
Efficient Representation of Very Large Linked Datasets as Graphs
Maria Krommyda, Verena Kantere and Yannis Vassiliou
School of Electrical and Computer Engineering, National Technical University of Athens, Athens, Greece
Keywords:
Big Data Exploration, Linked Datasets, Graphical Representation.
Abstract:
Large linked datasets are nowadays available on many scientific topics of interest and offer invaluable knowl-
edge. These datasets are of interest to a wide audience, people with limited or no knowledge about the
Semantic Web, that want to explore and analyse this information in a user-friendly way. Aiming to support
such usage, systems have been developed that support such exploration they impose however many limitations
as they provide to users access to a limited part of the input dataset either by aggregating information or by
exploiting data formats, such as hierarchies. As more linked datasets are becoming available and more people
are interested to explore them, it is imperative to provide an user-friendly way to access and explore diverse
and very large datasets in an intuitive way, as graphs. We present here an off-line pre-processing technique,
divided in three phases, that can transform any linked dataset, independently of size and characteristics to one
continuous graph in the two-dimensional space. We store the spatial information of the graph, add the needed
indices and provide the graphical information through a dedicated API to support the exploration of the infor-
mation. Finally, we conduct an experimental analysis to show that our technique can process and represent as
one continuous graph large and diverse datasets.
1 INTRODUCTION
The establishment of the RDF data model has made
available many large and diverse linked datasets from
a wide range of scientific areas
1
. Exploring and
querying these datasets as published, though, requires
extensive knowledge of the semantic web and expen-
sive infrastructure. As a result these datasets are inac-
cessible to a wider audience.
In order to alleviate this barrier, linked datasets
should be represented using a graph model, where
each entity is presented as a distinct object, and the in-
formation is explorable through multiple filtering cri-
teria. In order to implement a user-friendly system,
a technique should be developed that will meet some
important challenges. To begin with, the technique
should be able to handle a wide range of datasets that
differ in characteristics
2
. Available datasets may have
from a few thousands to millions of elements, they
may also be incomplete, have no semantic annotations
or not follow any data model or hierarchy
3
.
In addition, the technique should provide an easy
1
https://lod-cloud.net/
2
https://www.w3.org/wiki/TaskForces/Community
Projects/LinkingOpenData/DataSets/Statistics
3
https://datahub.io/collections/linked-open-data
and user-friendly way to support the exploration of
the information. As an example, entities should be
distinct object and connections between them clearly
defined, the complete dataset should be available and
the graph topology should be consistent to allow the
user to understand where the information is located.
The relevant but also the absolute position in the two-
dimensional space of the data objects should remain
the same regardless of the user actions. Furthermore,
plenty of functionalities should be available for the
exploration of the information. Keyword search, path
exploration, neighbour information, filtering and ag-
gregation, should be available in a user-friendly way
without requiring knowledge of querying or program-
ming languages from the user. Last but not least, the
information should be available for multiple users si-
multaneously through commodity hardware such as
laptops and tablets. Processing that requires expen-
sive infrastructure should be handled by the server
side of the system and not by the devices of the users.
An overview of the techniques and systems that
have been developed to address some of these chal-
lenges is presented here.
An initial approach to the problem of the explo-
ration of large linked datasets focused on generic vi-
sualization systems. These systems aim to extract
the information of linked, semantically annotated,
106
Krommyda, M., Kantere, V. and Vassiliou, Y.
Efficient Representation of Very Large Linked Datasets as Graphs.
DOI: 10.5220/0009389001060115
In Proceedings of the 22nd International Conference on Enterprise Information Systems (ICEIS 2020) - Volume 1, pages 106-115
ISBN: 978-989-758-423-7
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

datasets and present it through generic visualization
options. These visualization options are limited to
charts, such as column, line, pie, bar, scatter and
radar, that present the information in a summarized
way. Some systems (Voigt et al., 2012; Schlegel et al.,
2014) allow the dynamic choice of such visualization
types based on the user selection. The user can select
what information is of interest and the visualization
type to present it. Other systems (Brunetti et al., 2012;
Kl
´
ımek et al., 2013; Atemezing and Troncy, 2014)
utilize strict semantic rules and restrict the users to
predefined types of visualizations.
These systems handle data fitting specific crite-
ria regarding their structure and semantic information,
and present the information summarized in restrictive
forms. These approaches can give a general overview
of the information but cannot support the graphical
representation of the information. In addition, such
systems cannot handle datasets that are incomplete or
lack semantic annotations.
A large number of systems visualize linked
datasets adopting a graph-based approach (Mazumdar
et al., 2015). Some of these techniques (Abello et al.,
2006; Auber, 2004; Jr. et al., 2006) present small fil-
tered or aggregated graphs of the input dataset based
on hierarchical data models. These systems use ag-
gregation and filtering techniques to limit the size of
the represented graph and allow navigation only along
the hierarchy, making information inaccessible from
nodes at the same hierarchy level. While the naviga-
tion of highly aggregated data might help initially the
user to better understand the structure of the informa-
tion, at a deeper exploration of the dataset the loss of
information might be proven hindering to the useful-
ness of the representation.
The techniques discussed above meet only a sub-
set of the challenges. However, for many use cases,
such as road maps, communication networks and
blockchain transactions, it is of utmost importance to
meet all the challenges and not a subset of them.
Motivating Example. One of the most characteris-
tic examples of scientific data analysis that requires
a system that conforms to all the identified chal-
lenges, is the study and analysis of biological struc-
tures. Structural biology is a branch of molecular bi-
ology concerned with the molecular structure of bi-
ological macromolecules such as proteins, made up
of amino acids, how they acquire the structures they
have, and how alterations in their structures affect
their function. This subject is of great interest to bi-
ologists because macromolecules carry out most of
the functions of cells, and it is only by coiling into
specific structures that they are able to perform these
functions. The Protein Data Bank
4
is an archive
with information about the shapes of proteins, nucleic
acids, and complex assemblies. Most of the datasets
published there follow the Systems Biology Markup
Language
5
aiming to ensure model interoperability
and semantically correct information. As an example
we take the human cap-dependent 48S pre-initiation
complex
6
which represents 47 unique protein chains
with 116774 atom count.
Access to the Complete Information. Scientist aim-
ing to study and analyse this protein complex need to
have access to the complete information in a way that
will respect the initial spatial relations, without caus-
ing any alterations to connections between molecules.
Systems that use aggregation or summarization tech-
niques cannot be used for this analysis as they alter the
initial connections between atoms. Techniques that
exclude part of the input dataset if it does not follow
pre-defined data formats cannot be used either, as pro-
tein chains are not expected to fully comply with spe-
cific data formats and atoms that deviate from them
contain important information which should be avail-
able to the user. Systems that filter the input dataset
based on specific characteristics cannot support the
exploration of protein complexes as their analysis is
mostly based on low level connections.
Combination of Multiple Datasets. In addition, scien-
tists need to study the complete biological structures,
so the system should be able to combine multiple pro-
tein chains and complexes. As an example, the human
cap-dependent 48S pre-initiation protein complex has
a relatively small number of atoms as it contains only
47 protein chains. Such complexes, however, are
combined or expanded with additional protein chains
for analysis purposes. The system should be able to
incorporate any additional information and offer to
the user exploration and filtering services without any
limit to the size of the input dataset. Such dynamic
changes in the volume and content of the input dataset
is a challenge for all of the available systems. These
systems are based on complex pre-processing tech-
niques that aim to identify specific data models that
conform with the input dataset or perform a semantic
analysis and categorization. Due to their complexity,
such techniques fail to combine more than one dataset
or adapt to a large and complex one.
Multiple Filtering Criteria. Users interested in such
complex datasets also have high demands regarding
different and customizable filtering functions that can
be used simultaneously. Popular queries during such
data analysis include: “isolate specific molecules” ,
4
https://www.rcsb.org/
5
http://semanticsbml.org/semanticSBML/simple/index
6
http://www.rcsb.org/structure/6FEC
Efficient Representation of Very Large Linked Datasets as Graphs
107

“isolate one or more types of connections based on
their semantic annotations” , “identify the most/least
connected molecules” , “locate connections between
two molecules” , “find if two or more molecules are
independent to one other” and “identify specific con-
nections in parts of the biological structure that is re-
sponsible for a specific function”.
The system should offer such exploration queries
though a user-friendly interface, without asking the
user to write queries over the data. Depending on the
way the information is processed, however, systems
may not be able to offer such exploration queries.
Systems that aggregate the input dataset cannot offer
details for one molecule. Systems that present data
based on hierarchy levels or through semantic criteria
cannot present the connections between two or more
molecules or isolate types of connections that are of
interest.
Path Exploration. Finally, given the importance
of the connections between molecules it is crucial
to identify all the neighbors of a molecule or follow
paths of interest. All of the available systems, allow
either the exploration of the outgoing neighbors of
a node or the exploration of nodes along the hierar-
chy levels of the data model. Therefore, the system
does not allow the user to intuitively explore the in-
formation associated with a node. For datasets such
as the human cap-dependent 48S pre-initiation pro-
tein complex, users are very interested in locating all
the neighbors of a molecule, either incoming or out-
going, as these connections are valuable to the protein
structure analysis.
Contribution. We present here an off-line pre-
processing technique that enables the Efficient Rep-
resentation of Very Large Linked Datasets as Graphs.
Our technique has been carefully designed to alleviate
the restrictions of the above-mentioned approaches on
the input datasets, regarding accompanying metadata
related to the model, full or partial semantic anno-
tation, hierarchical structure, data completeness and
size limitation. It also meets the challenges of scal-
ability, consistent visualization and data exploration
through different filtering functions. The proposed
technique is split into three phases, designed to trans-
form any input dataset, such as highly connected, in-
complete or lacking semantic annotations, into one
continuous graph on the two-dimensional space.
• Pre-processing Technique.
Initially, the input
dataset is split into smaller parts, the number and
size of which is determined by the dataset size
and connectivity degree. Next, each part of the
input dataset is graphically represented as an in-
dependent sub-graph. Finally, the sub-graphs are
merged into one continuous graph, using a suit-
able cost function that ensures small navigation
paths and consistent exploration for millions of
nodes. Any information omitted during the cre-
ation of the sub-graphs is re-introduced to the
graph during this merging. The pre-processing
result is the representation as a graph of the com-
plete input dataset. We refer to the representation
of a dataset as one continuous graph in the two-
dimensional space as graphical representation.
Given that no part of the pre-processing phase is
based on specific data formats we can handle any
dataset including incomplete datasets, datasets
that are not annotated with respect to semantic
categories or datasets that do not comply with the
hierarchical model. Also, the modular design of
our technique allows us to handle real datasets,
with variable volume and connectivity degrees.
• Dedicated API. The graphical representation of
the input dataset is stored, in a distributed
database to ensure high performance and query
optimization, and indexed to facilitate the acces-
sibility and retrieval of the information. In or-
der to showcase the usability of our technique we
have implemented a dedicated API over the stor-
age schema that allows the exploration of the in-
formation. The API allows the retrieval of the
information using different spatial boundaries in-
cluding geometrical shapes, multiple filtering cri-
teria and keyword search.
• Experimental Analysis. We performed a thorough
experimental study on our technique. We em-
ployed our technique for the representation of
many real and synthetic datasets with as many as
150 million elements and an average node degree
as high as 20, in order to show that we can handle
large and diverse datasets without any limitations
to their characteristics.
The structure of the paper is as follows: In Section
2, we present the proposed technique for the graphi-
cal representation of the input datasets. In Section 3,
we present the review of the related work. Finally, in
Section 4, we present the experimental evaluation of
the proposed technique.
2 PROCESSING
METHODOLOGY
We present here the three phases of the off-line, pre-
processing technique that allow us to efficiently rep-
resent large linked dataset as one continuous graph.
ICEIS 2020 - 22nd International Conference on Enterprise Information Systems
108

The first phase of the technique ensures that we
are able to handle very large datasets. For this reason,
the input dataset is split in smaller parts. The number
and the size of these parts are decided based on the
size, number of entities and relationships, and charac-
teristics, connectivity degree and average label length,
of the input dataset.
The second phase of the technique is dedicated
to the proper representation of the parts of the input
dataset. Each part is transformed into an independent
graph, using the Scalable Force Directed Placement
algorithm (Kamada and Kawai, 1989). We chose this
algorithm due to its flexibility and robustness. We
carefully initialize the parameters of the algorithm
based on the characteristics of the parts. Our aim is
to achieve the right degree of compactness, avoid any
overlapping and ensure that each sub-graph has ap-
proximately the same horizontal and vertical span.
In the third phase of the technique, the sub-graphs
are merged into one continuous graph. It is important
to arrange the sub-graphs in a way that the connec-
tions between the sub-graphs, as identified and stored
at the first phase of the technique, are introduced here
as edges between the nodes in a way that minimizes
their total length. For this reason, we have imple-
mented a heuristic algorithm, which uses a cost func-
tion based on the length of the edges, to arrange the
sub-graphs.
Finally, the graphical information is translated to
a spatial storage schema and made accessible through
a flexible, complete and efficient API.
2.1 Phase A: Dataset Partitioning
This phase of the proposed technique is responsi-
ble for receiving as input the linked dataset, model
the information with respect to the graph model and
partition the graph into smaller sub-graphs. Linked
datasets can be mapped to a graph G = (V ,E) where
V is a set of nodes (v) that represent the entities of the
input dataset and E is a set of edges (e) that represent
the connections between the entities.
Formally, given a graph G = (V ,E) with |V | = n,
the k-way graph partitioning is defined as the par-
titioning of V into k subsets, V
1
,V
2
,...,V
k
where
V
i
∩ V
j
=
/
0 for i 6= j, |V
i
| ≈ n/k, and ∪
i
V
i
= V .
The k-way graph partitioning aims to minimize
the number of edges of E, the vertices of which be-
long to different subsets. Given a graph G = (V ,E)
and a k-way graph partitioning, the edges of the graph
G, the vertices of which belong to different subsets,
are called external edges. X
i j
⊂ E is the set of exter-
nal edges between two subsets V
i
and V
j
where i 6= j
and X is the set of all external edges between all pos-
sible pairs of V
i
∈ V .
In order to ensure that all datasets, no matter
their specific characteristics or volume, are parti-
tioned properly, we chose a robust algorithm with
the capability of handling datasets ranging from a
few thousands to millions, and from sparse to dense.
We use the k-way graph partitioning algorithm of G.
Karypis and V. Kumar (Karypis and Kumar, 1998) as
implemented in Metis
7
, as it meets the requirements
of handling any input dataset without any restrictions
on its size and properties.
Example 1. For the 48S pre-initiation protein com-
plex, the k-way partitioning divides the dataset into
nine sub-graphs, as shown in Figure 1. Given
the 116774 atoms of the dataset each partition has
13.000 atoms. The number of the partitions is se-
lected to support the second phase of the technique as
presented in 2.2 and create a continuous graph with
similar horizontal and vertical dimensions. Based on
the small size of the input dataset here, the possible
options are 4, 9 or 16 partitions. While with 4 parti-
tions the sub-graphs have too many elements for the
second phase, with 16 they have unnecessary few.
2.2 Phase B: Graphical Representation
of the Sub-graphs
This phase of our technique is dedicated to the trans-
formation of each part of the input dataset to an in-
dependent sub-graph. The main requirements of this
phase are two, both related with the spatial distribu-
tion of the nodes. To begin with, we need to achieve a
uniform density of the nodes in the two- dimensional
space for each sub-graph. This means that the nodes
of the sub-graph should be spaced out enough not to
have any overlaps between them while at the same
time as close as possible in order not to have a lot of
empty space. Moreover, we should have sub-graphs
requiring approximately the same horizontal and ver-
tical space. This is very important for the next phase
of the proposed technique, which connects all the sub-
graphs into one continuous graph by arranging them
in a two-dimensional grid. Given that the dimensions
of each grid cell are based on the width and height of
the included sub-graph, having sub-graphs with simi-
lar sizes ensures the lack of empty spaces.
Both requirements should be achieved indepen-
dently of the specific characteristics of the sub-
graphs. For this reason, we choose the Scalable Force
Directed Placement algorithm (Kamada and Kawai,
1989) for the graphical representation of the sub-
7
http://glaros.dtc.umn.edu/gkhome/metis/metis/
overview
Efficient Representation of Very Large Linked Datasets as Graphs
109
graphs. The algorithm is based on a physics ap-
proach, balancing a system where nodes are con-
sidered charged particles and edges are modeled as
springs. This allows for the parameterization of many
attributes for the output graph including the allowed
overlapping percentage, the size of the nodes and the
size of the graph.
2.3 Phase C: Merging the Sub-graphs
Into One Continuous Graph
In this section, the third phase of the technique that
connects the sub-graphs in one continuous graph is
described. Given that at we dynamically choose the
number of sub-graphs based on the input dataset, we
need a solution for the sub-graph arrangement that ac-
cepts as input any number of partitions and any set
of external edges between them and results to an ar-
rangement of the sub-graphs in the two-dimensional
space in a way that the length of long edges connect-
ing different sub-graphs is minimized.
Problem Definition. Let P = {P
1
,P
2
,...,P
k
} where
each P
i
for i = 1, 2,...,k corresponds to the i-th par-
tition and includes the vertices of the V
i
sub-graph
with their coordinates and the edges between them.
For each partition P
i
a CN
i
, a Complex Node (CN)
is defined. A Complex Node CN
i
refers to the
two-dimensional area with dimensions the width and
height of the P
i
and content all the graph elements,
nodes and edges, of the partition. The position of each
graph element in the continuous graph is relevant to
the position of the Complex Node that contains it to
the two-dimensional space.
Problem Transformation. Let CN denote the set of
all Complex Nodes for the partitions, with |C N | = k
and W the set containing the weights between the
Complex Nodes as defined by the number of exter-
nal edges between them, W (i, j) = X
i, j
. The process
of placing each CN
i
∈ C N into the two-dimensional
Cartesian integer grid by assigning grid positions to
each CN
i
, such that it minimizes the weights is defined
as Grid Arrangement Problem (GAP). The problem is
proven (Oswald et al., 2012) to be NP-hard for every
dimension of the grid.
Problem Solution. We present here a greedy heuris-
tic algorithm, as a solution for the Grid Arrangement
Problem that can be applies with large number of
nodes. The algorithm uses a weight-cost function to
calculate the cost of adding a CN to a specific posi-
tion given a grid arrangement. The algorithm, incre-
mentally creates the grid arrangement by selecting, in
each iteration, the CN which is going to be to placed
in the grid. The CN, with the highest cost as provided
by the weight-cost function, is selected on each itera-
tion. The weight-cost function is calculating the sum
of the weights between each examined CN to the ones
already placed on the grid.
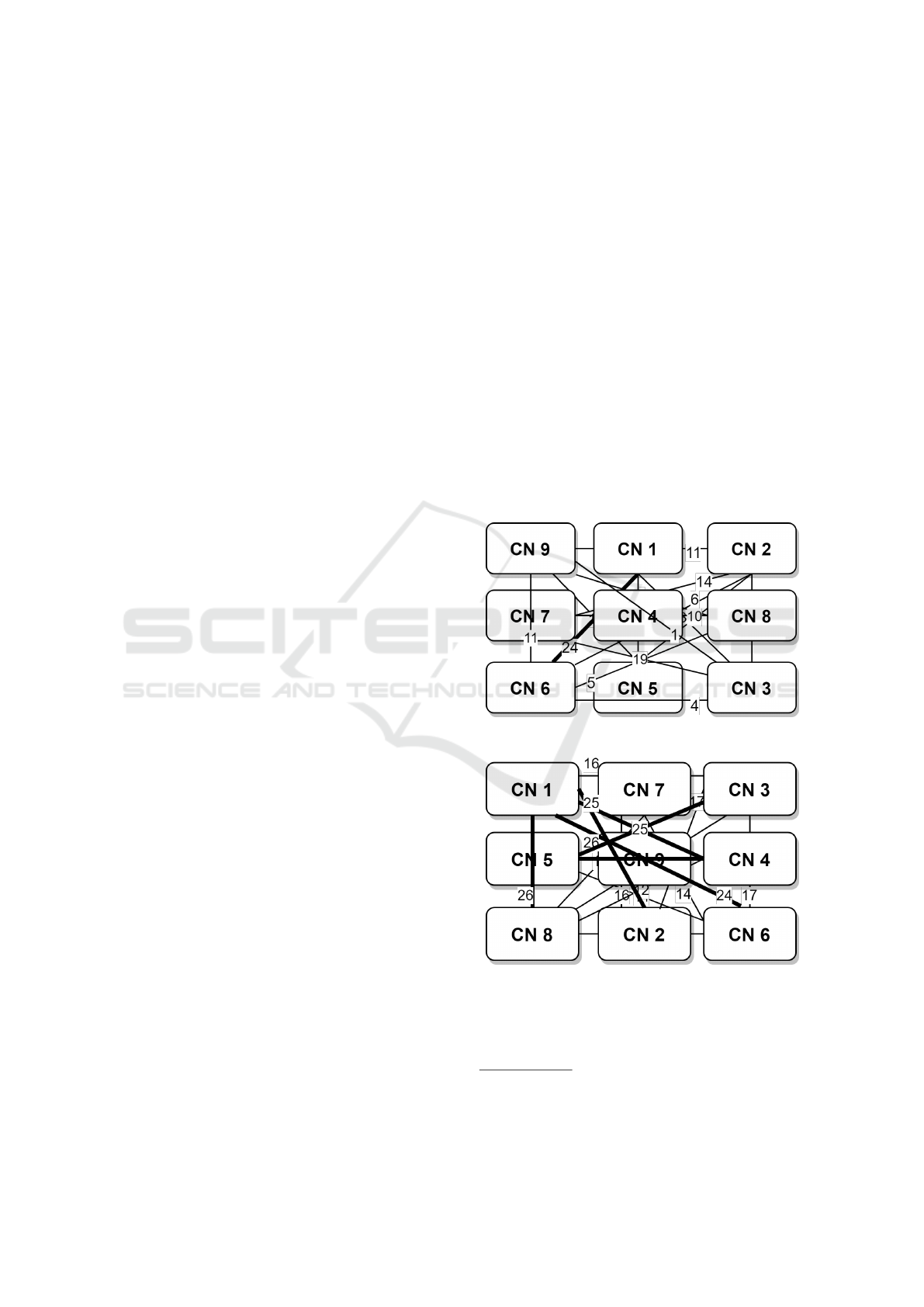
Example 2. We present the weights between the CNs,
of the human cap-dependent 48S pre-initiation com-
plex, in Table 1. To simplify the example the numbers
provided in the table have been scaled down, keeping
the initial ratio. First, we merge the sub-graphs into
one continuous graph, by following the steps of the
proposed algorithm. We present the steps followed as
well as the intermediate results of the cost function in
2. Then, we merge the sub-graphs into one continu-
ous graph in a random way. The results of the two
placements is presented in Figure 1. There is a 40%
decrease to the overall weight cost.
The merged information is stored in a distributed,
scalable Accumulo
8
key/value datastore where the
GeoMesa
9
XZ-ordering index is used for the spatial
information of the graph.
(a) Placement based on the proposed algorithm
(b) Random placement
Figure 1: CN Placement in the Two-Dimensional Cartesian
Integer Grid.
8
https://accumulo.apache.org/
9
https://www.geomesa.org/documentation/current/
index.html
ICEIS 2020 - 22nd International Conference on Enterprise Information Systems
110
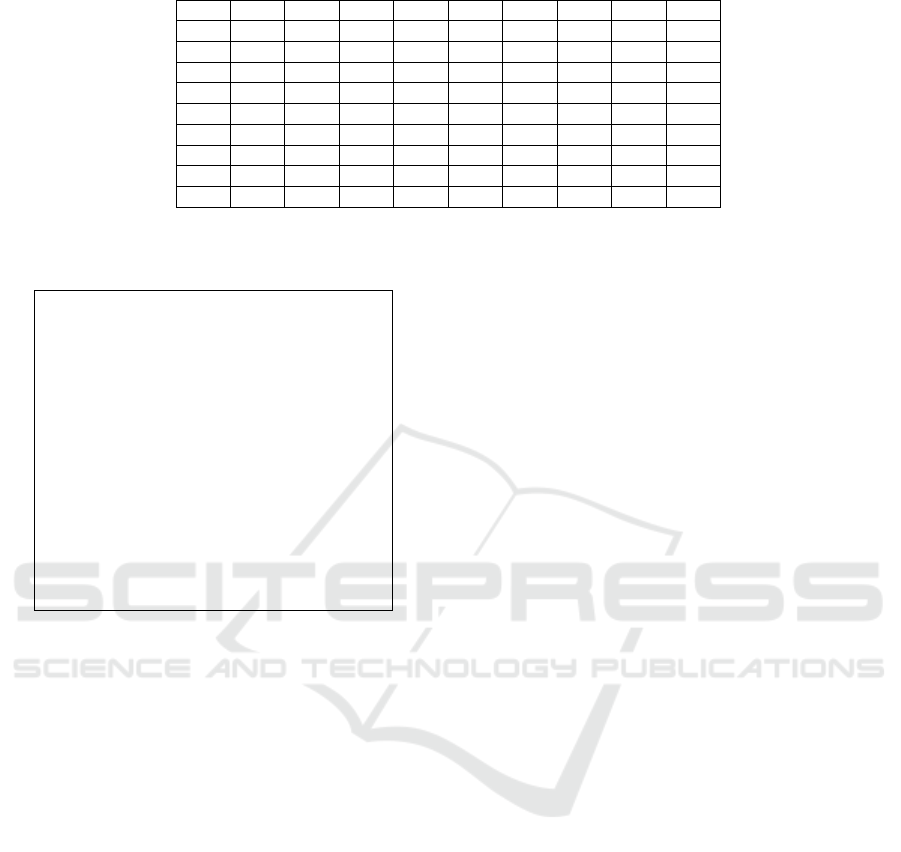
Table 1: Number of External Edges between the Nine CNs.
CN 1 CN 2 CN 3 CN 4 CN 5 CN 6 CN 7 CN 8 CN 9
CN 1 0 25 16 27 3 24 1 26 6
CN 2 25 0 17 24 4 6 14 25 11
CN 3 16 17 0 10 25 17 19 4 1
CN 4 27 24 10 0 26 18 13 12 7
CN 5 3 4 25 26 0 14 27 21 11
CN 6 24 6 17 18 14 0 14 5 11
CN 7 1 14 19 13 27 14 0 10 16
CN 8 26 25 4 12 21 5 10 0 9
CN 9 6 11 1 7 11 11 16 9 0
Table 2: Steps of Executing the Proposed Algorithm for the
Nine CNs.
Step 1 CN 4 is set at the center of the grid
Step 2 CNs are ranked based on the weight [1,5,2,6,7,8,3,9]
Step 3 CN 1 is placed next in the grid
Step 4 CNs are ranked based on the weight [2,6,8,5,3,7,9]
Step 5 CN 2 is placed next in the grid
Step 6 CNs are ranked based on the weight [8,6,3,5,7,9]
Step 7 CN 8 is placed next in the grid
Step 8 CNs are ranked based on the weight [3,6,5,7,9]
Step 9 CN 3 is placed next in the grid
Step 10 CNs are ranked based on the weight [5,6,7,9]
Step 11 CN 5 is placed next in the grid
Step 12 CNs are ranked based on the weight [6,7,9]
Step 13 CN 6 is placed next in the grid
Step 14 CNs are ranked based on the weight [7,9]
Step 15 CN 7 is placed next in the grid
Step 16 CN 9 is placed next in the grid
2.4 Use Cases
In order to provide access to the graph of the input
dataset in an efficient and user-friendly way, we have
implemented a dedicated API that allows the retrieval
of the information in different ways, including spa-
tial and filtering queries and keyword search. We
have implemented a standalone Tomcat application,
written using Java servlets,that receives user requests
along with multiple parameters and returns the result
as an XML file, that follows the GraphML format and
includes the proper geometry parameters that spec-
ify the position of the nodes in the two-dimensional
space. The API is designed to receive any Common
Query Language function, including all the spatial
functions and geometries. We are going to showcase
the usability of the API through a series of use cases
as identified in the presented motivating example.
Example 3. Keyword Search. The initial exploration
of the dataset begins when the user wants to locate
information of interest. The intuitive way for this is
to use a keyword and search among the entities of the
dataset. For a protein complex the user may search
the identifier of an atom or a group of atoms. The API
will return a list containing all the entities containing
the given keyword along with their spatial informa-
tion.
Example 4. Spatial Queries. After locating an entity
of interest, the user can use its spatial information and
query the API over the two-dimensional space, using
any Common Query Language spatial function and
geometry. As an example, a user interested in iden-
tifying specific connections in parts of the biological
structure, responsible for a specific function, can per-
form spatial queries and retrieve this information.
Example 5. Filtering based on Connection Types.
The API enables the retrieval of the information us-
ing one or multiple filtering criteria at the same time.
The user can easily select to isolate one or more types
of connections between atoms based on their seman-
tic annotations, and can combine this filtering with a
spatial query for a specific area of the graph.
Example 6. Path Exploration. After locating an en-
tity that is of interest, the user is able to retrieve all the
paths of the graph that include this node, up to a cus-
tomisable length. In contrast to most approaches in
which a path can be traversed only with respect to the
edge direction, in our API when a node is selected, all
neighbors, either incoming or outgoing are retrieved.
3 RELATED WORK
In this section we describe with further details related
work. To begin with, systems that aim to overcome
the restriction of the size limitation based on the ex-
traction of the included information and its represen-
tation through generic visualization options will be
presented.
Facet Graphs (Heim et al., 2010a) allows humans
to access information contained in the Semantic Web
according to its semantics and thus to leverage the
specific characteristic of this Web. To avoid the am-
biguity of natural language queries, users only select
already defined attributes organized in facets to build
their search queries. The facets are represented as
Efficient Representation of Very Large Linked Datasets as Graphs
111

nodes in a graph visualization and can be interactively
added and removed by the users in order to produce
individual search interfaces. This provides the possi-
bility to generate interfaces in arbitrary complexities
and access arbitrary domains.
SemLens (Heim et al., 2011) is a visual tool that
arranges objects on scatter plots and them using user-
defined semantic lenses, offering visual discovery of
correlations and patterns in data. Haystack (Huynh
et al., 2002) is a platform for creating, organizing and
visualizing information using RDF. It is based on the
idea that aggregating various types of users’ data to-
gether in a homogeneous representation, agents can
make more informed deductions in automating tasks
for users. LODeX (Benedetti et al., 2014) is a tool
that generates a representative summary of a linked
data source. The tool takes as input a SPARQL end-
point and generates a visual summary of the linked
data source, accompanied by statistical and struc-
tural information of the source. Polaris (Stolte and
Hanrahan, 2000) is a visual query language for de-
scribing table-based graphical presentations of data
which compiles into both the queries and the visual-
ization,enabling users to integrate analysis and visu-
alization.
A large number of systems visualize linked
datasets adopting a graph-based approach (Mazumdar
et al., 2015). Most systems, limit the displayed infor-
mation by enforcing a path-based navigation of the
data to the user. Such examples include the RelFinder
(Heim et al., 2010b) which is a web-based tool that
offers interactive discovery and visualization of re-
lationships between selected linked data resources.
Similary, Fenfire (Hastrup et al., 2008) and Lodlive
(Camarda et al., 2012) are exploratory tools that allow
users to browse linked data using interactive graph
navigation. Starting from a given URI, the user can
explore linked data by following the links. Zoom-
RDF (Zhang et al., 2010) employs a space-optimized
visualization algorithm in order to display additional
resources with respect to the user navigation choises
while Trisolda (Dokulil and Katreniakov
´
a, 2009) pro-
poses a clustered RDF graph visualization in which
input information is merged to graph nodes. FlexViz
(Falconer et al., 2010) offers node and edge specific
filters that are based on search and navigation crite-
ria aiming to reduce the amount of handled data and
provide to a meaningful subset to the user.
The Tabulator (Berners-Lee et al., 2006) project
is an attempt to demonstrate and utilize the power of
linked RDF data with a user-friendly Semantic Web
browser that is able to recognize and follow RDF links
to other RDF resources based on the user’s explo-
ration and analysis. It is a generic browser for linked
Table 3: Synthetic Datasets.
Dataset #Edges #Nodes Degree
1M D5 5.1M 1M 5.09
10M D5 50.2M 10M 5.02
10M D10 103M 10M 10.3
10M D15 157M 10M 15.7
50M D5 259M 50M 5.18
data on the web without the expectation of provid-
ing as intuitive an interface as a domain-specific ap-
plication but aiming to provide the sort of common
user interface tools used in such applications, and
to allow domain-specific functionality to be loaded
transparently from the web and be instantly applica-
ble to any new domain of information. Explorator
(De Araujo and Schwabe, 2009) is an open-source ex-
ploratory search tool for RDF graphs, implemented
in a direct manipulation interface metaphor. It imple-
ments a custom model of operations, and also pro-
vides a Query-by-example interface. Additionally, it
provides faceted navigation over any set obtained dur-
ing the operations in the model that are exposed in the
interface. It can be used to explore both a SPARQL
endpoint as well as an RDF graph in the same way as
“traditional” RDF browsers.
Another approach to the problem of linked data
visualization is the visualization of the information
based on hierarchical models. GrouseFlocks (Ar-
chambault et al., 2007) works only on datasets struc-
tured over well-defined hierarchies. It requires com-
plete data and can handle up to 220K elements when
the hierarchy is predefined. Tulip (Auber, 2004) de-
velops a technique to solve the problem of presenting
to the users the data in only one predetermined way,
regardless of the features of the input dataset. This
tool stores the input data once and extracts the infor-
mation with different techniques providing the user
with multiple views/clusters over the data. GMine (Jr.
et al., 2006) uses the state-of-the-art partitioning algo-
rithm METIS (Karypis and Kumar, 1998) to split the
input dataset in reasonable partitions. Each partition
is visualized as one node, resulting in the loss of the
information included in each partition.
4 EXPERIMENTAL ANALYSIS
In order to evaluate the proposed technique and carry
out an experimental analysis on the efficiency of the
information retrieval through the API we developed a
web tool. We used the tool to spatially query the API
and evaluate the response time. The experiments were
conducted using a laptop with an Intel(R) Core(TM)
ICEIS 2020 - 22nd International Conference on Enterprise Information Systems
112
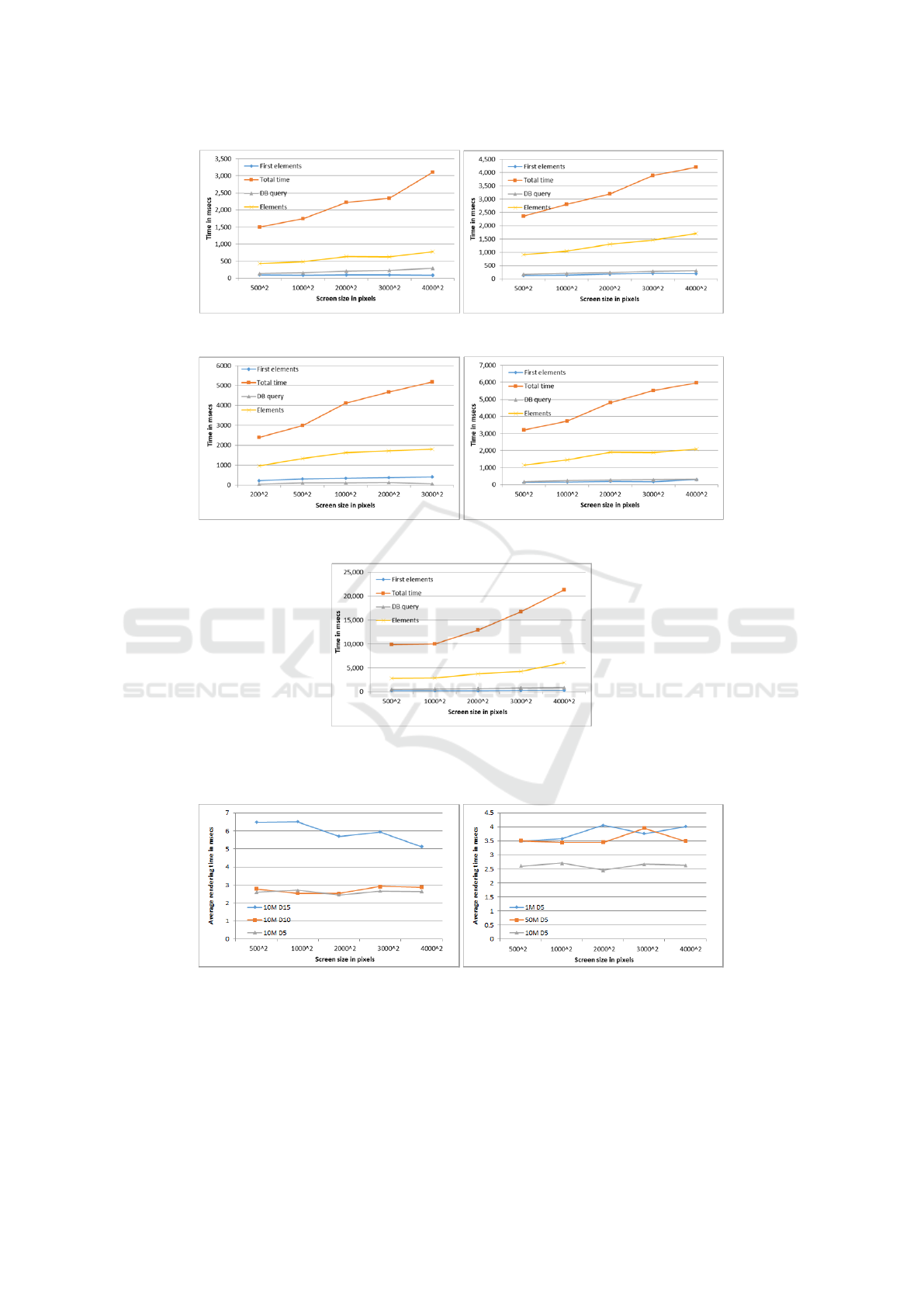
(a) 1M nodes, degree 5 (b) 10M nodes, degree 5
(c) 10M nodes, degree 10 (d) 10M nodes, degree 15
(e) 50M nodes, degree 5
Figure 2: Response Time for Synthetic Datasets.
(a) 10M nodes, all degrees (b) degree 5, synthetic datasets
Figure 3: Comparison of Average Response Time per Graph Element.
i7-4500U CPU at 1.80GHz, 4GB RAM memory over
a 12Mbps network connection.
Datasets. In order to test our system with many di-
verse datasets with a wide range of characteristics we
used synthetic datasets. The synthetic datasets, pre-
sented in Table 3, were created to comply with spe-
cific requirements for number of nodes and node de-
gree to showcase the scalability of the technique to
the size and density as well as the adaptation to any
dataset regardless of its characteristics.
Efficient Representation of Very Large Linked Datasets as Graphs
113

Web Tool Over the Dedicated API. We have imple-
mented a basic web tool for the experimental analysis
of the presented technique. The web tool is based on
the client-server architecture. The server side of the
system is the implementation of the pre-processing
technique as described above, where the input dataset
is transformed into one continuous graph, stored,
indexed and made available through the API. The
client side consists of simple user interface that di-
rectly utilize the API, and depicts the user navigation
actions, such as panning and scrolling, into spatial
queries with respect to the two-dimensional space be-
fore sending the request to the server.
4.1 API Efficiency
Metrics. We evaluate the efficiency of the technique
based on the time needed to retrieve the nodes from
the API and present them at the web tool after a spatial
query, measured in msecs. The time presented is the
figures is the time needed for the query execution, the
rendering time for the first elements to appear on the
screen and the total response time of the system when
all the elements are rendered.
Methodology. We render randomly selected parts
of the datasets using spatial queries with rectan-
gular bounding boxes ranging from 500x500 px to
4000x4000 px. As the size of the area increases the
spatial queries on the dataset match larger number of
graph elements, allowing us to examine the response
time over a variation of total rendered graph elements.
The experiments present the average results of a series
of one hundred repetitions of the graph rendering for
each rectangle size.
Results. In Figure 2 we present the results for the
synthetic datasets. In all cases the total time is closely
connected to the number of rendered elements. The
system renders more than 500 graph elements in less
than two seconds and up to 5200 graph elements with-
out causing lagging, performance issues or hindering
the user experience, as shown in 2 (e). The fact that
so many graph elements can be rendered smoothly,
is of high importance, as similar systems have lim-
its to the number of presented elements. In Figure 3
we examine the average time needed for the render-
ing of one graph element. In Figure 3 (a) we show
that the average rendering time is not dependent on
the density of the input dataset, while in Figure 3 (b)
we show that it is not dependent on the size of the
input dataset. These experiments prove that our tech-
nique scales efficiently for any size or density of the
input dataset and supports the exploration of the in-
formation for datasets with millions of nodes without
any performance issues.
5 CONCLUSIONS
In this paper, we present a novel technique for the pre-
processing very large datasets with hundreds of mil-
lions of elements and their representation as graphs in
the two-dimensional space. Our technique has been
designed in a way to meet all the identified challenges
regarding exploration needs and user experience. The
presented technique process large real datasets with
millions of elements as well as dense graphs with
high node degree. The technique does not impose
any restrictions on the dataset while the information
is offered through a dedicated API that supports many
functionalities, including keyword search, path explo-
ration and neighbor information.
REFERENCES
Abello, J., van Ham, F., and Krishnan, N. (2006). ASK-
GraphView: A Large Scale Graph Visualization Sys-
tem. TVCG, 12(5).
Archambault, D., Munzner, T., and Auber, D. (2007).
Grouse: Feature-Based, Steerable Graph Hierarchy
Exploration. In EuroVis.
Atemezing, G. A. and Troncy, R. (2014). Towards a linked-
data based visualization wizard. In COLD.
Auber, D. (2004). Tulip - A Huge Graph Visualization
Framework. In Graph Drawing Software.
Benedetti, F., Po, L., and Bergamaschi, S. (2014). A Visual
Summary for Linked Open Data sources. In ISWC.
Berners-Lee, T., Chen, Y., Chilton, L., Connolly, D., Dha-
naraj, R., Hollenbach, J., Lerer, A., and Sheets, D.
(2006). Tabulator: Exploring and analyzing linked
data on the semantic web. In Proceedings of the
3rd international semantic web user interaction work-
shop, volume 2006, page 159. Citeseer.
Brunetti, J. M., Gil, R., and Garc
´
ıa, R. (2012). Facets and
Pivoting for Flexible and Usable Linked Data Explo-
ration. In Interacting with Linked Data Workshop.
Camarda, D. V., Mazzini, S., and Antonuccio, A.
(2012). LodLive, exploring the web of data. In I-
SEMANTICS.
De Araujo, S. F. and Schwabe, D. (2009). Explorator: a
tool for exploring rdf data through direct manipula-
tion. In Linked data on the web WWW2009 workshop
(LDOW2009).
Dokulil, J. and Katreniakov
´
a, J. (2009). Using Clusters in
RDF Visualization. In Advances in Semantic Process-
ing.
Falconer, S., Callendar, C., and Storey, M.-A. (2010). A Vi-
sualization Service for the Semantic Web. In Knowl-
edge Engineering and Management by the Masses.
Hastrup, T., Cyganiak, R., and Bojars, U. (2008). Browsing
Linked Data with Fenfire. In WWW.
Heim, P., Ertl, T., and Ziegler, J. (2010a). Facet graphs:
Complex semantic querying made easy. In Extended
Semantic Web Conference, pages 288–302. Springer.
ICEIS 2020 - 22nd International Conference on Enterprise Information Systems
114

Heim, P., Lohmann, S., and Stegemann, T. (2010b). Inter-
active Relationship Discovery via the Semantic Web.
In ESWC.
Heim, P., Lohmann, S., Tsendragchaa, D., and Ertl,
T. (2011). SemLens: visual analysis of semantic
data with scatter plots and semantic lenses. In I-
SEMANTICS.
Huynh, D., Karger, D. R., Quan, D., et al. (2002). Haystack:
A platform for creating, organizing and visualizing in-
formation using rdf. In Semantic Web Workshop, vol-
ume 52.
Jr., J. F. R., Tong, H., Traina, A. J. M., Faloutsos, C., and
Leskovec, J. (2006). GMine: A System for Scalable,
Interactive Graph Visualization and Mining. In VLDB.
Kamada, T. and Kawai, S. (1989). An algorithm for draw-
ing general undirected graphs. Information Process-
ing Letters.
Karypis, G. and Kumar, V. (1998). A fast and high qual-
ity multilevel scheme for partitioning irregular graphs.
SIAM J. Sci. Comput., pages 359–392.
Kl
´
ımek, J., Helmich, J., and Necask
´
y, M. (2013). Payola:
Collaborative Linked Data Analysis and Visualization
Framework. In ESWC.
Mazumdar, S., Petrelli, D., Elbedweihy, K., Lanfranchi, V.,
and Ciravegna, F. (2015). Affective graphs: The visual
appeal of Linked Data. Semantic Web, 6(3).
Oswald, M., Reinelt, G., and Wiesberg, S. (2012). Exact
solution of the 2-dimensional grid arrangement prob-
lem. Discrete Optimization.
Schlegel, K., Weißgerber, T., Stegmaier, F., Seifert, C.,
Granitzer, M., and Kosch, H. (2014). Balloon Syn-
opsis: A Modern Node-Centric RDF Viewer and
Browser for the Web. In ESWC.
Stolte, C. and Hanrahan, P. (2000). Polaris: A Sys-
tem for Query, Analysis and Visualization of Multi-
Dimensional Relational Databases. In InfoVis.
Voigt, M., Pietschmann, S., Grammel, L., and Meißner, K.
(2012). Context-aware Recommendation of Visual-
ization Components. In eKNOW.
Zhang, K., Wang, H., Tran, D. T., and Yu, Y. (2010). Zoom-
RDF: semantic fisheye zooming on RDF data. In
WWW.
Efficient Representation of Very Large Linked Datasets as Graphs
115
