
Enhancing Decision-making Systems with Relevant Patient Information
by Leveraging Clinical Notes
Jo
˜
ao Rafael Almeida
1,2,∗ a
, Jo
˜
ao Figueira Silva
1,∗ b
, Alejandro Pazos Sierra
2 c
, Sergio Matos
1 d
and Jos
´
e Lu
´
ıs Oliveira
1 e
1
DETI/IEETA, University of Aveiro, Aveiro, Portugal
2
Department of Information and Communications Technologies, University of A Coru
˜
na, A Coru
˜
na, Spain
Keywords:
EHR, CDSS, NLP, Clinical Notes, Clinical Decision-making, Treatment Guidance.
Abstract:
Hospitalised patients suffering from secondary illnesses that require daily medication typically need per-
sonalised treatment. Although clinical guidelines were designed considering those circumstances, existing
decision-support features fail in assimilating detailed relevant patient information, which opens up opportu-
nities for systems capable of performing a real-time evaluation of such data against existing knowledge and
providing recommendations during clinical treatments. In this paper, we present a proposal for a new feature
to integrate with electronic health record (EHR) systems that enriches the health treatment process by auto-
matically extracting information from patient medical notes and aggregating it in clinical protocols. Our goal
is to leverage the historical component of the patient trajectory to improve clinical decision support systems
performance.
1 INTRODUCTION
Progress in technology has proved fruitful for the field
of medicine and health care throughout the years,
leading to an enhanced quality of life for the general
population. Tools and data sources originated from
the merging of these two fields have fostered improve-
ments in disease prevention, diagnosis and treatment,
and can play an important role in clinical pipelines by
assisting physicians in tasks such as clinical decision
making and patient follow-up. Furthermore, by pro-
viding access to increasing amounts of medical data,
it is possible to shift towards the more patient-centric
view of personalised medicine.
Aside from scale, technology also brought diver-
sity to medical data, comprising various data types
such as medical imaging, genomic, signal, or labora-
tory data, which must be stored and organised. Elec-
tronic health records (EHRs) provide electronic sup-
port to agglutinate administrative and medical data
a
https://orcid.org/0000-0003-0729-2264
b
https://orcid.org/0000-0001-5535-754X
c
https://orcid.org/0000-0003-2324-238X
d
https://orcid.org/0000-0003-1941-3983
e
https://orcid.org/0000-0002-6672-6176
∗
Both authors contributed equally to this work.
from various sources and to centralise information
at the patient level (Katehakis and Tsiknakis, 2006;
Costa, 2004), enabling the documentation of a pa-
tient’s health status throughout time and represent-
ing the patient trajectory. By having a longitudinal
view of the patient medical history accessible in a sin-
gle structure, the EHR can provide physicians with
important contextual information therefore rendering
EHRs as an important piece to support the medical
act.
EHR information can be analysed regarding data
type and structure. From a structural perspective,
EHRs can contain structured and unstructured data.
Structured data can be found in forms, being com-
mon in patient demographics data where patient in-
formation is organised in form fields. It is also found
in certain medical reporting forms, where codes from
coding standards such as ICD (International Classi-
fication of Diseases) (WHO, 2018), SNOMED-CT
(Systematized Nomenclature of Medicine - Clinical
Terms) (Stearns et al., 2001) or RxNorm (Nelson
et al., 2011) can be used. These standards attempt
to structure text data by mapping medical concepts
regarding topics such as symptoms, diagnosis, treat-
ments and procedures, to codes which act as unique
identifiers and that can be easily processed. How-
ever, coding standards also pose certain limitations
254
Almeida, J., Silva, J., Sierra, A., Matos, S. and Oliveira, J.
Enhancing Decision-making Systems with Relevant Patient Information by Leveraging Clinical Notes.
DOI: 10.5220/0009166902540262
In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2020) - Volume 5: HEALTHINF, pages 254-262
ISBN: 978-989-758-398-8; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

that hinder their use, namely the time required by clin-
ical staff to sift through the standards to select the
most adequate code, or the ambiguity or lack of speci-
ficity in certain terms.
On the other hand, unstructured data such as free
text, is typically present in clinical notes such as clin-
ical appointment reports or patient discharge reports.
Since free text is written in natural language, it over-
comes the limitations of structured text by providing
physicians with a flexible convoy to document com-
plete descriptions of the patient health status, contain-
ing the context and rationale for a certain diagnosis or
treatment procedure, thus containing valuable infor-
mation for the processes of clinical decision making
and patient follow-up.
Owing to the aforementioned reasons, free text
present in clinical notes represents a large amount
of the data contained in EHRs. In fact, the impor-
tance of free text extends beyond that as it is ac-
knowledged that clinical free text can often encom-
pass information otherwise not obtainable from other
data sources (Jensen et al., 2017). However, much
of this potential remains underexplored mainly due
to the nature of clinical text, which makes it very
challenging to process and explore (Neustein et al.,
2014). Nonetheless, interest in this domain has shown
continuous growth during the past years, with some
research efforts having already been made on fields
such as clinical natural language processing (NLP) to
develop solutions for annotating and summarising rel-
evant data in clinical notes (Pivovarov and Elhadad,
2015).
The increased involvement of technology in
healthcare has opened up not only opportunities but
also challenges. For instance, the increased availabil-
ity of medical data, that can aid physicians in their
decisions, also resulted in a heavier burden for physi-
cians who must sift through larger amounts of less rel-
evant data to find information of their actual interest.
An illustrative example of this can be found in clini-
cal notes, where the increased easiness of replicating
information through copy-paste actions has resulted
in more redundant information (sometimes even erro-
neous), and consequently on lower data quality which
can ultimately compromise the quality of the medical
act (Cohen et al., 2013; Singh et al., 2013).
It comes naturally that solutions such as a clinical
decision support system (CDSS) can play an impor-
tant role by leveraging the large amounts of existing
EHR data to provide physicians with only key infor-
mation. However, when doing so, these systems must
deal with the natural challenges associated with medi-
cal data, namely its high heterogeneity and poor qual-
ity (data is frequently incomplete, noisy and sparse)
which are worsened by non-standardized physician
practices (Hripcsak and Albers, 2012).
Regarding data sources, these systems can explore
single and multi modality - the latter combining vari-
ous sources and being reckoned as a particularly chal-
lenging task (Miotto et al., 2017) - and also structured
and/or unstructured data. Structured data is in general
more straightforward to exploit, thus being used more
frequently. However, the inclusion of unstructured
data can provide key content to improve the perfor-
mance of previously existing systems. For instance,
structured text has already been used for various pur-
poses such as prediction modelling (Wu et al., 2010;
Ferr
˜
ao et al., 2013; Ferr
˜
ao et al., 2016) whereas appli-
cations with free text are relatively scarce, despite the
acknowledgement that free text holds great potential
as a source of relevant data (Jensen et al., 2017).
Progressing from a data to a medical process per-
spective, clinical practice guidelines (CPGs) consist
of systematically developed statements that were cre-
ated to assist physicians by providing recommen-
dations for diagnosis and treatment guidance (NC-
CIH, 2017). Despite the relevance of their objec-
tive, CPGs did not have the expected impact on health
care, which can be explained by factors such as the
lack of time by physicians to learn them, or the fact
that CPGs lack manageable workflows that could ef-
fectively help putting recommended tasks to prac-
tice. EHR-based CDSSs can provide a contribution
in treatment guidance, but to succeed it is vital that
they incorporate detailed relevant patient data, per-
form an on the fly evaluation against prior knowledge,
and provide recommendations which physicians can
act upon (Stewart et al., 2007).
Moreover, to make their impact in health care
more significant, CDSSs should be deployed along
with slight adjustments in clinic workflow and staff
duty, and should be further explored for appointment
planning instead of only providing information to-
wards the end of clinical appointments. Several EHR-
based CDSSs have already been tested in the past for
patients with illnesses such as diabetes, hypertension
and others, with the objective of improving key in-
termediate clinical outcomes of chronic disease care.
However, the majority of these systems failed because
they missed many of the above-mentioned key as-
pects (O’Connor et al., 2011).
In this paper, we present a proposal for a system
that enriches the health treatment process by auto-
matically extracting information from patient medical
notes and aggregating it in clinical protocols, with the
goal of leveraging the historical component of the pa-
tient trajectory to improve CDSS performance. Our
main contributions in this paper are the following:
Enhancing Decision-making Systems with Relevant Patient Information by Leveraging Clinical Notes
255

• We create new opportunities mainly related to
EHR exploration, enabling the enhancement of
decision-making processes based on new relation-
ships and pathways between diseases and parental
phenotypes;
• The proposed methodology implements different
strategies to automatically extract relevant patient
clinical information during medical treatments;
• The methodology was integrated in an existing
open-source clinical decision support tool to ex-
plore newly extracted information in clinical pro-
tocols, with the objective of providing better treat-
ment guidance.
2 GATHERING RELEVANT
INFORMATION FROM
CLINICAL NOTES
Clinical notes are an important “tool” for physicians
as they keep record of patient trajectories in a readily
accessible format, making them suitable for aiding in
clinical decision making and patient follow-up. The
trajectory component in clinical notes can be partic-
ularly evident as these notes can be produced in dif-
ferent stages of health care (e.g. patient admission,
discharge, clinical appointment). Despite constituting
a big source of relevant patient information (e.g. di-
agnosis, recommended or followed procedure, medi-
cation, family history), many clinical notes are stored
and unexplored due to the intricacies of processing
free text.
When considering the process of information ex-
traction (IE) from clinical free text, a similar pipeline
is used as that explored for common text. This
pipeline typically involves two steps: NER (Named
Entity Recognition) where entities such as drugs or
diseases are identified in the text, and NEN (Named
Entity Normalization) where identified entities can be
disambiguated and normalised to unique identities. In
clinical text, NEN can explore coding standards such
as ICD, RxNorm or UMLS (Unified Medical Lan-
guage System) - a metathesaurus that aggregates mul-
tiple lexicons - so as to obtain normalised text which
can be stored as structured data. Furthermore, these
steps can explore different approaches to process text,
namely heuristic and NLP approaches.
Structured text is easier to integrate in clinical de-
cision support systems, therefore it is vital to extract
relevant patient information from medical narratives
and store it as structured data. Taking that into ac-
count, different types of information were extracted
from clinical notes. Firstly, heuristics and NLP tech-
niques were combined to extract entities related with
several classification criteria, namely cardiovascular
diseases, medication taken to prevent a given disease,
HbA1c values, among others (Antunes. et al., 2019).
These criteria were also mapped to possible ICD-9
codes as these enable the augmentation of the dataset
with related clinical notes from the MIMIC-III critical
care database (Johnson et al., 2016).
Then, focusing on the family history component
of clinical notes, a methodology based on heuristics
and NLP was used to extract information regarding
family members, their association to diseases and liv-
ing status. For that, clinical text was firstly prepro-
cessed with the Stanford CoreNLP (Manning et al.,
2014) dependency parsing and co-reference resolu-
tion steps. A lexicon with possible family members
was compiled, and the co-reference graph was used
along with a set of rules to identify family mem-
ber mentions. Disease mentions were identified us-
ing Neji, a biomedical text annotation server (Matos,
2018), with a disease dictionary compiled from the
UMLS. The shortest path in the dependency graph
was used to associate disease mentions to family
members as well as to determine the living status.
Finally, shifting from NER to NEN tasks, a sys-
tem was developed targeting clinical concept normal-
isation. This system uses dictionary matching ap-
proaches, with exact and partial matching mecha-
nisms, combined with word embedding similarity to
normalise relevant entities in clinical notes. With
this approach, identified entities are mapped to their
respective concept unique identifier (CUI) from the
UMLS metathesaurus.
The above mentioned extraction methodologies
were developed under the scope of several research
challenges focused on leveraging clinical text, and
were validated with datasets from the 2018 n2c2
track on cohort selection for clinical trials, 2019
n2c2/OHNLP track on family history extraction and
2019 n2c2/OHNLP track on clinical concept normal-
isation (HMS, 2018; HMS, 2019a; HMS, 2019b).
All relevant patient information resulting from the
combined use of these strategies was organised in a
data structure ready to be supplied to the CDSS during
clinical treatments.
3 COMBINING PROTOCOLS
WITH TEXT DATA
Clinical guidelines have been created to help health
professionals during the treatment of specific patholo-
gies. These guidelines consist of rules and procedures
that should be followed during patient evaluation and
HEALTHINF 2020 - 13th International Conference on Health Informatics
256
treatment, and can be described in paper format with-
out any digital system. However, due to the number
of treatment protocols and their potential complexity,
the use of CDSSs is a valuable resource to simplify
and optimise health care professionals’ tasks.
Therefore, to process clinical guidelines in
CDSSs, it is necessary to convert treatment path-
ways into a digital format. This process can al-
ready be performed with existing methodologies
such as the Guideline Interchange Format version
3 (GLIF3), which is a model designed to repre-
sent shareable computer-interpretable guidelines in
the medical field. This model intends to represent dif-
ferent types of guidelines by specifying them follow-
ing some low-level primitives, which could be applied
in screening, diagnosis, and treatment in primary or
speciality unit care (Boxwala et al., 2004).
Based on the previous description, we followed a
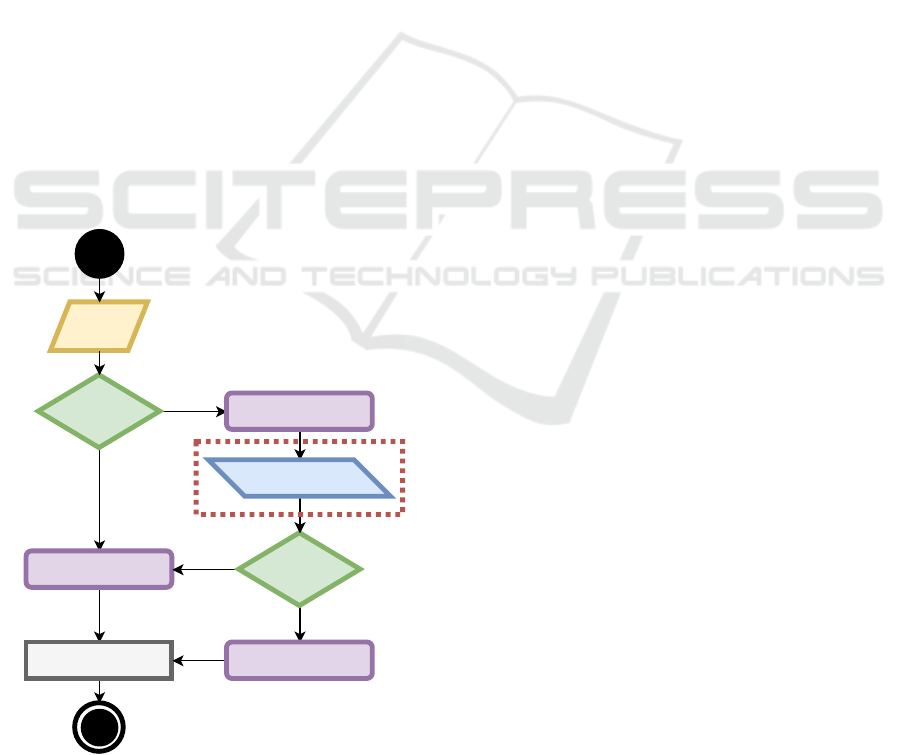
more straightforward approach, as represented in Fig-
ure 1. As observable on the left side of Figure 1,
the process of converting treatment guidelines into a
digital format can be accomplished using only four
elements (Almeida et al., 2018). Action elements
are responsible for providing treatment recommenda-
tions to physicians, which are typically procedures to
be executed during treatment. These recommenda-
tions are influenced by the Decision elements, which
Inquiry
element
Decision
element
Action element
Action element
True
False
Scheduling element
Decision
element
NLP element
Action element
False
True
Figure 1: Protocol components for the digital format repre-
sentation. Surrounded by the red box is the main element
for the proposed methodology.
are the flow changers in the protocol, mainly because
they define the actions that must be done in the treat-
ment based on conditions. Typically, these condi-
tions are constituted by boolean operations that can
be represented in the IF-THEN format. Data used
in those operations is received from the Inquiry ele-
ments that gather patient information before and dur-
ing treatment. Finally, protocols are executed follow-
ing a schedule, which is defined using Scheduling el-
ements.
To improve this approach, a fifth protocol element
(NLP element) was added that was designed to ac-
cess information from clinical notes (marked with a
red box on the right side of Figure 1). This element
identifies information in the patient history that can be
relevant to mention when the system gives a treatment
recommendation. This type of protocol element also
provides suggestions of which would be the informa-
tion for a specific variable, avoiding inquiry elements.
Therefore it simplifies the execution of complex pro-
tocols, mainly because it reduces the number of in-
quiry elements required during treatment.
The use of the NLP element can automatically
provide information to the system that is relevant to
the treatment. A possible scenario can be that of pa-
tients that are taking medication at home and inform
the clinical staff during the admission stage about
their situation. Commonly, the medications being
used and their dosages can have an impact on the
treatments that are prescribed. For instance, type 2 di-
abetic inpatients may need to be medicated daily with
insulin dosages, but depending on the insulin product
used at home, the treatment in the hospital must be
adjusted.
Another scenario is the history of relatives with
certain hereditary diseases. With this information, the
system can alert the physician to inform the patient
about several risks based on their current condition.
For instance, patients with irregular blood pressure
and diabetic family members are more prone to suffer
from diabetes or cardiac diseases in the future. This
information can be provided when measurements are
done combined with the patient family history.
A final possible scenario concerns protocols for
surgery preparation that, depending on the patient
state, can lead to a surgery postponement. The sys-
tem can detect that the patient took a specific med-
ication in past visits and forgot to mention that in a
more recent appointment. However, the medication
described in their history combined with their current
clinical state could indicate that the patient is unable
to withstand surgery.
Enhancing Decision-making Systems with Relevant Patient Information by Leveraging Clinical Notes
257

4 DECISION SUPPORT TOOL
FOR TREATMENT GUIDANCE
The use of CDSSs for treatment guidance has become
an essential piece in continuing care units, as it im-
proves the quality of patient care, helps disease pre-
vention, and supports scientific discoveries. These
systems are typically part of the EHR features, some
standalone solutions also exist. However, all CDSSs
are only able to manage and provide guidance on the
medical treatment when clinical protocols are repre-
sented in a digital format.
Therefore, in our proposal, we used a non-
integrated CDSS to put the proposed methodology
in practice. We used GenericCDSS, an open-source
web-based solution that is prepared to create and ex-
ecute protocols (Almeida and Oliveira, 2019). The
tool provides an editor to create and manage clini-
cal protocols in digital format, which we used and
improved for our goal, and alerts health profession-
als about treatment schedules and when the proto-
col needs to be executed. During protocol execution,
the tool requests information about the patient clini-
cal state and then provides several recommendations
about the treatment in question.
The typical behaviour of this tool begins by pro-
cessing the protocol following the workflow structure
defined by physicians specialised in a given disease.
Then, it considers the clinical information requested
during the treatment, and at the end it provides a
set of treatment suggestions. The original system in-
put required structured information that could be pro-
vided in run-time, however, the same information can
be found in the patient diary (in free text) and could
be aggregated in the protocol workflow during treat-
ments. Therefore, we created a new feature in the
tool to access information extracted from the clinical
reports.
These improvements allowed us to assess the im-
pact of enhancing treatment protocols with relevant
information from clinical notes on treatment recom-
mendations. We observed that patient clinical state
can be complemented with patient history informa-
tion to obtain better treatments. Therefore, by using
the patient history in GenericCDSS, we were able to
enrich the protocols and provide more accurate sug-
gestions.
5 RESULTS AND DISCUSSION
The previous sections described in detail the differ-
ent pieces of the proposed methodology. Figure 2
presents the different stages of this workflow. The
pipeline starts with the patient admission to the health
institution. This is done by a physician after the ini-
tial triage when it is defined that the patient will be
hospitalised. During patient admission the physician
collects patient information such as medication taken
at home, family history information and other clini-
cal information relevant to the admission. This data is
inserted into the EHR and can be stored in structured
format and free-text clinical notes.
After the patient admission and all additional ex-
ams are performed, the patient is hospitalised if nec-
essary. In parallel, the system processes the clinical
notes to supply the CDSS with additional relevant in-
formation. In this stage of the pipeline, notes relevant
to the protocol are made available to the CDSS to op-
timise the process, for when a treatment is required
later on.
The next step of the proposed methodology is the
treatment guidance supported by the CDSS. Here,
the nurse or physician can provide the system with
the necessary measurements taken from the patient,
which are mostly vital signals and variables that
change over time. Clinical information extracted from
the admission notes is also provided to the CDSS, re-
ducing considerably the information requested by the
system and warning for possible associated risks.
5.1 Use Case Overview
The proposed methodology was designed to improve
clinical treatments in a general way without specifi-
cally following the requirements for a given disease.
However, we decided to explore the potential of this
proposal in the diabetes scenario. Our main motiva-
tion was the lack of effective treatments in health in-
stitutions for patients with this disease, mainly due to
insufficient exploitation of decision-making systems.
Hyperglycemia is a health condition characterised
by abnormally high blood glucose, typically caused
by a deficient usage of insulin. Due to the metabolic
derangements of this clinical condition, regular mon-
itoring as well as administration of the most effective
treatment are major concerns for healthcare institu-
tions. Inpatient hyperglycemia is an event that oc-
curs frequently, with a rate of approximately 40% of
all hospitalisations, rendering it as a metric that de-
serves special attention from health care institutions
and public health services (Inzucchi, 2006).
Basal-bolus insulin therapy is the generally rec-
ommended treatment to manage the hyperglycemia
of hospitalised diabetic patients (Umpierrez et al.,
2012). However, this therapy is also related to high
rates of hypoglycemia, reaching values up to 32%,
with the main reason for this occurrence being the
HEALTHINF 2020 - 13th International Conference on Health Informatics
258

Clinical NLP TreatmentsAdmission
Figure 2: Methodology overview, from admission stage to treatment guidance using the GenericCDSS tool.
meal insulin and food intake mismatch (Umpierrez
et al., 2013). As a result, it is possible to recognise
that most of the adverse medication occasions and
blunders happen when insulin is prescribed or admin-
istered. These cases of hypoglycemia in non-intensive
care unit settings are a concern because they have
been associated with increased length of stay, hospi-
tal complications, and mortality (Kim et al., 2014).
Several protocols were proposed for glycemic admin-
istration to reduce these high rates (Neinstein et al.,
2014). However, these procedures are usually avail-
able on paper and difficult to follow, which hinders its
regular use by non-trained professionals. The goal of
the proposed methodology is to reduce this handicap
using a system to support the execution of the clinical
protocols by considering the information in clinical
notes.
5.2 Protocol Discussion
Diabetic patients have several base treatments defined
depending on their clinical state. Different protocols
exist for hospitalised diabetic patients, hypoglycemic
patients, diabetic surgical patients, diabetes in preg-
nant patients, ketoacidosis and hyperosmolar hyper-
glycemia syndrome in adults and children, among
other more specific protocols. However, to describe
our methodology more in-depth, we describe the im-
pact of the most common protocol, which is used in
hospitalised diabetic patients. This protocol is ap-
plied in type 2 diabetic inpatients and has two differ-
ent stages: the admission moment where the medica-
tion taken at home is converted to the one used in the
hospital; and then during the patient’s stay after the
initial set up.
Insulin dosages of patients are defined as Total
Daily Dosage (TDD) and calculated based on the pa-
tients’ information. This value is used as a reference
for the basal or long-acting insulin dosages. However,
when the patient is taking insulin before the admis-
sion, i.e. daily dosages at home, this information must
be considered and protocols may need to be changed.
Typically, this information is provided during the ad-
mission stage and kept in clinical notes, which end up
not being adequately considered in the protocols.
Insulin taken by patients at home can contain a
mix of long and fast-acting insulin in the same drug,
and the percentages of each vary depending on the
drug. Therefore, interpreting which drug and dosages
are being taken during the day is essential to optimise
TDD calculation. Table 1 presents the most common
insulin products that patients use at home. LA Insulin
and SA Insulin columns represent the percentages of
long and short-acting insulin in each product, respec-
tively. With this information, it is possible to deter-
mine the total of both insulins taken by the patient
Table 1: Percentage of long (LA) and short-acting (SA) in-
sulins from the most common insulin products used by pa-
tients in a domestic setting.
Insulin products LA Insulin SA Insulin
Mixtard 30 Penfill 70% 30%
Insuman Comb 25 75% 25%
Humulin M3 70% 30%
NovoMix 30 70% 30%
Humalog Mix 25 75% 25%
Humalog MIx 50 50% 50%
Enhancing Decision-making Systems with Relevant Patient Information by Leveraging Clinical Notes
259

and split them as the protocol recommends.
In order to simplify the description of the system
execution, the following example is provided:
Example: A patient is taking Mixtard 30 Penfill at
home, 30 units before breakfast and 15 units be-
fore dinner. This insulin product contains 70% of
long-acting and 30% of short-acting insulin as it is
described in Table 1. Therefore, this patient has a
TDD of 45 unit, more precisely 31,5 and 13,5 units
of long and short-acting insulin, respectively. Based
on the protocol, this patient needs to reduce the to-
tal amount of administered insulin, taking only
2
3
of
31,5 units of long-acting insulin at breakfast during
the internment, and
2
3
of the remaining daily dosage
in short-acting insulin.
Moreover, this information is spread over product
manuals, patient diaries and clinical guidelines. The
system can gather all this information, and following
the provided example, it can also recognise that this
patient is taking the Mixtard 30 Penfill at home along
with the respective dosages. This section describes
some relevant aspects that the proposed methodology
solves. However, in addition to what has been de-
scribed, the protocol also considers patient insulin re-
sistance and the different sliding scales present in the
protocol must be adjusted depending on patient re-
sponses and their plasma glucose values.
5.3 Validation
Methodology validation was performed in a con-
trolled environment using a public dataset. The 2014
n2c2 track 2 on de-identification and heart disease
risk factors challenge consisted on identifying risk
factors for heart disease over time. The dataset used
in this challenge contained 1,304 clinical narratives
from 296 diabetic patients (2-5 records per patient).
These narratives contain information about heart dis-
ease risk factors such as high blood pressure and
cholesterol levels (Stubbs et al., 2015).
Therefore, we selected 25 patients whose notes
contained more information to create some difficul-
ties in the NLP methodologies (e.g. more redundant
and less concise information). Additionally, we ran-
domly added sentences indicating that the patient is
taking insulin products at home, and following some
criteria, we also added information about dosages that
they administered during the day (before breakfast,
lunch, dinner, meals or bedtime).
Then, we manually simulated the physician work
and protocol execution during treatments. Altogether,
the proposed methodology produced positive results.
However, we noticed that the system faced some com-
plications in the clinical notes analysis stage. The
system had issues when randomly inserted sentences
were too complex, referencing past medication that
is currently not being taken by the patient. However,
we solved this issue by giving the possibility to the
physician to consult the clinical report in run-time,
identifying which were the sentences that originated
that recommendation.
6 CONCLUSION
The secondary use of clinical notes is a subject that
has been under study over the past years. In the
medical field, the use of systems for decision-making
and treatment guidance is a subject of much research.
Moreover, we detected an opportunity to enrich med-
ical treatments by combining both topics with the
objective of reducing existing gaps in treatment pre-
scriptions.
The proposed methodology was integrated and
validated with an open-source CDSS due to its au-
tonomy and ease of development. However, the goal
was to demonstrate the positive impact of combining
these subjects and define a supportive approach. The
methodology can be applied in the different decision-
making features existent in the EHR systems available
in the market.
As future work, and to better understand possi-
ble improvements, we expect to apply this method-
ology with different diseases beyond diabetes. Addi-
tionally, to validate the protocol recommendations, as
well as the extracted clinical concepts, we intend to
incorporate a classification feature. With this feature,
the physician can evaluate the accuracy of the system,
which may thus help to increase the performance and
discover possible gaps to be addressed.
ACKNOWLEDGEMENTS
This work has received support from the EU/EFPIA
Innovative Medicines Initiative 2 Joint Undertaking
under grant agreement No 806968 and from the
NETDIAMOND project (POCI-01-0145-FEDER-
016385), co-funded by Centro 2020 program, Por-
tugal 2020, European Union. Jo
˜
ao Figueira Silva
and Jo
˜
ao Rafael Almeida are funded by the FCT
- Foundation for Science and Technology (national
funds) under the grants PD/BD/142878/2018 and
SFRH/BD/147837/2019 respectively.
HEALTHINF 2020 - 13th International Conference on Health Informatics
260

REFERENCES
Almeida, J. R., Guimar
˜
aes, J., and Oliveira, J. L. (2018).
Simplifying the digitization of clinical protocols for
diabetes management. In 2018 IEEE 31st Interna-
tional Symposium on Computer-Based Medical Sys-
tems (CBMS), pages 176–181. IEEE.
Almeida, J. R. and Oliveira, J. L. (2019). GenericCDSS-
a generic clinical decision support system. In 2019
IEEE 32nd International Symposium on Computer-
Based Medical Systems (CBMS), pages 186–191.
IEEE.
Antunes., R., Silva., J. F., Pereira., A., and Matos., S.
(2019). Rule-based and machine learning hybrid sys-
tem for patient cohort selection. In Proceedings of
the 12th International Joint Conference on Biomed-
ical Engineering Systems and Technologies - Volume
2: HEALTHINF,, pages 59–67. INSTICC, SciTePress.
Boxwala, A. A., Peleg, M., Tu, S., Ogunyemi, O., Zeng,
Q. T., Wang, D., Patel, V. L., Greenes, R. A., and
Shortliffe, E. H. (2004). Glif3: a representation for-
mat for sharable computer-interpretable clinical prac-
tice guidelines. Journal of biomedical informatics,
37(3):147–161.
Cohen, R., Elhadad, M., and Elhadad, N. (2013). Re-
dundancy in electronic health record corpora: analy-
sis, impact on text mining performance and mitigation
strategies. BMC Bioinformatics, 14(10).
Costa, C. M. A. (2004). Concepc¸
˜
ao, desenvolvimento e
avaliac¸
˜
ao de um modelo integrado de acesso a reg-
istos cl
´
ınicos electr
´
onicos. PhD thesis, University of
Aveiro.
Ferr
˜
ao, J. C., Janela, F., Oliveira, M. D., and Martins, H.
M. G. (2013). Using structured EHR data and SVM
to support ICD-9-CM coding. In 2013 IEEE Interna-
tional Conference on Healthcare Informatics, pages
511–516, Philadelphia, PA, USA. IEEE.
Ferr
˜
ao, J. C., Oliveira, M. D., Janela, F., and Martins, H.
M. G. (2016). Preprocessing structured clinical data
for predictive modeling and decision support. Applied
Clinical Informatics, 07(04):1135–1153.
HMS (2018). 2018 n2c2 - Track 1: Cohort Selection for
Clinical Trials.
HMS (2019a). 2019 n2c2 Shared-Task and Workshop,
Track2: n2c2/OHNLP Track on Family History Ex-
traction.
HMS (2019b). 2019 n2c2 Shared-Task and Workshop,
Track3: n2c2/UMass Track on Clinical Concept Nor-
malization.
Hripcsak, G. and Albers, D. J. (2012). Next-generation
phenotyping of electronic health records. Journal
of the American Medical Informatics Association,
20(1):117–121.
Inzucchi, S. E. (2006). Management of hyperglycemia in
the hospital setting. New England journal of medicine,
355(18):1903–1911.
Jensen, K., Soguero-Ruiz, C., Oyvind Mikalsen, K., Lind-
setmo, R.-O., Kouskoumvekaki, I., Girolami, M.,
Olav Skrovseth, S., and Augestad, K. M. (2017).
Analysis of free text in electronic health records for
identification of cancer patient trajectories. Scientific
Reports, 7(46226).
Johnson, A. E. W., Pollard, T. J., Shen, L., Lehman, L.-
w. H., Feng, M., Ghassemi, M., Moody, B., Szolovits,
P., Anthony Celi, L., and Mark, R. G. (2016). MIMIC-
III, a freely accessible critical care database. Scientific
Data, 3.
Katehakis, D. G. and Tsiknakis, M. (2006). Electronic
health record. In Wiley Encyclopedia of Biomedical
Engineering. Wiley.
Kim, Y., Rajan, K. B., Sims, S. A., Wroblewski, K. E., and
Reutrakul, S. (2014). Impact of glycemic variabil-
ity and hypoglycemia on adverse hospital outcomes
in non-critically ill patients. Diabetes research and
clinical practice, 103(3):437–443.
Manning, C. D., Surdeanu, M., Bauer, J., Finkel, J.,
Bethard, S. J., and McClosky, D. (2014). The Stan-
ford CoreNLP natural language processing toolkit. In
Association for Computational Linguistics (ACL) Sys-
tem Demonstrations, pages 55–60.
Matos, S. (2018). Configurable web-services for biomedi-
cal document annotation. Journal of cheminformatics,
10(1):68.
Miotto, R., Wang, F., Wang, S., Jiang, X., and Dudley, J. T.
(2017). Deep learning for healthcare: review, oppor-
tunities and challenges. Briefings in Bioinformatics.
NCCIH (2017). Clinical Practice Guidelines.
Neinstein, A., MacMaster, H. W., Sullivan, M. M., and
Rushakoff, R. (2014). A detailed description of the
implementation of inpatient insulin orders with a com-
mercial electronic health record system. Journal of
diabetes science and technology, 8(4):641–651.
Nelson, S. J., Zeng, K., Kilbourne, J., Powell, T., and
Moore, R. (2011). Normalized names for clinical
drugs: RxNorm at 6 years. Journal of the American
Medical Informatics Association, 18(4):441.
Neustein, A., Imambi, S. S., Rodrigues, M., Teixeira, A.,
and Ferreira, L. (2014). Application of text mining to
biomedical knowledge extraction: analyzing clinical
narratives and medical literature. In Text Mining of
Web-based Medical Content, pages 3–32. De Gruyter.
O’Connor, P. J., Sperl-Hillen, J. M., Rush, W. A., Johnson,
P. E., Amundson, G. H., Asche, S. E., Ekstrom, H. L.,
and Gilmer, T. P. (2011). Impact of electronic health
record clinical decision support on diabetes care: a
randomized trial. Annals of family medicine, 9(1):12–
21.
Pivovarov, R. and Elhadad, N. (2015). Automated meth-
ods for the summarization of electronic health records.
Journal of the American Medical Informatics Associ-
ation, 22(5):938–947.
Singh, H., Giardina, T. D., Meyer, A. N. D., Forjuoh, S. N.,
Reis, M. D., and Thomas, E. J. (2013). Types and
origins of diagnostic errors in primary care settings.
JAMA Internal Medicine, 173(6):418–425.
Stearns, M. Q., Price, C., Kent A. Spackman, and Wang,
A. Y. (2001). SNOMED clinical terms: overview of
the development process and project status. In Pro-
ceedings of the AMIA Symposium, pages 662–666,
Enhancing Decision-making Systems with Relevant Patient Information by Leveraging Clinical Notes
261

Washington, DC, USA. American Medical Informat-
ics Association.
Stewart, W. F., Shah, N. R., Selna, M. J., Paulus, R. A.,
and Walker, J. M. (2007). Bridging the inferential
gap: The electronic health record and clinical evi-
dence. Health Affairs, 26(Supplement 1):w181–w191.
Stubbs, A., Kotfila, C., Xu, H., and Uzuner,
¨
O. (2015).
Identifying risk factors for heart disease over time:
Overview of 2014 i2b2/uthealth shared task track 2.
Journal of biomedical informatics, 58:S67–S77.
Umpierrez, G. E., Gianchandani, R., Smiley, D., Jacobs, S.,
Wesorick, D. H., Newton, C., Farrokhi, F., Peng, L.,
Reyes, D., Lathkar-Pradhan, S., et al. (2013). Safety
and efficacy of sitagliptin therapy for the inpatient
management of general medicine and surgery patients
with type 2 diabetes: a pilot, randomized, controlled
study. Diabetes Care, 36(11):3430–3435.
Umpierrez, G. E., Hellman, R., Korytkowski, M. T., Kosi-
borod, M., Maynard, G. A., Montori, V. M., Seley,
J. J., and Van den Berghe, G. (2012). Management of
hyperglycemia in hospitalized patients in non-critical
care setting: an endocrine society clinical practice
guideline. The Journal of Clinical Endocrinology &
Metabolism, 97(1):16–38.
WHO (2018). World Health Organization: International
classification of diseases, 11th Revision (ICD-11).
Wu, J., Roy, J., and Stewart, W. F. (2010). Prediction mod-
eling using EHR data: challenges, strategies, and a
comparison of machine learning approaches. Medical
Care, 48(6):S106–S113.
HEALTHINF 2020 - 13th International Conference on Health Informatics
262
