
Multiple Instance Learning for Detection of Polyps in Computed
Tomographic Colonography Images
Yunshen Xie
1
, Jianqiang Li
1 a
and Yan Pei
2 b
1
Faculty of Information, Beijing University of Technology, Beijing, 100124, China
2
Computer Science Division, University of Aizu, Aizu-wakamatsu, 965-8580, Japan
Keywords:
Machine Learning, Computed Tomographic Colonography, Computer Aid Diagnosis, Polyps, Bioinformatics.
Abstract:
Colorectal cancer(CRC) is a significant health problem in the world, the incidence of CRC can be largely
preventable by early detection and removal of the polyps before they turn into the malignant structure. Most
existing CAD system for polyps detection rely on fully supervised learning which requires the tedious manual
annotation and precise colon segmentation. This paper proposed a method based on multiple instance learning
and transfer learning. Our scheme firstly extracts many small patches from CTC images by using threshold
segmentation method, then a pre-trained model was applied for feature extracting of instances, next pooling
operator was used to aggregating these instance features into a bag, finally, classification result was obtained
by a classifier. Our proposed method does not rely on accurate colon segmentation and the result show that it
can achieve a high accuracy rate.
1 INTRODUCTION
According to the recent statistics from the American
Cancer Society, both incidence and mortality of col-
orectal cancer(CRC) rank the third among all kinds
of cancers in 2019 (DeSantis et al., 2019). The
majority of CRCs are thought to arise from polyps,
and the process can take 5-15 years for malignant
transformation into cancer. Thus, the incidence of
CRC can be largely preventable by early detection
and removal of the polyps before they turn into the
malignant structure. Nowadays, computed tomogra-
phy colonoscopy(CTC) provides a non-invasive tech-
nique for colorectal cancer screening. However, it
is a time-consuming task to review the result of the
colonoscopy, furthermore, different radiologists often
have different opinions, even for the same patient. To
overcome the limitations, various computer-aided di-
agnosis (CAD) systems were developed for the detec-
tion of polyps in CTC images.
Generally speaking, the CAD systems consist of
three main components: colon segmentation, feature
extraction and classification. Polyp candidates on
the colon surface are identified in colon segmenta-
tion step. Li et al. performed colon segmentation
a
https://orcid.org/0000-0003-1995-9249
b
https://orcid.org/0000-0003-1545-9204
using a two-dimensional region growing algorithm
on each CT slice image(Li et al., 2009). Chowdury
and Whelan developed a method for colon segmenta-
tion using geometric features(Chowdhury and Whe-
lan, 2011). Masutani et al. proposed a method to
realize colon segmentation through thresholding of
CT values and gradient magnitude values(Masutani
et al., 2001). Subsequently, a centerline-based seg-
mentation method was presented and improved the
preformance(Frimmel et al., 2005). Moreover, a
knowledge-based method was used for colon segmen-
tation(Manjunath et al., 2015), and Wyatt et al. ap-
plied 3-D region growing technique to achieve the
goal(Wyatt et al., 2000).
For feature extraction, the distinguishing features
of polyps which are malignant are curvature, size,
haustral folds, shape, colour and texture(Mittal et al.,
2016). Hu et al. used Haralick’s texture features
for 3D space. They applied the Karhunen-Loeve(KL)
transformation on these features to obtain new fea-
tures and classified by the random forest algorithm.
The volumetric curvedness and shape index is used
for polyps detection based on colon segmentation
(Zhu et al., 2009; Wang et al., 2008). Besides, Xu
and Zhao developed an algorithm based on comple-
mentary geodesic distance transformation in consid-
eration of challenges for polyps detection due to haus-
tral folds(Xu and Zhao, 2014). The morphological
236
Xie, Y., Li, J. and Pei, Y.
Multiple Instance Learning for Detection of Polyps in Computed Tomographic Colonography Images.
DOI: 10.5220/0009352002360240
In Proceedings of the 6th International Conference on Information and Communication Technologies for Ageing Well and e-Health (ICT4AWE 2020), pages 236-240
ISBN: 978-989-758-420-6
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

features, statistical and textural features of polyps in
CT images are extracted and classified by the differ-
ent classification algorithm.
Most existing CAD systems comprise of three
stages: identify polyps candidates in images; extract
features for each candidate; classify each candidate
as negative or positive. These approaches rely on
fully supervised learning, which requires the tedious
manual annotation of object location in a training
set. Moreover, there does not exist any public CT
colonography dataset with annotated polyps.
Because of polyps are too small relative to the im-
age’s size, and many noises in CT images, classify
for the whole images do not perform well. To over-
come the limitations, we proposed a MILTL method
based on multiple instance learning(MIL) and transfer
learning for CT images.
In remainder of this paper is organized as follows.
We describe our method in section II and report the
experiments and results in section III.Section IV pro-
vides the discussions and conclusions.P
2 METHOD
In this section, we will firstly introduce the formu-
lation of MIL, then define transfer learning, finally
show the structure of our proposed system.
2.1 Multiple Instance Learning
Here we review the definition of MIL Formally,
the task is to learn f : X 7−→ Y from a train-
ing data set D =
{
(x
1
, y
1
), ..., (x
m
, y
m
)
}
, where
X
i
=
{
x
i
1, ..., x
i
m
}
⊆ X is called a bag, x
i j
∈
X ( j ∈
{
1, ..., m
i
}
) is an instance, m
i
is the number of
instances in X
i
, and y
i
∈ Y =
{
Y, N
}
. X
i
is a positive
bag, i.e. y
i
= Y , if there exists x
ip
that is positive,
while p ∈
{
1, ..., m
i
}
is unknown.The goal is to pre-
dict labels for unseen bags (Zhou, 2017).
2.2 Transfer Learning
In recent years, deep convolutional neural net-
works(DCNN) have rapidly become a methodology
of choice for analyzing medical images. However, ro-
bust supervised training of a DCNN by making use of
a large amount of annotated training images(LeCun
et al., 2015). Transfer learning is essentially the use
of pre-trained networks to try to work around the re-
quirement of large data sets for deep network train-
ing(Litjens et al., 2017). Two transfer learning strate-
gies were used for medical images classification, the
first is using a pre-trained network as a feature extrac-
tor and the second is fine-tuning a pre-trained network
on training data.
2.3 Proposed System
The overall structure of the training of our proposed
system is shown in Figure.1 Firstly, the colon lumen
is segmented from the CT images. Secondly, many
small patches are extracted from an image. In our
task, an image is natural to regard as a bag and patches
which are extracted from the image as its instances.
After that, we used the pre-trained network to learn
these instance features, then a pooling layer to aggre-
gate these instance scores into bag score. Finally, we
initialize the classification layer with random weights
and configure it for CT images classification.
2.3.1 Colon Segmentation
First, threshold segmentation has been used for the
detection of colon lumen, then the morphological op-
eration is applied for noise elimination.
2.3.2 Instance Identified
The CT images are divided into several parts based
on the result of colon segmentation. This image is
viewed as a bag, and each part is treated as an instance
in the bag. The colon segmentation and instance iden-
tified can be seen in the Figure2.
2.3.3 Feature Extraction
In feature extraction, we use a VGG-Net trained on
the ImageNet dataset as a fixed feature extractor. We
first extract the features of instance through the fea-
ture extractor, then a pooling layer is used to aggre-
gate these instance features into a bag. We look at
three pooling method(max pooling, mean pooling and
log pooling) in our proposed system.
2.3.4 Classification
When we performed the classification step, we build a
classifier from three fully connected layers which use
the cross-entropy to calculate the cost.
3 EXPERIMENTS AND RESULTS
3.1 Materials
The CTC data used in this study include 67 cases from
The Cancer Imaging Archive(TCIA), which consisted
Multiple Instance Learning for Detection of Polyps in Computed Tomographic Colonography Images
237
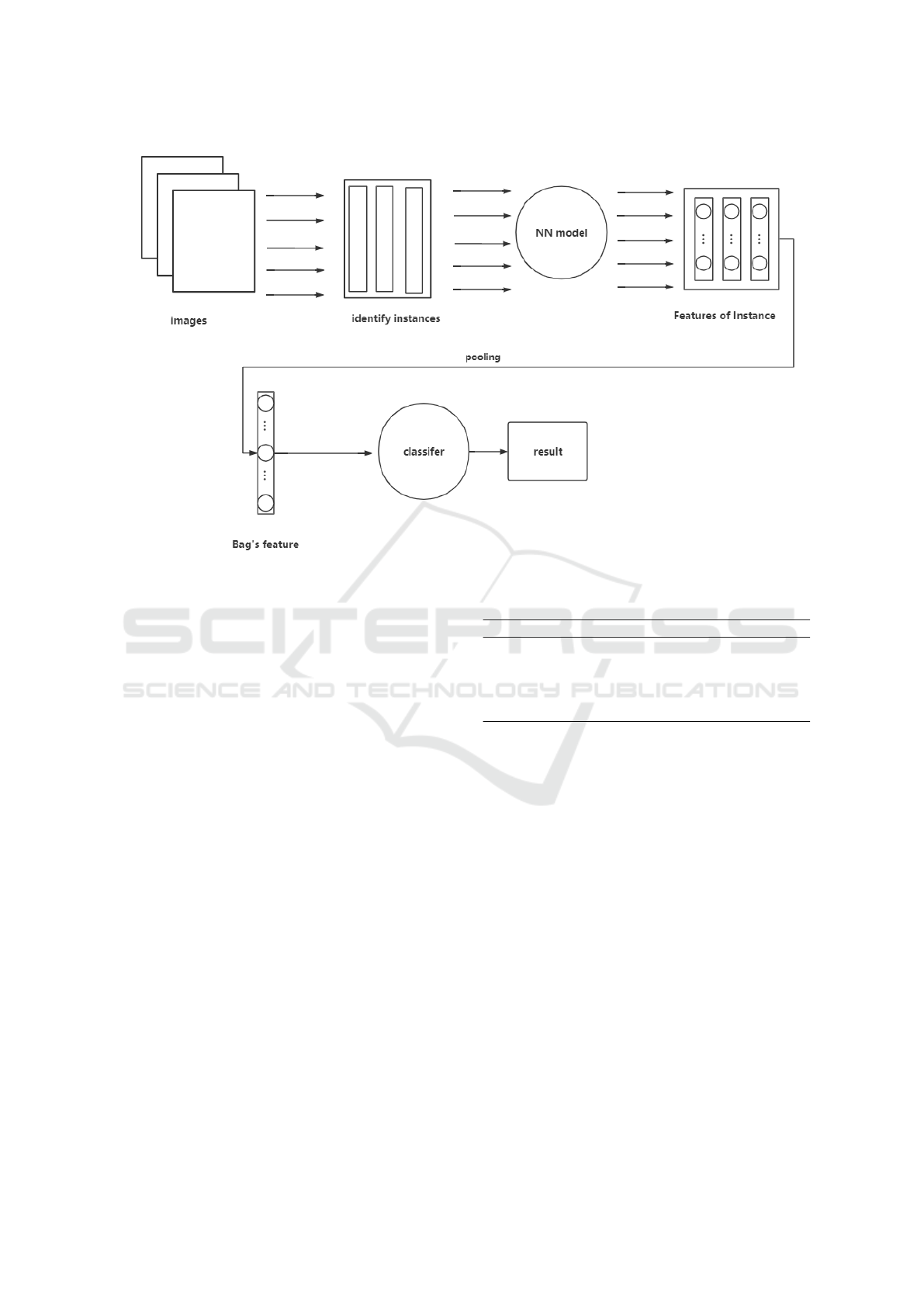
Figure 1: Proposed System.
of 39 cases with 6-9 mm polyps and 28 cases which
have at least one 10 mm or larger size polyp was
found. Because each patient case includes two scans,
supine and prone, there are in total 180 positive im-
ages (with polyp). And we random sampled 180 neg-
ative images from the case with no polyp found.
The TCIA CTC datasets were acquired by using
at least a 16 slice CT scanner with 0.5-1.0 mm col-
limation, 0.98-1.5 pitch, 0.8 mm reconstruction in-
terval, 1-1.25 mm slice thickness, 50 effective mAs,
120 kVp, CT slice size of 512*512 pixels(Ren et al.,
2018).
3.2 Experimental Setting and
Evaluation
In this part, we make two comparison experiment
by using pre-trained VGG-16 and ResNet50, respec-
tively. The VGG-16 and ResNet50 are available for
use in the TensorFlow models repository. In the eval-
uation of classification performance, a ten-fold cross-
validation method was used to minimize the evalua-
tion bias. The accuracy, recall, precision and AUC
are evaluation metrics in our study. Also, the evalua-
tion is conducted with 10 trails running for statistical
results.
Table 1: Comparisions of Algorithms.
methods accuracy recall precision AUC
VGG 0.7778 0.8056 0.7838 0.7778
ResNet 0.8333 0.8611 0.7750 0.8333
max-MILTL 0.9028 0.9167 0.8250 0.9167
mean-MILIL 0.8472 0.9167 0.8049 0.8333
log-MILIL 0.8750 0.8889 0.8049 0.8858
3.3 Result
It can be seen from Table I that the models we con-
structed are better than the existing pre-trained VGG-
16 and ResNet-50. Moreover, compared with the
other two, MILTL with max-pooling layer was the
best with the accuracy of 0.9028 and AUC of 0.9167.
4 CONCLUSION
The framework provided by MIL is particularly suit-
able for CTC image classification. In this paper, we
proposed a new method for the automatic detection
of colon polyps based on CTC images. This method
includes colon segmentation, instance identified, fea-
ture extraction and classification. Due to the nature of
MIL method, the colon segmentation does not require
precise segmentation results, which undoubtedly pro-
vides convenience and saves time for the polyps de-
tection. According to our experiments, the proposed
ICT4AWE 2020 - 6th International Conference on Information and Communication Technologies for Ageing Well and e-Health
238
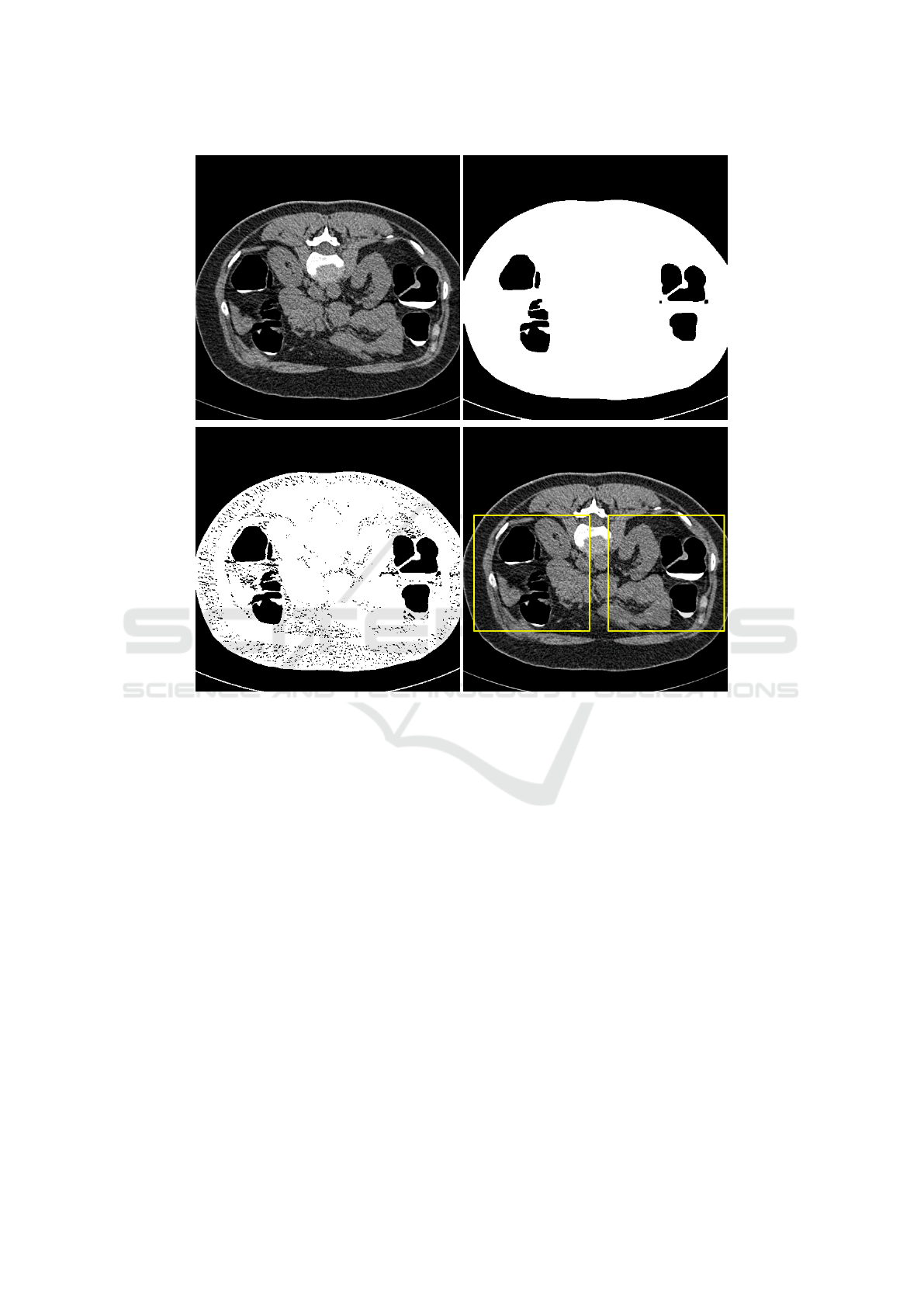
Figure 2: Colon Segmentation and Instance Identified.
method can improve the accuracy of classification. In
the future, we will focus on the probability relation
between bag and instances, to make sure the label for
instances, especially for positive instances.
ACKNOWLEDGEMENTS
This work was supported by Beijing Natural Science
Foundation under Grant 4184082, in part by the Na-
tional Natural Science Foundation of China under
Grant 61806014.
REFERENCES
Chowdhury, T. A. and Whelan, P. F. (2011). A fast and ac-
curate method for automatic segmentation of colons at
ct colonography based on colon geometrical features.
In 2011 Irish Machine Vision and Image Processing
Conference, pages 94–100. IEEE.
DeSantis, C. E., Miller, K. D., Goding Sauer, A., Jemal, A.,
and Siegel, R. L. (2019). Cancer statistics for african
americans, 2019. CA: a cancer journal for clinicians,
69(3):211–233.
Frimmel, H., N
¨
appi, J., and Yoshida, H. (2005). Centerline-
based colon segmentation for ct colonography. Medi-
cal Physics, 32(8):2665–2672.
LeCun, Y., Bengio, Y., and Hinton, G. (2015). Deep learn-
ing. nature, 521(7553):436.
Li, J., Huang, A., Yao, J., Liu, J., Van Uitert, R. L.,
Petrick, N., and Summers, R. M. (2009). Opti-
mizing computer-aided colonic polyp detection for ct
colonography by evolving the pareto front a. Medical
physics, 36(1):201–212.
Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A.,
Ciompi, F., Ghafoorian, M., Van Der Laak, J. A.,
Van Ginneken, B., and S
´
anchez, C. I. (2017). A survey
on deep learning in medical image analysis. Medical
image analysis, 42:60–88.
Manjunath, K., Prabhu, K. G., and Siddalingaswamy, P.
(2015). A knowledge based approach for colon seg-
mentation in ct colonography images. In 2015 IEEE
International Conference on Signal and Image Pro-
cessing Applications (ICSIPA), pages 65–70. IEEE.
Multiple Instance Learning for Detection of Polyps in Computed Tomographic Colonography Images
239

Masutani, Y., Yoshida, H., MacEneaney, P. M., and
Dachman, A. H. (2001). Automated segmentation of
colonic walls for computerized detection of polyps in
ct colonography. Journal of Computer Assisted To-
mography, 25(4):629–638.
Mittal, A., Kaur, M., et al. (2016). Computer-aided-
diagnosis in colorectal cancer: A survey of state of
the art techniques. In 2016 International Conference
on Inventive Computation Technologies (ICICT), vol-
ume 1, pages 1–6. IEEE.
Ren, Y., Ma, J., Xiong, J., Chen, Y., Lu, L., and Zhao, J.
(2018). Improved false positive reduction by novel
morphological features for computer-aided polyp de-
tection in ct colonography. IEEE journal of biomedi-
cal and health informatics, 23(1):324–333.
Wang, S., Zhu, H., Lu, H., and Liang, Z. (2008). Volume-
based feature analysis of mucosa for automatic initial
polyp detection in virtual colonoscopy. International
journal of computer assisted radiology and surgery,
3(1-2):131–142.
Wyatt, C., Ge, Y., and Vining, D. (2000). Automatic seg-
mentation of the colon for virtual colonoscopy. Com-
puterized medical imaging and graphics, 24(1):1–9.
Xu, Y.-r. and Zhao, J. (2014). Segmentation of haustral
folds and polyps on haustral folds in ct colonography
using complementary geodesic distance transforma-
tion. Journal of Shanghai Jiaotong University (Sci-
ence), 19(5):513–520.
Zhou, Z.-H. (2017). A brief introduction to weakly super-
vised learning. National Science Review, 5(1):44–53.
Zhu, H., Duan, C., Pickhardt, P., Wang, S., and Liang, Z.
(2009). Computer-aided detection of colonic polyps
with level set-based adaptive convolution in volu-
metric mucosa to advance ct colonography toward a
screening modality. Cancer Management and Re-
search, 1:1.
ICT4AWE 2020 - 6th International Conference on Information and Communication Technologies for Ageing Well and e-Health
240
