
Theoretical Study of the Fidelity of Transcription
Yao-Gen Shu
1,2
, Ming Li
3
and Zhong-Can Ou-Yang
2
1
Bioinformatics Laboratory of Yishang Innovation Technology Co., Ltd, Beijing 100081, China
2
Institute of Theoretical Physics, Chinese Academy of Sciences, Beijing 100190, China
3
School of Physical Sciences, University of Chinese Academy of Sciences, Beijing 100049, China
Keywords:
Gene Transcription Fidelity, First-passage Approach, First-order Neighbor Effects.
Abstract:
This year we celebrate the 50
th
anniversary of the discovery of the three eukaryotic RNA polymerases. Ever
since this seminal event was uncovered by Robert Roeder in 1969(Roeder and Rutter, 1969), researchers
have investigated the intricate mechanisms of gene transcription with great dedication. However, there is not
breakthrough in study of the fidelity of transcription still. Here, we propose a simplest model with first-order
neighbor effects, a first-passage approach, to theoretically investigate the gene transcription fidelity.
1 INTRODUCTION
Transcription is the process in which a gene’s DNA
sequence is transcribed by an RNA polymerase
(RNAp). RNAp uses one of the DNA strands as
a template to make a new, complementary RNA
molecule. The transcription cycle includes three
phases: initiation, elongation, and termination. The
initiation phase involves recognition of promoter
DNA, DNA opening, and synthesis of a short ini-
tial RNA oligomer. During the elongation phase,
the polymerase uses the DNA template to extend the
growing RNA chain in a processive manner. Finally,
DNA and RNA are released during termination, and
the polymerase can then be recycled and re-initiate
transcription(Cramer, 2019).
The probability of mismatch during transcription
is more than that during replication of the DNA. It is
because errors in transcription may not be fatal due to
its non-inherited. When any mismatched nucleotide
is added to the template DNA, the RNAp will halt
and then it will either proofread the nascent chain
or continue without correcting. There are two ma-
jor ways of proofreading: pyrophosphorolytic editing
and hydrolytic one. Once the incorrect nucleotide is
added, the very first way to rectify that mistake is
by pyrophosphorolytic editing. In this, a pyrophos-
phate (PPi) will enter the active cleft and attack the
wrong nucleotide added, i.e. NMP (such as AMP,
GMP, CMP or UMP). It would lead to the conversion
of NMP to NTP. Thus the incorrect nucleotide is re-
moved, and the RNAp will add a correct nucleotide. If
pyrophosphorolytic editing can not rectify the mRNA
sequence, then another method called hydrolytic edit-
ing is activated. In this proofreading, certain proteins
belonging to Gre and Nus protein family gets acti-
vated. If the correct base pair is not added, the RNAp
will halt, and these proteins will enter the active cleft
of the core enzyme. These proteins would remove 4
Nucleotides from the nascent RNA chain and then the
RNAp will add the correct nucleotides again(Libby
and Gallant, 1991; von Hippel, 1998).
It’s now widely acknowledged that match (A→U,
T→A and G↔C, denoted as Right(R) pairs) play
a dominate role in the transcription, while the mis-
match (denoted as Wrong(W) pairs) occur with very
low probability. This is not due to the difference be-
tween the free energy of R and W pairs in the dou-
ble chains: in fact, this free energy difference is only
about 2 ∼ 4k
B
T (where k
B
and T are Boltzmann con-
stant and absolute temperature respectively.), which
cannot account for such low mismatch probability
if it is estimated by Boltzmann factor. As pointed
out by J.Hopfield(Hopfield, 1974) and J.Ninio(Ninio,
1975) in study of the fidelity DNA polymerases, the
low mismatch probability may originate from the
huge difference of replication kinetics between R and
W(I. R. Lehman and Kornberg, 1958; Kunkel and
Bebenek, 2000).
The fidelity of DNA replication has been inves-
tigated in kinetics recently(Y. G. Shu and Li, 2015;
Y. S. Song and Li, 2017; Gaspard, 2017; Q. S. Li and
Li, 2019; M. Li and Shu, 2018). However, the fidelity
of transcription is rarely studied. Here, we propose a
Shu, Y., Li, M. and Ou-Yang, Z.
Theoretical Study of the Fidelity of Transcription.
DOI: 10.5220/0009181002390241
In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2020) - Volume 3: BIOINFORMATICS, pages 239-241
ISBN: 978-989-758-398-8; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
239
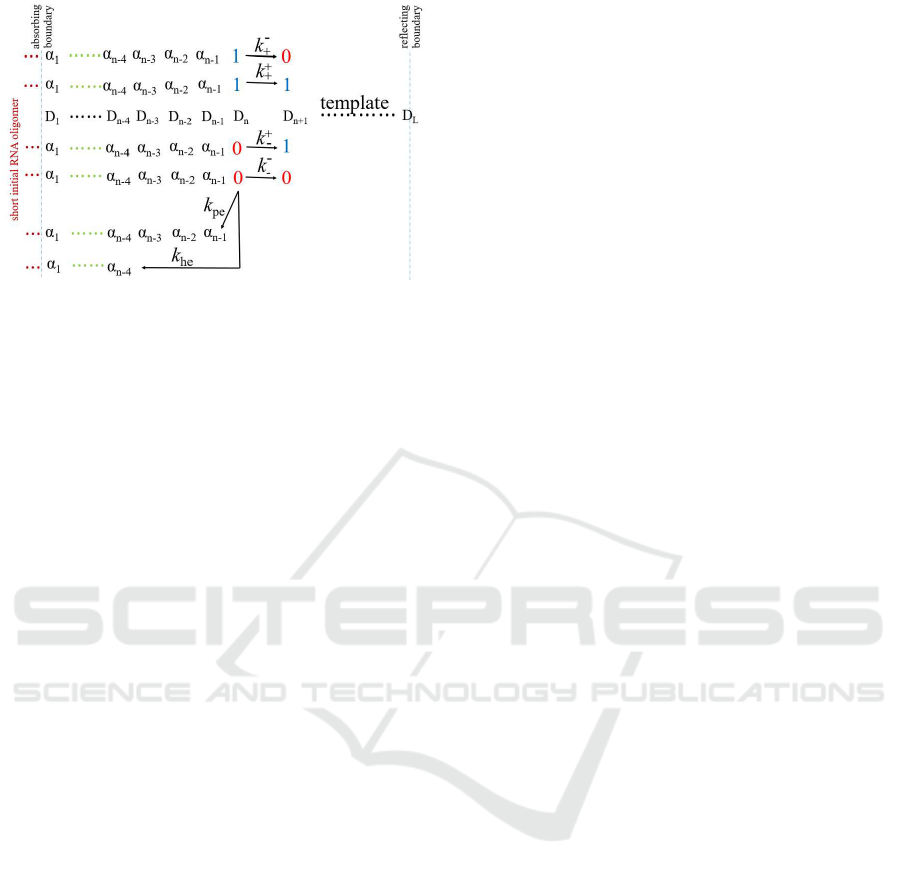
Figure 1: Kinetics of transcription in our model. At the end
of initiation phase (after the synthesis of a short initial RNA
oligomer, that is the correct nucleation), there is a reflect-
ing boundary at starting of elongation, which corresponds to
Eqs.(1) or (2). At i ∈ [2, 4], there is no hydrolytic editing if
incorrect nucleotide is added, which corresponds to Eqs.(3)
or (4). At i ∈ [5, L − 6], a normal elongation phase, which
can be described by Eqs.(5) or (6). There is no hydrolytic
editing for i ∈ [L − 5, L − 2] due to absorbing boundary at
termination as presenting in Eqs.(7) or (8). At absorbing
boundary (termination, i = L − 1), the kinetics is described
by Eqs.(9) or (10).
simplest model to theoretically try the fidelity of tran-
scription.
2 FIRST-PASSAGE MODEL
For brevity and not losing generality, we suppose that
the gene consists of two kinds of units A (denotes A or
T) and G (denotes G or C), and correspondingly two
kinds of monomers U (denotes U or A) and C (denotes
C or G) are to be added to the end of the growing chain
and paired with A or G to transcript the gene with L
bases. Since U pairs with A much more probably than
with G, we denote (
A
U
) as the match 1 pair and (
A
G
) as
the mismatch 0 pair. Similarly, we denote (
G
C
) as 1
and (
G
U
) as 0. Besides, we only consider the first-order
copolymeriztion processes as below: the adding rate
of a correct nucleotide at match end is k
+
+
; that of a
correct nucleotide at mismatch end is k
+
−
, while the
adding rate of an incorrect nucleotide at match end is
k
−
+
; that of a incorrect nucleotide at mismatch end is
k
−
−
.
Since transcription proceeds unidirectionally, we
assume that the nascent chain initiates from a pre-
existing seed (a short initial RNA oligomer) after ini-
tiation phase, then elongates as a template-directed
binary copolymer, and terminates at when its length
reaches L. In the elongation phase, the monomer U or
C can be added to or deleted from the growing end due
to proofreading, where k
pe
and k
he
denote the rate of
pyrophosphorolytic editing and that of hydrolytic one
with removing 4 bases respectively. In contrast, the
initial seed and the lastly-added monomer can not be
deleted. In other words, this is a first-passage process
from a reflecting boundary at the first position to an
absorbing boundary at the last position. It’s worth to
note that the initiation and termination here are purely
imaginary to simplify the mathematical treatments
and do not imply the fact of initiation and termination
of transcription. The first-passage treatment largely
simplifies the calculations by introducing a closed set
of kinetic equations, so it is more convenient to be
chosen for approximate calculations. Above kinetic
framework is shown in Fig.1.
The probability of the growing RNA sequence
α
1
...α
i
(where 1 ≤ i ≤ L, and α
i
= 1 denotes pairs
of (
A
U
), (
T
A
), (
G
C
), or (
C
G
). Otherwise, α
i
= 0.) appearing
in the transcription process along gene D
1
· · · D
L
(D
i
denotes A/T/C/G) at time t is denoted as ρ
D
1
···D
L
α
1
···α
i
(t).
Now we have the following master equations.
At the end of initiation phase (after the synthesis
of a short initial RNA oligomer, that is the correct nu-
cleation), there is a reflecting boundary at starting of
elongation.
˙
ρ
D
1
···D
L
1
= k
+
+
+ k
pe
ρ
D
1
···D
L
10
+ k
he
ρ
D
1
···D
L
1α
2
α
3
α
4
0
(1)
˙
ρ
D
1
···D
L
0
= k
−
+
+ k
pe
ρ
D
1
···D
L
00
+ k
he
ρ
D
1
···D
L
0α
2
α
3
α
4
0
(2)
There is no hydrolytic editing if incorrect nu-
cleotide is added at 2 ≤ i ≤ 4,
˙
ρ
D
1
···D
L
α
1
···α
i
1
= k
pe
ρ
D
1
···D
L
α
1
···α
i
10
+ k
he
ρ
D
1
···D
L
α
1
···α
i
1α
i+2
···α
i+4
0
+k
+
+
ρ
D
1
···D
L
α
1
···α
i−1
1
+ k
+
−
ρ
D
1
···D
L
α
1
···α
i−1
0
−
k
+
+
+ k
−
+
ρ
D
1
···D
L
α
1
···α
i
1
(3)
˙
ρ
D
1
···D
L
α
1
···α
i
0
= k
pe
ρ
D
1
···D
L
α
1
···α
i
00
+ k
he
ρ
D
1
···D
L
α
1
···α
i
0α
i+2
···α
i+4
0
+k
−
−
ρ
D
1
···D
L
α
1
···α
i−1
0
+ k
−
+
ρ
D
1
···D
L
α
1
···α
i−1
1
−
k
pe
+ k
+
−
+ k
−
−
ρ
D
1
···D
L
α
1
···α
i
0
(4)
At elongation phase for 5 ≤ i ≤ L − 5
˙
ρ
D
1
···D
L
α
1
···α
i
1
= k
pe
ρ
D
1
···D
L
α
1
···α
i
10
+ k
he
ρ
D
1
···D
L
α
1
···α
i
1α
i+2
···α
i+4
0
+k
+
+
ρ
D
1
···D
L
α
1
···α
i−1
1
+ k
+
−
ρ
D
1
···D
L
α
1
···α
i−1
0
−
k
+
+
+ k
−
+
ρ
D
1
···D
L
α
1
···α
i
1
(5)
˙
ρ
D
1
···D
L
α
1
···α
i
0
= k
pe
ρ
D
1
···D
L
α
1
···α
i
00
+ k
he
ρ
D
1
···D
L
α
1
···α
i
0α
i+2
···α
i+4
0
+k
−
−
ρ
D
1
···D
L
α
1
···α
i−1
0
+ k
−
+
ρ
D
1
···D
L
α
1
···α
i−1
1
−
k
he
+ k
pe
+ k
+
−
+ k
−
−
ρ
D
1
···D
L
α
1
···α
i
0
(6)
There is not item of hydrolytic editing for L − 4 ≤
BIOINFORMATICS 2020 - 11th International Conference on Bioinformatics Models, Methods and Algorithms
240

i ≤ L − 2 due to absorbing boundary at termination.
˙
ρ
D
1
···D
L
α
1
···α
i
1
= k
pe
ρ
D
1
···D
L
α
1
···α
i
10
+ k
+
+
ρ
D
1
···D
L
α
1
···α
i−1
1
+ k
+
−
ρ
D
1
···D
L
α
1
···α
i−1
0
−
k
+
+
+ k
−
+
ρ
D
1
···D
L
α
1
···α
i
1
(7)
˙
ρ
D
1
···D
L
α
1
···α
i
0
= k
pe
ρ
D
1
···D
L
α
1
···α
i
00
+ k
−
−
ρ
D
1
···D
L
α
1
···α
i−1
0
+ k
−
+
ρ
D
1
···D
L
α
1
···α
i−1
1
−
k
he
+ k
pe
+ k
+
−
+ k
−
−
ρ
D
1
···D
L
α
1
···α
i
0
(8)
At absorbing boundary (termination, i = L − 1),
˙
ρ
D
1
···D
L
α
1
···α
i
1
= k
+
+
ρ
D
1
···D
L
α
1
···α
i−1
1
+ k
+
−
ρ
D
1
···D
L
α
1
···α
i−1
0
(9)
˙
ρ
D
1
···D
L
α
1
···α
i
0
= k
−
−
ρ
D
1
···D
L
α
1
···α
i−1
0
+ k
−
+
ρ
D
1
···D
L
α
1
···α
i−1
1
(10)
One of our major concerns is the final distribu-
tion of the nascent RNA sequence, i.e, the long-time
limit P
D
1
···D
L
α
1
···α
L
= ρ
D
1
···D
L
α
1
···α
L
(t → ∞). To calculate it, we
assume the initial conditions ρ
D
1
···D
L
α
1
(t = 0) ≡ q
α
1
,
where q
1
+ q
0
= 1. q
α
1
can be arbitrarily chosen be-
cause it has negligible impacts on the fidelity pro-
file except few positions near the reflecting boundary.
Thus, the P
D
1
···D
L
α
1
···α
L
can be calculated by simulation with
known parameters L, k
+
−
, k
−
−
, k
+
+
, k
−
+
, k
pe
and k
he
.
We define the fidelity of limited length transcrip-
tion in percent as
fidelity of transcription ≡
∑
L
i=1
α
i
L
× 100%, (11)
which is different from the definition of that of DNA
replication.
3 DISCUSSION
Though mismatch in transcription is not fatal, it is
tightly related to cancer. In this manuscript, we pro-
posed a general approach, based on the first-passage
description of the transcription process, to calculate
the positional fidelity for any given gene. The math-
ematical treatment is shown in Eqs.(1)-(10) with 6
parameters. Although we only consider the first-
order copolymeriztion processes, it can be extended
to higher-order copolymeriztion as Ref(Q. S. Li and
Li, 2019).
We neither do the simulation nor solve analyt-
ically Eqs. by iteration as Refs.(Gaspard, 2017;
Q. S. Li and Li, 2019) because of the lack of experi-
mental data such as parameters and fidelity. Neverthe-
less, this is a preliminary theoretical investigation. We
hope it will motivates experimenter to quantitatively
measure parameters and the fidelity defined above.
ACKNOWLEDGEMENTS
The authors thank the financial support by Key Re-
search Program of Frontier Sciences of CAS (No.
Y7Y1472Y61), National Natural Science Founda-
tion of China (No.11574329, 11774358, 11675180),
CAS Strategic Priority Research Program (No.
XDA17010504), and CAS Biophysics Interdisci-
plinary Innovation Team Project (No.2060299).
REFERENCES
Cramer, P. (2019). Eukaryotic transcription turns 50. Cell,
179:808.
Gaspard, P. (2017). Iterated function systems for DNA
replication. Phys. Rev. E, 96:042403.
Hopfield, J. J. (1974). Kinetic proofreading: A new mecha-
nism for reducing errors in biosynthetic processes re-
quiring high specificity. PNAS, 71:4135.
I. R. Lehman, M. J. Bessman, E. S. S. and Kornberg,
A. (1958). Enzymatic synthesis of deoxyribonucleic
acid: I. preparation of substrates and partial purifi-
cation of an enzyme from escherichia coli. J. Biol.
Chem., 233:163.
Kunkel, T. A. and Bebenek, K. (2000). DNA replication
fidelity. Ann. Rev. Biochem., 69:497.
Libby, R. T. and Gallant, J. A. (1991). The role of rna poly-
merase in transcriptional fidelity. Mol. Micro., 5:999.
M. Li, Z. C. O.-Y. and Shu, Y. G. (2018). Study
on the fidelity of biodevice T7 DNA polymerase.
BIOSTEC2018, Volume3: BIOINFORMATICS:135.
Ninio, J. (1975). Kinetic amplification of enzyme discrimi-
nation. Biochimie, 57:587.
Q. S. Li, P. D. Zheng, Y. G. S. Z. C. O.-Y. and Li, M.
(2019). Template-specific fidelity of DNA replication
with high-order neighbor effects: A first-passage ap-
proach. Phys. Rev. E, 100:012131.
Roeder, R. G. and Rutter, W. J. (1969). Multiple forms
of DNA-dependent RNA polymerase in eukaryotic or-
ganisms. Nature, 224:234.
von Hippel, P. H. (1998). An integrated model of the tran-
scription complex in elongation, termination, and edit-
ing. Science, 281:660.
Y. G. Shu, Y. S. Song, Z. C. O.-Y. and Li, M. (2015). A gen-
eral theory of kinetics and thermodynamics of steady-
state copolymerization. J. Phys.:Condens. Matter,
27:235105.
Y. S. Song, Y. G. Shu, X. Z. Z. C. O.-Y. and Li, M.
(2017). Proofreading of DNA polymerase: a new
kinetic model with higher-order terminal effects. J.
Phys.:Condens. Matter, 29:025101.
Theoretical Study of the Fidelity of Transcription
241
