. It is trained to
reconstruct similar input at output layer effectively.
One of autoencoders disadvantages is the limited
number of hidden units. The Spar Autoencoder
(SAE) (Makhzani & Frey., 2013; fgJanowczyk, A..,
et al., 2017; Hou, L., et al., 2019) tried to solve this
by adding a sparsity constraint, to tune a large
number of neurons with low average output and
hence neurons appear schematic inactive most of the
time. This can be implemented by setting a loss
function during training. Assume hidden neurons
activation function = haj, then average activation of
it is in Eq.1
A
∑
h
x
(1)
The objective of sparsity constraints is to minimize
A
f
so that A
f
=A , where A is a sparsity constraint
between 0 such as 0.05 , A ̂
j
, the average activation
of hidden unit j (in the sparse autoencoder), p
t
is the
penalty term and N= number of neurons in the
hidden layers
p
∑
KLA||A
(2)
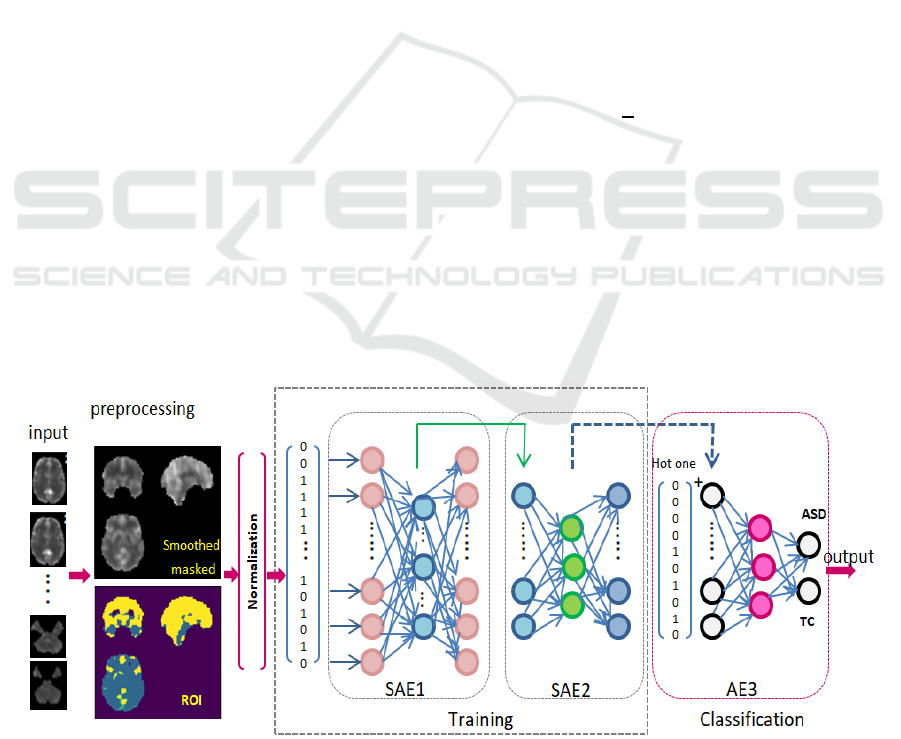
Figure 1: Proposed Sparse autoencoder based deep learning model for medical data classification.
ICEIS 2020 - 22nd International Conference on Enterprise Information Systems
542
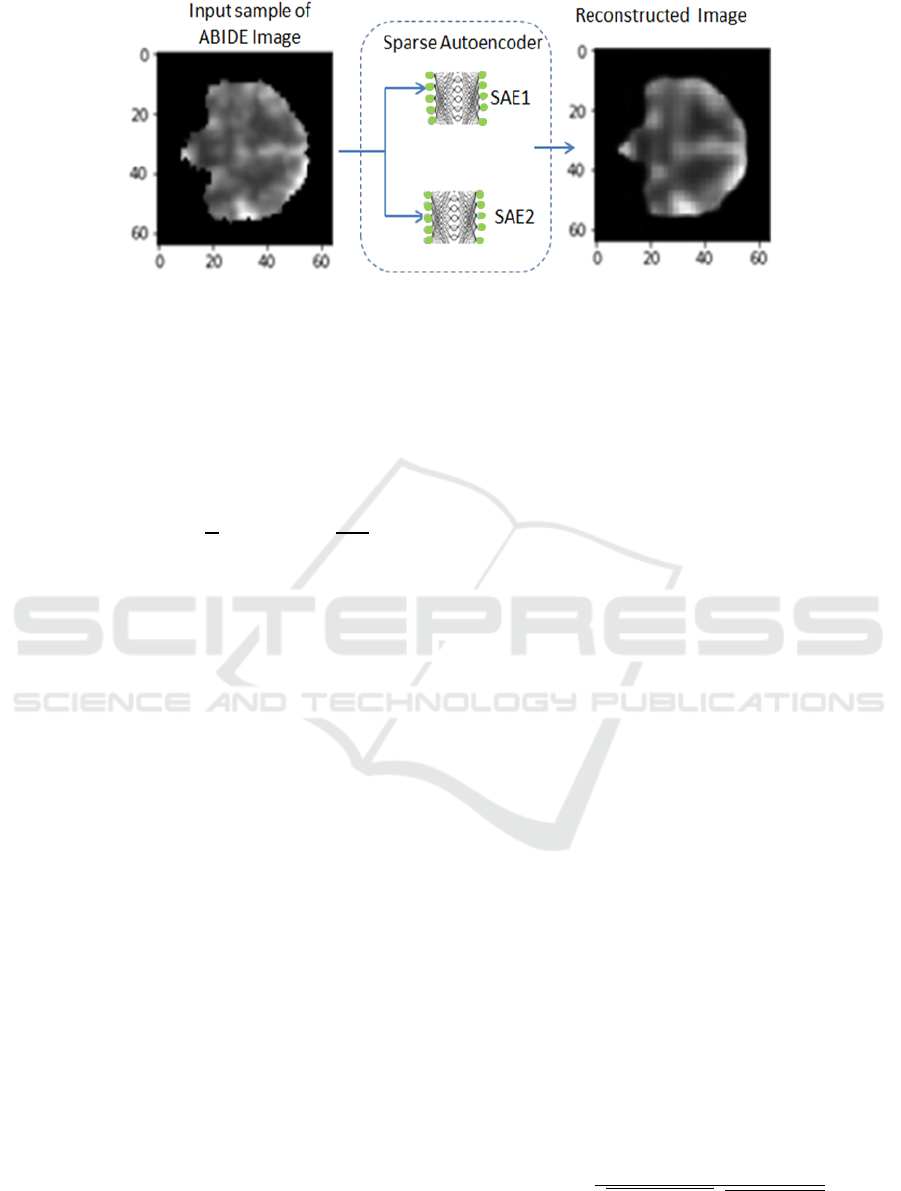
Figure 2: Un-supervised sparse autoencoders for feature extraction.
To enforce sparsity constraints regularizes
complexity and prevent over-fitting, a penalty term
is added to cost function which penalizes A ̂, de-
weighting significantly from A. Since A and A ̂, can
be seen as the probabilities of Bernoulli random
variables, a Kullback-Leibler (KL) divergence (Shin
et al.,2013), represented by Eq.3.
KLA|A
A log
1A
log
(3)
3.2 Proposed Architecture &
Extracting Features
The proposed architecture is shown in Figure 1. It is
composed of two Sparse AEs as unsupervised
learning for feature extraction and followed by AE
for classification task:
Input layer: receiving the pre-processed fMRI
scans.
Two Sparse AEs: are two unsupervised
autoencoders; for deeper feature extraction and
refinement of highly important set of unique
features. Figure 2, shows the unsupervised role
of the two SAE to reconstruct input while
learning most valuable feature.
Output layer: is a supervised autoencoder to
classify cases into ASD or TC.
The unsupervised Sparse AEs receives data without
labels. Each Sparse autoencoder contains two
convolutional (CNN) layers. Normalization and
Max-pooling are necessary for smoothing the
learning process. Then the identical set of
parameters are kept for decoding phase (eg: kernel
size (3,3,3) for dimensionality reduction). To reach
the nearest reconstruction of its input at the output
layer, the AE is forced to infer major information
preserving a reduction while representing the input
in the hidden layer, then mapped to the output layer.
Therefore, each hidden node represents a feature of a
reduced but accurate copy and this can be evaluated
through visualization, See figure 2. SAE has a
sparsity constraint that is imposed on the mean
activity of hidden layer (Shin et al.,2013) to
overcome overfitting.
The input and output layers for the first
autoencoder have 17668 features fully connected to
hidden bottleneck of 3015 units from the hidden
layer. The second autoencoder maps 3015 inputs
from the output of the previous autoencoder to
outputs through a hidden layer of 983 units and then
to 271 units. The batch size=256 and epoch=80. To
classify the individuals with ASD, we used
supervised autoencoder, inserted in the last layer
(output layer) of the proposed neural network. The
discriminative features were injected to last
autoencoder, in addition to the correspondence
vector of numbers for each class of output (one-hot).
The vector consists of only 1 for the class it
represents and all others are zeros. Softmax function
is used for regression of output. Batch size=1024
and epoch=100, are used.
3.3 Training, Validation and Testing
First, each raw rs-fMRI data was preprocessed, and
the whole-brain function connectivity patterns (FCP)
were obtained by calculating the Pearson’s
Correlation coefficient (CC) of Time series (TSs)
from any pair of ROIs. Given two times series, T
s1
and T
s2
, each of length T
L
, the Pearson’s correlation
can be computed Eq.4
𝜌
,
∑
∑
∑
(4)
An Effective Sparse Autoencoders based Deep Learning Framework for fMRI Scans Classification
543
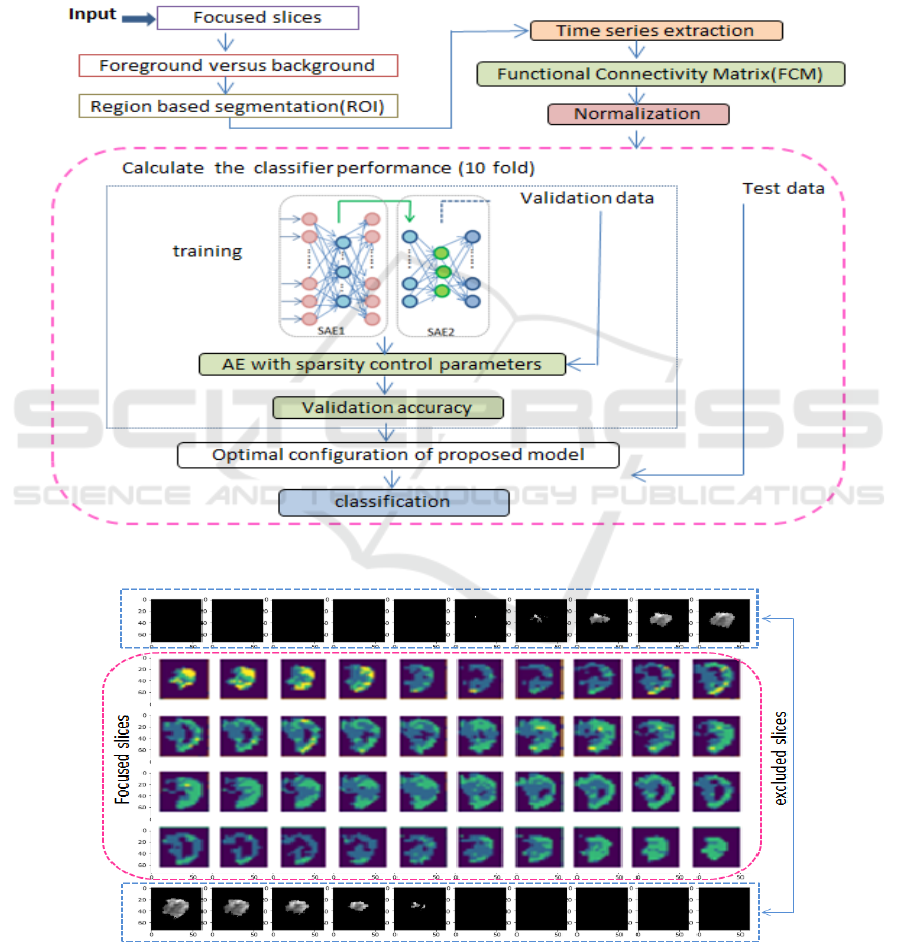
Multiple SAEs were used in training phase to
conclude low but valuable and most discriminative
representations of data. In fact single SAE usually
gives less expectation, however two SAEs deeper
the learning and enrich result. These in turn will
affect the classifier positively. Training, testing,
optimizing parameters, and 10-fold cross validation
flow of the proposed model, is shown in Figure 3.
First, the original fMRI is consisting of a number of
slices. Applying region segmentation on each slice,
then calculating functional connectivity analysis.
The normalization of input is necessary for
autoencoder in general.
Figure 3: The proposed Deep framework for training, validation, and testing.
Figure 4: Sample of focused and excluded slices during training.
ICEIS 2020 - 22nd International Conference on Enterprise Information Systems
544
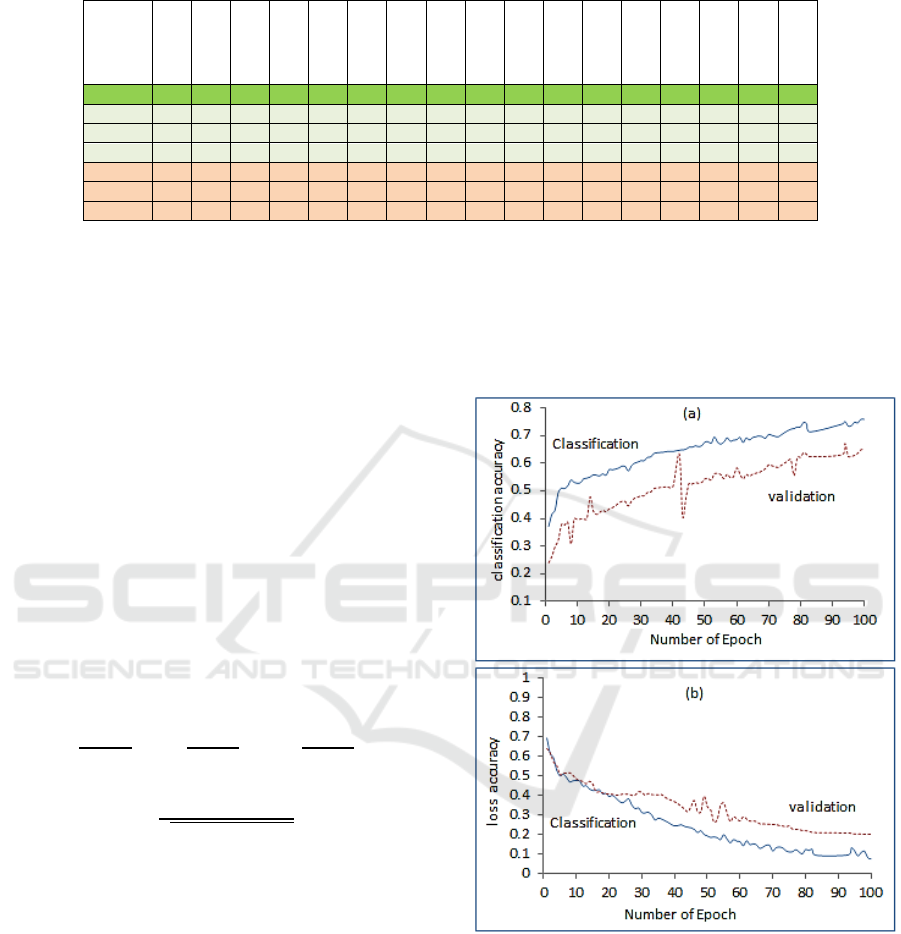
Table 1: ABIDE-I providers class distribution of genders, ASD and TC.
Site
Caltech
CMU
KKI
Leuven
MaxMun
NYU
OHSU
OLIN
PITT
SBL
SDSU
STANFORD
TRINITY
UCLA
UM
USM
YALE
Ts 37 27 48 63 52 175 26 34 56 30 36 39 47 98 140 71 56
ASD 19 14 20 29 24 75 12 19 29 15 14 19 22 54 66 46 28
Male 15 11 16 25 21 65 12 15 25 15 13 15 22 48 57 46 20
Female 4 3 4 3 3 10 0 3 4 0 1 4 0 6 9 0 8
TC 18 13 28 34 28 100 14 15 27 15 22 20 25 44 74 25 28
Male 14 9 20 29 27 73 14 13 23 15 16 16 25 38 57 25 20
Female 4 4 8 5 1 27 0 2 4 0 6 4 0 6 17 0 8
Accuracy versus number of epoch (1-100) (b)
classification loss and validation loss versus number
of epoch (1-100).
After training and validation are accomplished, an
optimized set of parameters are saved for later
testing. The data were divided 70% training and
30% testing.
The performance of the system was evaluated
based on the four criteria: Sensitivity (SE),
Specificity (SP), Accuracy (ACC) and Matthew’s
Correlation Coefficient (MCC). These are calculated
based on the accurate identification of positive
(ASD) or negative (TC) samples. A True Positive
(TP) would indicate that autism fMRI labeled and
identified correctly through data description, while a
False Positive (FP) indicates that fMRI is identified
as normal individual. Conversely, True Negatives
and False Negatives (FN) are calculated for
controlled individual. The metrics are defined by the
following equations:
A
,S
,S
(5)
𝑀
(6)
Where N=T
N
+T
P
+F
N
+F
P
, S=(T
P
+F
N
)/N and
P=(T
P
+F
P
)/N.
4 DATA ACQUISITION &
EXPERMINTES
The Autism data ABIDE I used in this paper was
acquired public through request for Autism Brain
Imaging Data Exchange to use it in research purpose
only. Table 1, shows the class and gender from 17
sites participation of (ABIDE I), (ADi. Martino, et
al., 2014) Pre-processed Connectomes Project
(http://preprocessed-connectomesproject.org/). C-
PAC pipeline was chosen for pre-processed version
(Y. Behzadi, et al., 2007). Actually, the project
offers four pipelines to download data all provide
basic requirement of handling data but they are
different in the pre-processing algorithm. C-PAC
was chosen to compare our results with other
research paper that used same pipeline data.
Figure 5: (a) Classification accuracy and validation, (b)
loss accuracy and validation.
For more details about pre-processing the raw data
please read (ADi. Martino, et al., 2014). 1035
samples were obtained after excluding the corrupted
samples, 505 ASD and 530 TC. Technical
Implementation All aspects of data pre-processing,
analysis, feature extraction and building the
classifier are implemented using python and
colaboratory resources. The used lab top is with intel
An Effective Sparse Autoencoders based Deep Learning Framework for fMRI Scans Classification
545
(R) core (TM) i5, 7200 U processor 2.5 GHz and 8
GB of RAM specification.
The data 3D volume is 61×73 ×61. First, during
visualization, the centred slices provides clearer and
consistent information while the beginning 10 slices
and last as well seems to be a burden while training,
validating and testing and provide no valuable
information from our prospective. Therefore, the
centred 40 slice were chosen as in figure 4.
Additionally, the data was smoothed and masked,
and ROI are calculated from time difference in slices
as seen in figure 1. Sparse autoencoder like the basic
autoencoder needs normalization (scaling issues and
vectors flatten) for data to be suitable for neural
network. The model was trained over 70% of the
data and 30% for testing and was used in a cross-
validation 10-fold schema.
The main performance measure is the accuracy
of correctly classify ASD from TC patients.
Therefore the proposed network training accuracy
and validation accuracy, training loss and validation
loss, versus epochs is depicted in Figure 5. The
mean values are higher with increase of epoch
number. The results are reasonable as more training
epoch increase the classification and validation
accuracy and reduce the loss as in Figure 5(b).
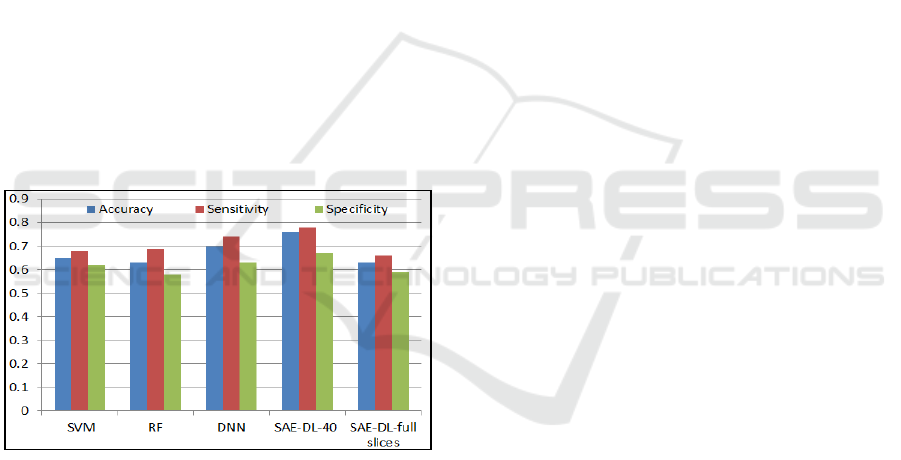
Figure 6: Results comparison of Techniques in (A.
Heinsfeld, et al., 2018) and SAE-DL.
Also, a comparison of accuracy, sensitivity and
specificity with author that applied Support Vector
Machine (SVM, Acc=0.65, Se=0.68, Sp=0.62),
Random Forest (RF, Acc=0.63, Se=0.69, Sp=0.58)
and Deep Neural Network (DNN, Acc=0.70,
Se=0.74, Sp=0.63), on the full data set (A.
Heinsfeld, 2018). Figure 6, shows these values in
comparison with our method in two cases. Case1:
selecting 40 centred slices (SAE-DL-40, Acc=0.76,
Se=0.78, Sp=0.67), and Case2: using full slices
(SAE-DL-full slices(73), Acc=0.65, Se=0.67,
Sp=0.57),). Based on mentioned literatures, and the
depicted similar case of using full database sample,
the proposed classification accuracy in case1,
reported higher classification accuracy due to
removing the burden of useless slices of patients that
provide poor information and reduces the learning
capabilities.
5 CONCLUSIONS
In the present paper, a two sparse autoencoders deep
learning framework was developed for classifying
ASD individuals and TD controls based on fMRI
brain scans. The first contribution of this work is the
proposed architecture for feature selection based on
multiple sparse AEs to improve the quality of the
extracted features and treats issues like over fitting
and generalization. The second contribution is using
full data sets with accompany complexities, where
many researchers avoid full set and preferred a
partial sub set of ABIDE. The proposed framework
trained and evaluated by the 10-fold evaluation was
implemented. An accuracy of 76% was achieved
with fMRI data, thus achieving higher predicative
performance than the literatures of techniques
applied on the same data.
REFERENCES
A. Abraham, MP. Milham, A. Martino, RC. Craddock, D.
Samaras, B. Thirion and G. Varoquaux. 2017,
Deriving reproducible biomarkers from multi-site
resting-state data: An Autism-based example,
NeuroImage vol.147, pp.736–745
ADi. Martino, et al., 2014, The autism brain imaging
data exchange: towards large-scale evaluation of the
intrinsic brain architecture in autism. vol.6, pp.659.-
667.
A. Heinsfeld, et al., 2018, Identification of autism
spectrum disorder using deep learning and the ABIDE
dataset. NeuroImage: Clinical. vol.17, pp. 16-23.
Bengio, Y., Courville, A., Vincent, P., 2013.
Representation learning: A review and new
perspectives. IEEE Trans Pattern Anal Mach Intell,
vol.35 (8), pp.1798–1828.
Bi Xia-an, et al, 2018, Analysis of Asperger Syndrome
Using Genetic-Evolutionary Random Support Vector
Machine Cluster, Frontiers in Physiology journal,
vol.9, pp:1646.
C. J. Brown, J. Kawahara, and G. Hamarneh, 2018,
Connectome priors in deep neural networks to predict
autism, in Biomedical Imaging (ISBI 2018), IEEE 15th
International Symposium on. IEEE, pp. 110–113
Eslami, T., Mirjalili, V., Fong, A., Laird, A., and Saeed,
F., 2019, ASD-DiagNet: A hybrid learning approach
ICEIS 2020 - 22nd International Conference on Enterprise Information Systems
546

for detection of Autism Spectrum Disorder using
fMRI data , arXiv preprint arXiv:1904.07577
G. Chanel, et al., 2016, Classification of autistic
individuals and controls using cross-task
characterization of fMRI activity. NeuroImage:
Clinical, vol.10, pp. 78-88.
G Litjens, T Kooi, BE Bejnordi, AAA Setio, F Ciompi, M
Ghafoorian, 2017, A survey on deep learning in
medical image analysis, Medical image analysis, vol,
42, pp.60-88
H. Chen, X. Duan, F. Liu, F. Lu, X. Ma, Y. Zhang, L. Q.
Uddin, and H. Chen, 2016, Multivariate classification
of autism spectrum disorder using frequency-specific
resting-state functional connectivity—a multicenter
study, Progress in Neuro-Psychopharmacology and
Biological Psychiatry, vol. 64, pp. 1–9.
H. Li, N. Parikh and L. He. , 2018, A Novel Transfer
Learning Approach to Enhance Deep Neural Network
Classification of Brain Functional Connectomes.
Front. Neurosci. vol.12, pp.491.
Krizhevsky, A., Sutskever, I., Hinton, G.E., 2012. Image-
net classification with deep convolutional neural
networks, in: Advances in neural information
processing systems, pp. 1097–1105.
Makhzani, Alireza and Frey, Brendan. 2013, k-sparse
autoencoders. CoRR, abs/1312.5663.
M. Khosla, K. Jamison, A. Kuceyeski and M. Sabuncu.,
2018, 3D Convolutional Neural Networks for
Classification of Functional Connectomes, arXiv:
1806.04209
MN. Parikh, H. Li and L. He. 2019, Enhancing Diagnosis
of Autism With Optimized Machine Learning Models
and Personal Characteristic Data. Frontiers in Human
Neuroscience. vol.13, pp.1-5.
M. Plitt, KA. Barnes and A. Martin., 2015, Functional
connectivity classification of autism identifies highly
predictive brain features but falls short of biomarker
standards. NeuroImage: Clinical, vol.9, pp. 359-366.
Najafabadi M.M., Villanustre F., Khoshgoftaar T.M.,
Seliya N., Wald R., and Muharemagic E., 2015. Deep
learning applications and challenges in big data
analytics. Journal of Big Data, vol. 2, no. 1, pp. 1-21.
Ravì D., et al., 2017. Deep learning for health informatics.
IEEE Journal of Biomedical and Health Informatics,
vol. 21, pp. 4-21.
SE. Schipul, DL. Williams, TA. Keller, NJ. Minshew &
MA.Just, 2012. Distinctive Neural Processes during
Learning in Autism, Cereb Cortex. vol, 22(4), pp.937.
Shin, H.-C., Orton, M. R., Collins, D. J., Doran, S. J., and
Leach, M. O. 2013. Stacked autoencoders for
unsupervised feature learning and multiple organ
detection in a pilot study using 4D patient data.
Pattern Anal.Mach. Intell. IEEE Trans. vol.35,
pp.1930–1943.
XA. Bi, Y. Wang, Q. Shu, Q. Sun, and Q. Xu. , 2018,
Classification of Autism Spectrum Disorder Using
Random Support Vector Machine Cluster. Frontiers in
Genetics. vol.9.
XA. Bi, Y. Liu, Q. Jiang, Q. Shu, Q. Sun and J Dai., 2018,
The Diagnosis of Autism Spectrum Disorder Based on
the Random Neural Network Cluster. Frontiers in
Human Neuroscience, vol.12, pp. 257.
Xi. Li, N. Dvornek, et al., 2018, 2-Channel Convolutional
3d Deep Neural Network (2CC3D) For fMRI
Analysis: ASD Classification and Feature Learning.
IEEE 15th International Symposium on Biomedical
Imaging (ISBI): Washington, D.C., USA.
Xi. Li, N. Dvorneky, J. Zhuang, P. Ventolaz and J.
Duncan. 2018, Brain Biomarker Interpretation in ASD
Using Deep Learning and fMRI. Int. Conference on
Medical Image Computing and Computer-Assisted
Intervention, MICCAI, pp. 206-214,.
X. Guo, KC. Dominick, AA. Minai, H. Li, CE. and LJ. Lu,
2017, Diagnosing Autism Spectrum Disorder from
Brain Resting-State Functional Connectivity Patterns
Using a Deep Neural Network with a Novel Feature
Selection Method, Front. Neurosci. vol.11, pp 460.
Y. Behzadi, K. Restom K, J. Liau, TT. Liu. , 2007, A
component based noise correction method (CompCor)
for BOLD and perfusion based fMRI. NeuroImage,
vol.37 (1), pp. 90- 101.
An Effective Sparse Autoencoders based Deep Learning Framework for fMRI Scans Classification
547
