
Process Mining of Disease Trajectories: A Feasibility Study
Guntur P. Kusuma
1,2 a
, Samantha Sykes
3b
, Ciarán McInerney
1c
and Owen Johnson
1d
1
School of Computing, University of Leeds, Leeds, U.K.
2
School of Applied Science, Telkom University, Bandung, Indonesia
3
School of Medicine, University of Leeds, Leeds, U.K.
Keywords: Disease Trajectories, Process Mining, Electronic Health Records.
Abstract: Modelling patient disease trajectories from evidence in electronic health records could help clinicians and
medical researchers develop a better understanding of the progression of diseases within target populations.
Process mining provides a set of well-established tools and techniques that have been used to mine electronic
health record data to understand healthcare care pathways. In this paper we explore the feasibility for using a
process mining methodology and toolset to automate the identification of disease trajectory models. We
created synthetic electronic health record data based on a published disease trajectory model and developed a
series of event log transformations to reproduce the disease trajectory model using standard process mining
tools. Our approach will make it easier to produce disease trajectory models from routine health data.
1 INTRODUCTION
Diseases occur at various points during a person’s
life-course and impact on health, lifestyle, quality of
life, morbidity and mortality. Disease can be seen as
a pathological process that requires judgement from a
clinician to objectify its occurrence (Boyd, 2000).
The record of disease occurrences over time become
the “footprints” that can tell the story of how diseases
have progressed for each individual. This type of
historic patient information is vital evidence that can
help clinicians to diagnosis appropriately and to
decide on appropriate interventions (Muhrer, 2014;
World Health Organization, 2016). More generally,
medical research recognises common patterns of
diseases where one disease is often found to precede
others. These commonly-found patterns for disease
progression are sometimes referred to as disease
trajectories (A. B. Jensen et al., 2014).
The temporal record of diseases can be observed
within electronic healthcare records (EHR) and can
be used to understand the occurrence and behaviour
of diseases. The trajectories of diseases can be
identified by observing the sequence of disease
a
https://orcid.org/0000-0002-0208-125X
b
https://orcid.org/0000-0001-7098-3928
c
https://orcid.org/0000-0001-7620-7110
d
https://orcid.org/0000-0003-3998-541X
diagnoses and the time intervals between them.
Investigating disease trajectories has the potential to
provide personalised medical treatment (P. B. Jensen
et al., 2012) and to understand the potential cause-
and-effect association between diseases (Hanauer &
Ramakrishnan, 2013; Rothman & Greenland, 2005).
In A. B. Jensen et al.'s (2014) widely cited work,
the authors produced a number of disease trajectories
based on EHR data from the population of Denmark.
Disease trajectories were defined as the time-ordered
sequence of diagnoses observed in the patients. An
example of a disease trajectory model is presented in
Figure 1. The model consist of nodes representing the
diseases and directed arcs representing the common
trajectories between diseases with the thickness of the
arcs representing the relative number of patients.
In many countries, healthcare providers are now
supported by EHR systems containing episodic and
longitudinal data of a patient’s medical history,
diagnosis and treatment (Hemingway et al., 2018).
The World Health Organisation had introduced
standards for medical records (World Health
Organization, 2002) and the European Medicines
Agency suggests that clinicians use of medical record
information is good clinical practice (European
Kusuma, G., Sykes, S., McInerney, C. and Johnson, O.
Process Mining of Disease Trajectories: A Feasibility Study.
DOI: 10.5220/0009166607050712
In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2020) - Volume 5: HEALTHINF, pages 705-712
ISBN: 978-989-758-398-8; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
705
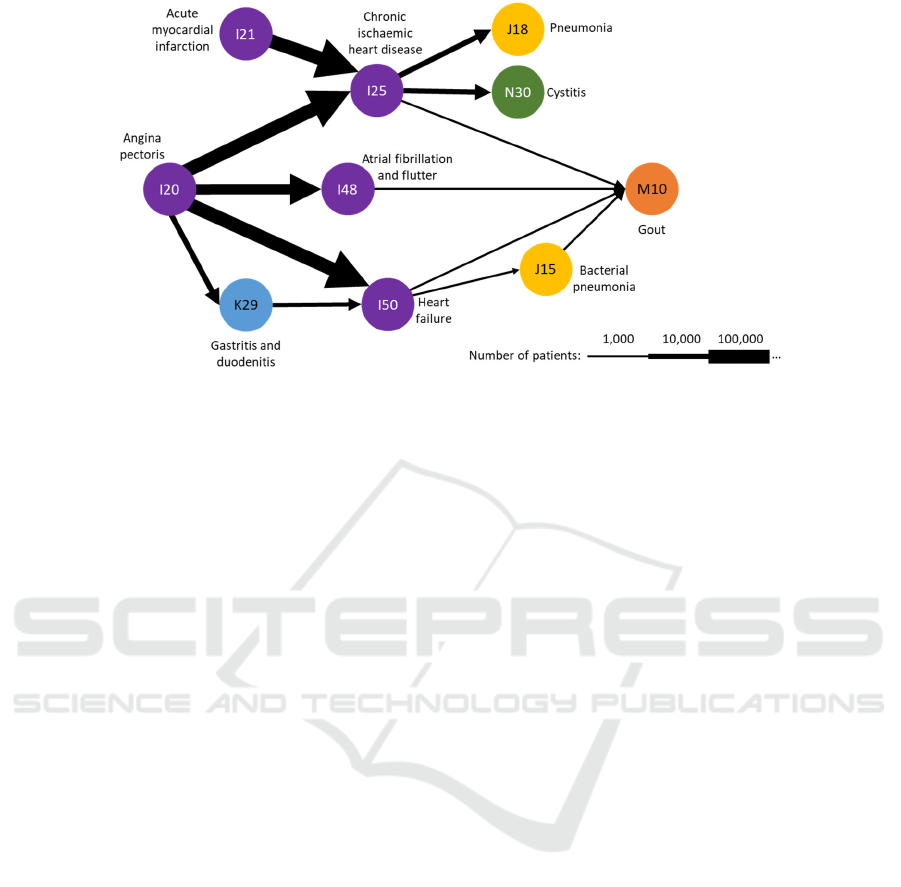
Figure 1: Example of a disease trajectory model, adapted from Figure 4.b of A.B. Jensen et al. (2014).
Medicines Agency, 2002). In some countries, the
initial motivation of developing EHR was for billing
purpose but in many countries EHR use is now
comprehensive and includes records of disease
diagnoses, with use being expanded for clinical and
research purposes (Casey et al., 2016). The World
Health Organisation provides the International
Classification of Diseases (ICD) with the purpose of
standardising the coding of diseases within EHR to
support evidence-based decision making, sharing and
comparing of health information, monitoring the
incidence of disease, and helping healthcare
organisations in managing disease related billing
(WHO, 2019). Disease codes in EHR are commonly
used, but there are known data quality issues. For
example, in the ICD-10 standard, the code I20 is used
for angina pectoris. This structured encoding
framework has facilitated the construction of disease
trajectories using EHRs (A. B. Jensen et al., 2014)
and for process mining of care pathways (Rojas et al.,
2016).
Process mining is a data mining approach that
examines temporal and sequential data to analyse
processes including the discovery of process models,
conformance checking and process enhancement
(van der Aalst, 2011). Process mining provides a
holistic view, end-to-end analysis, and generates
easy-to-read models and simulations (van der Aalst,
2011). The input of process mining is an event log
detailing who did what and when. More formally, the
event log is a collection of time stamped events
containing at least a case, an activity, and a
timestamp. The output of process mining is often
graphical, producing visual models of processes and
pathways. Conformance of event logs to expected
models can be measured and a process modelling
project may involve multiple iterations of data
extraction, transformation, modelling, measuring and
refinement to construct valuable process models and
process insights.
In our review of the literature, we found that
process mining techniques have not yet been utilised
to extract patients’ disease trajectory models from
health data despite the many similarities between
process models and disease-trajectory models. This is
despite the data required for such an approach often
being available within EHRs.
The aim of this study, therefore, was to assess the
feasibility of using process mining methods and tools
with EHR data to construct disease trajectory models.
In our study, we simulated a scenario from A. B.
Jensen et al.’s trajectories that were centred on
chronic ischaemic heart disease (the example in
Figure 1). Our hypothesis was that by treating the
entry of a disease code in the EHR as the equivalent
of an activity in process mining we could exploit the
rich toolset of process mining for the mining of
disease trajectories.
2 BACKGROUND
2.1 Disease Trajectory
2.1.1 Definitions
Multiple definitions of disease trajectory have been
proposed. Murray et al (2005) defined a disease
trajectory as the progressiveness of physical health
deterioration over time. They described three types of
trajectories: the short period, where the decline of
physical health happens within a few months or a few
HEALTHINF 2020 - 13th International Conference on Health Informatics
706

years; long-term limitations, where the decline
happens between 2 to 5 years; and prolonged
dwindling, where the decline happens in 6 to 8 years.
A disease trajectory is also defined as the progression
of a specific disease by observing a clinical measure
of the severity of a disease. An example of work
which follow this definition is a study observing the
progression of chronic kidney disease by measuring
the estimated glomerular filtration rate (eGFR)
(Sumida & Kovesdy, 2017). Finally, A. B. Jensen et
al. (2014) proposed a definition of disease trajectory
as the sequence of diseases that are ordered by the
time of the occurrence. Our work follows this
definition from A. B. Jensen et al. We note that this
definition of disease trajectory is similar to definitions
of patient trajectories found in other literature
(Pavalko, 1997; Pescosolido, 2013). Specifically, we
take the first occurrence of a new disease code as it is
recorded in the EHR.
2.1.2 Disease Trajectory Modelling
Disease trajectory models are typically represented as
acyclic graphs, where each node represents a disease
and each directed arc represents a progression from
one disease to another. Reducing the graph to remove
cycles presents a stronger representation of a general
trajectory (progression) of diseases at the cost of
simplifying the reality of complex real-world cases.
Constructing disease trajectories from EHR data
is a challenging process. Various techniques have
been used to investigate and model disease
trajectories, including a data-driven approach
(Glicksberg et al., 2016; Hanauer & Ramakrishnan,
2013; Hidalgo et al., 2009; A. B. Jensen et al., 2014),
data-mining (Giannoula et al., 2018; Ji et al., 2016;
Wang et al., 2014), network-based (Steinhaeuser &
Chawla, 2009), free-text analysis (K. Jensen et al.,
2017), and more recently by implementing a deep-
learning techniques (Beaulieu-Jones et al., 2018;
Futoma et al., 2015; Pham et al., 2017).
The disease trajectories modelled by A. B. Jensen
et al. (2014) were constructed by joining overlapping
pairs of diagnoses (bi-grams) to form longer
trajectory chains. For example, the A. B. Jensen et al.
method might have identified a bi-gram pair of
diseases I21 (acute myocardial infarction) I25
(chronic ischaemic heart disease), where I21 is a
disease code that is recorded against a patient some
time before I25. To construct a longer trajectory, A.
B. Jensen et al. combined multiple bi-grams such as
I21I25 and I25N30 to form a longer sequence of
three diagnoses, I21I25N30. Although appealing
in its simplicity, the concatenated trajectory might not
be evidenced in any patient’s record nor does it
consider the conditional likelihood of the latter bi-
gram given the former. No standard tools for disease
trajectory modelling are evident in the literature. In
contrast, process mining methods and tool are well
established and have the potential to efficiently define
such longer trajectories and also provide diagnostics
to evaluate representativeness.
2.2 Process Mining in Healthcare
Process mining in healthcare is now well established
with strong support from commercial and open-
source tools, for example ProM Framework (Process
Mining Group, 2010), Celonis (Celonis GmbH,
2019), or Disco (Fluxicon BV, 2019). Process mining
also boasts a growing body of literature (Rojas et al.,
2016) and an international research community
Process Oriented Data Science for Healthcare
(“PODS4H,” 2019).
The implementation of process mining in
healthcare has been proven applicable to analyse care
pathways (Mans et al., 2015; Rojas et al., 2016).
Process mining is commonly used for mining the
sequence of activities but the time between activities
can also be analysed. Process mining has been used
in cancer (Kurniati et al., 2016), cardiovascular
disease (Kusuma et al., 2017), dentistry (Fox et al.,
2018; R S Mans et al., 2012), frailty (Farid et al.,
2019), sepsis (Mannhardt & Blinde, 2017), and in
primary care (Williams et al., 2018). Process mining
is suitable for answering frequently posed questions
by extracting information from an EHR (Mans et al.,
2013). Unlike disease trajectory models which use the
first occurrence of a disease, process mining is able to
model multiple simultaneous and recurrent activity.
3 MATERIALS AND METHOD
The goal of this exploratory data-driven study is to
explore the feasibility of producing disease
trajectories using a process mining approach. This
study uses synthetic data and has been made available
on GitHub (Kusuma et al., 2019) and therefore useful
for reproducibility. We examined A. B. Jensen et al.’s
(2014) trajectories to simulate a set of EHR data that
reflected a subset of the disease trajectories shown in
Figure 1. Table 1 summarises the variables simulated
that contained 50 patients with 146 diagnosis codes
from 10 distinct diagnosis, using the first three
characters of ICD-10 format. We treat each patient as
a case and use the diagnosis codes in place of the
standard process mining event-log activity names.
Process Mining of Disease Trajectories: A Feasibility Study
707

The extracted data was formatted following the
structure of a process mining event log, see Figure
2(a).
The synthetic event log was created by
constructing event data for each case (patient) with
the minimum of two events to form a diagnoses pair
(D1D2). We created the time of D1 earlier than the
time of D2, to ensure D1 occurred as an antecedent of
D2. We created some cases with 3 or more events to
represent a sequence of diagnoses D1D2D3…
which follow trajectories recognised by A. B. Jensen
et al. as a collection of diagnoses pairs (D1D2,
D2D3, D3…). To make the event log even look
similar with the real-life EHR, then we added some
repeating events as noise in the event log.
Table 1: The sources of required data from the synthetic
dataset.
Variables Data Field name
Case
identifier
Patient identifier subject_id
Event Diagnosis code diagnosis
Time
stamps
Time when the
diagnosis was
recorded
admittime
For conducting the process mining experiments,
we followed the Process Mining Project
Methodology, PM
2
(van Eck et al., 2015). Our use of
the PM
2
method is summarised below.
In Stage-1: Planning, research questions were
identified from a literature review and confirmed by
the project team during study planning. The team
included a clinician, epidemiologist and computer
scientists.
In Stage-2: Extraction, we defined the scope of
the extraction by determining the granularity of the
data, the time period, and selected the related
attributes. We used the synthetic dataset as the input
for creating an event log in the next stage. Only the
first of any recurring diagnosis codes for each patient
were used to create an acyclic, disease-trajectory
model. We treated each patient as a case and used the
diagnosis codes in place of the standard process
mining event-log activity names.
In Stage-3: Data Processing, we followed the
activities to create the event log as defined in PM
2
by
creating views, filtering logs, and event log
transformation into pair log, the collection of event
data in the form of pairs. The filtering was done by
(a)
(b)
(c)
Figure 2: The filtering and transformation steps of event
log: (a) the extracted event log from simulated data; (b) the
recurrent diagnoses for each patient were filtered; and (c)
the pair log with duplicate diagnoses removed.
removing the recurring diagnoses for each patient and
keep the first occurrence.
In Stage-4: Mining and Analysis, the event log
was analysed by applying process discovery and
conformance-checking methods using process
mining tools Disco and plugins in the ProM
Framework. The discovered model was evaluated
using the measures of fitness, precision and
generalisability (van der Aalst, 2011). Fitness is a
measure of how many traces in the event log can be
replayed through the discovered model. Precision is a
measure of how much the discovered model over
estimates the traces in the event log; Low precision,
or under-fitting, indicates that the model can
represent traces that never occur in the event log,
while high precision, or over-fitting, indicates that the
model can represent the traces in the event log, only.
Generalisation is a measure of how often activities in
the model occur in the event log.
Disease trajectories were ‘discovered’ using
Disco, conformance-checking and the measurement
of precision and generalisation were done in ProM
Framework using the plugin Replay a Log on Petri
Subject_id Diagnosis Time
3 I21 01/01/2100
3 I25 02/03/2100
4 I21 21/02/2100
4 I25 14/06/2100
6 I21 01/01/2100
6 I25 02/01/2100
6 J18 03/01/2100
Subj ect _i d Ant ecedent Subsequent T i me1 T i me2
3 I21 I25 01/01/2100 02/03/2100
3 I25 I25 02/03/2100 12/05/2100
4 I21 I25 21/02/2100 14/06/2100
6 I21 I25 01/01/2100 02/01/2100
6 I25 J18 02/01/2100 03/01/2100
HEALTHINF 2020 - 13th International Conference on Health Informatics
708
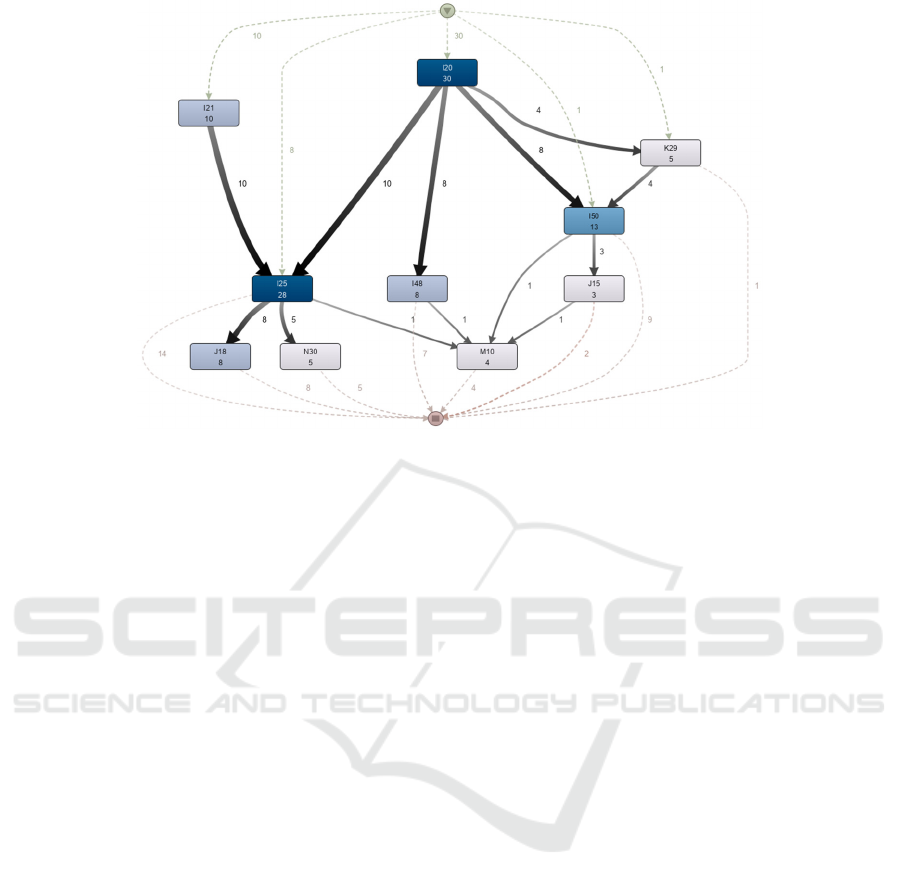
Figure 3: Disease trajectory model using process mining (the model generated from Disco).
Net for Conformance Analysis (Ardiansyah, 2012)
and Measure Precision/Generalization respectively.
Because our discovered process model and the
trajectories of A. B. Jensen et al. (2014) can both be
considered directed graphs, we were able to assess
agreement between them by checking if both were
isomorphic. Two graphs are said to be isomorphic if
they have the same (“iso-“) structure (“-morph”),
where structure is defined (Goldberg, 2003). In our
case, by the count of diseases, the count of disease-
pair connections and pattern of connected nodes and
arcs is identical. More formally, two graphs are
isomorphic if there exists a mapping that is a bijective
function that preserves the branch structure of the
graphs. We applied Cordella et al.'s (2001) method to
check for isomorphism using the NetworkX Python
library.
All processing other than discovery and
conference checking were conducted in Python
through Jupyter Notebook (Kluyver et al., 2016).
4 RESULTS
By following the PM
2
, the result of each stage is
described. In Stage-1,we aimed to mine the disease
trajectory agnostically without any specific selection
of diagnosis and time window. We defined the main
research question as Can disease trajectories be
identified from an EHR, using a process mining
approach?
In Stage-2, the synthetic data was formatted to
follow the structure of a process mining event log
(Figure 2a). In Stage-3, the recurring diagnoses for
each patient were filtered out and we kept the first
occurrence. The filtering step reduce the total number
of events from 126 to 117 for the next stage.
In Stage-4, using the process-mining tool Disco a
disease trajectory model was discovered (Figure 3).
The trajectory model shows the same characteristics
as the sub-trajectory of A. B. Jensen et al. (2014) in
Figure 1. Both models have 10 nodes and 13 arcs
including the thickness representation despite the
difference on the scale and the addition of the case
frequency (which is represented in Figure 3 by darker
shades of nodes colour for more frequently
occurring). The application of an isomorphic checker
(following Cordella et al., 2001) determined that the
two graphs could be considered isomorphic.
One benefit of our process-mining approach is
that the temporal information about the time elapsed
between disease occurrences is preserved and can be
examined using standard tools. For example, the
median duration between events can be displayed by
process mining software such as Disco and ProM.
The preservation of temporal data is a significant
improvement over the simple models of disease
sequence produced by the trajectory method of A, B.
Jensen et al. (2014). Further, we can use process
mining tools to measure the quality of the discovered
model. In our experiment: the fitness value is 0.961,
precision 0.79 and generalisation 0.963.
Process Mining of Disease Trajectories: A Feasibility Study
709

5 DISCUSSION
In this work we sought to assess the feasibility of
process mining to identify disease trajectories in a
simulated dataset representative of a published
disease trajectory. We applied the Process Mining
Project Methodology PM
2
to discover and
conformance check a process model that was
qualitatively similar to the sub-graph of trajectories
from A. B. Jensen et al. (2014). In combination with
good estimates for fitness, precision and
generalisability, we conclude that process mining is a
feasible approach for identifying disease trajectories
using data similar to that found in EHRs.
The originality of this study is around the method
where disease trajectories can be discovered using
process mining techniques. We further suggest that
our process mining method is an improvement on the
disease trajectory method of A. B. Jensen et al.
(2014). The high fitness, precision and generalisation
scores permit us to make the follow statement:
process models discovered using our methods on data
similar to ours would permit the behaviour seen in the
event log, would be precise enough to not allow
behaviour unrelated to what was seen in the event log,
and would be general enough to reproduce future
behaviour of the trajectories. This is in contrast to the
concatenation of bi-grams approach of A. B. Jensen
et al. (2014), which implies the existence of long
trajectories of diseases based on combining direct
disease pairs, end to end, without being in a position
to validate from the data.
By default, process mining methods also provide
additional information not found in the purely-
sequential output provided by A. B. Jensen et al.
(2014). The output of some process mining
algorithms present the durations and counts of cases
that follow the trajectories. For example, the median
duration among the patients is one day, while the
longest duration is 150 days. Following our approach
disease trajectory models can be automatically
visualised in keeping with the graphical and
exploratory ethos of the process mining. A major
benefit of process mining is that its application is
supported by commercial and open-source software
(Fluxicon BV, 2019; Process Mining Group, 2010), a
healthcare-specific literature base (Rojas, Munoz-
Gama, Sepúlveda, & Capurro, 2016) and an
international research community.
A limitation identified by our particular
implementation of PM
2
was the decision to include
only the first occurrence of the primary diagnoses as
the main event. This step promotes a representational
bias that cannot be avoided in the study of model
discovery. It is possible that different trajectories
exist for recurrent diagnoses but we constrained our
investigations for the purpose of demonstrating
feasibility.
For future work, process mining should be
applied to real-life EHRs to identify disease
trajectories. Using high-volume datasets is necessary
to evaluate could evaluate the scalability of the
method. The role of a clinical domain expert could
help to limit the number of variables of interest to
build a more tightly-focussed model examining
specific disease trajectory patterns and this approach
would also make the discovery task more efficient.
Despite the limitations, our approach has
demonstrated that we can use process mining tools to
mine disease trajectories and has opened up an
interesting field for further work.
6 CONCLUSION
In this paper we have demonstrated the feasibility of
mining of disease trajectories using process mining.
The mining was conducted using a synthetic dataset
which is similar to the data available from many EHR
systems. Our study included the use of the PM
2
framework to mine a representative disease
trajectories model from EHR format data and
addressed several quality dimension standards.
This feasibility study opens opportunities for
future works in implementation the technique using
population sized EHR data. The application of
different discovery algorithms to mine the disease
trajectory model may improve the conformance
measurement and the disease trajectory model’s
quality dimension. Our approach will be of interest to
the wide range of multi-disciplinary researchers
interested in exploring healthcare record data for
identifying disease trajectories to improve medicine
and health.
ACKNOWLEDGEMENTS
This research is part of the first author’s PhD study
funded by the Indonesia Endowment Fund for
Education (LPDP). The research was supported by
the National Institute for Health Research (NIHR)
Yorkshire and Humber Patient Safety Translational
Research Centre (NIHR YH PSTRC). The views
expressed in this article are those of the author(s) and
not necessarily those of the NHS, the NIHR or the
Department of Health and Social Care.
HEALTHINF 2020 - 13th International Conference on Health Informatics
710

REFERENCES
Ardiansyah, A. (2012). Replay a Log on Petri Net for
Conformance Analysis-plugin.pdf.
Beaulieu-Jones, B. K., Orzechowski, P., & Moore, J. H.
(2018). Mapping patient trajectories using longitudinal
extraction and deep learning in the MIMIC-III critical
care database. Pacific Symposium on Biocomputing,
0(212669), 123–132. https://doi.org/10.1142/978981
3235533_0012
Boyd, K. M. (2000). Disease, illness, sickness, health,
healing and wholeness: Exploring some elusive
concepts. Medical Humanities, 26(1), 9–17.
https://doi.org/10.1136/mh.26.1.9
Casey, J. A., Schwartz, B. S., Stewart, W. F., & Adler, N.
E. (2016). Using Electronic Health Records for
Population Health Research: A Review of Methods and
Applications. Annual Review of Public Health, 37(1),
61–81. https://doi.org/10.1146/annurev-publhealth-
032315-021353
Celonis GmbH. (2019). Celonis. Retrieved from
https://www.celonis.com/
Cordella, L. P., Foggia, P., Sansone, C., & Vento, M.
(2001). An improved algorithm for matching large
graphs. 3rd IAPR-TC15 Workshop on Graph-Based
Representations in Pattern Recognition, 149–159.
Retrieved from https://citeseerx.ist.psu.edu/viewdoc/
summary?doi=10.1.1.101.5342
European Medicines Agency. (2002). Note for the
Guidance on Good Clinical Practice. Retrieved from
https://www.ema.europa.eu/en/documents/scientific-
guideline/ich-e6-r1-guideline-good-clinical-practice_
en.pdf
Farid, N., De Kamps, M., & Johnson, O. (2019). Process
Mining in Frail Elderly Care: A Literature Review.
Biomedical Engineering Systems and Technologies, 5,
332–339. https://doi.org/10.5220/0007392903320339
Fluxicon BV. (2019). Disco. Retrieved from https://flux
icon.com/disco/
Fox, F., Aggarwal, V. R., Whelton, H., & Johnson, O.
(2018). A data quality framework for process mining of
electronic health record data. Proceedings - 2018 IEEE
International Conference on Healthcare Informatics,
ICHI 2018, 12–21. https://doi.org/10.1109/ICHI.
2018.00009
Futoma, J., Morris, J., & Lucas, J. (2015). A comparison of
models for predicting early hospital readmissions.
Journal of Biomedical Informatics, 56, 229–238.
https://doi.org/10.1016/j.jbi.2015.05.016
Giannoula, A., Gutierrez-Sacristán, A., Bravo, Á., Sanz, F.,
& Furlong, L. I. (2018). Identifying temporal patterns
in patient disease trajectories using dynamic time
warping: A population-based study. Scientific Reports
2018 8:1, 8(1), 4216. https://doi.org/10.1038/s41598-
018-22578-1
Glicksberg, B. S., Li, L., Badgeley, M. A., Shameer, K.,
Kosoy, R., Beckmann, N. D., … Dudley, J. T. (2016).
Comparative analyses of population-scale phenomic
data in electronic medical records reveal race-specific
disease networks. Bioinformatics, 32(12), i101–i110.
https://doi.org/10.1093/bioinformatics/btw282
Goldberg, M. (2003). The graph isomorphism problem. In
J. L. Gross & J. Yellen (Eds.), Handbook of graph
theory (2nd ed., pp. 68–78). Boca Raton, FL: CRC
Press.
Hanauer, D. A., & Ramakrishnan, N. (2013). Modeling
temporal relationships in large scale clinical
associations. Journal of the American Medical
Informatics Association, 20(2), 332–341. https://doi.
org/10.1136/amiajnl-2012-001117
Hemingway, H., Asselbergs, F. W., Danesh, J., Dobson, R.,
Maniadakis, N., Maggioni, A., … Denaxas, S. (2018).
Big data from electronic health records for early and
late translational cardiovascular research: Challenges
and potential. European Heart Journal, 39(16), 1481–
1495. https://doi.org/10.1093/eurheartj/ehx487
Hidalgo, C. A., Blumm, N., Barabási, A.-L., & Christakis,
N. A. (2009). A Dynamic Network Approach for the
Study of Human Phenotypes. PLoS Computational
Biology, 5(4), e1000353.
https://doi.org/10.1371/journal.pcbi.1000353
Jensen, A. B., Moseley, P. L., Oprea, T. I., Ellesøe, S. G.,
Eriksson, R., Schmock, H., … Brunak, S. (2014).
Temporal disease trajectories condensed from
population-wide registry data covering 6.2 million
patients. Nature Communications, 5(May), 1–10.
https://doi.org/10.1038/ncomms5022
Jensen, K., Soguero-Ruiz, C., Oyvind Mikalsen, K.,
Lindsetmo, R. O., Kouskoumvekaki, I., Girolami, M.,
… Magne Augestad, K. (2017). Analysis of free text in
electronic health records for identification of cancer
patient trajectories. Scientific Reports, 7. https://doi.
org/10.1038/srep46226
Jensen, P. B., s, L. J., & Brunak, S. (2012). Mining
electronic health records: Towards better research
applications and clinical care. Nature Reviews
Genetics, Vol. 13, pp. 395–405. https://doi.org/
10.1038/nrg3208
Ji, X., Chun, S. A., & Geller, J. (2016). Predicting
Comorbid Conditions and Trajectories Using Social
Health Records. IEEE Transactions on Nano
bioscience, 15(4), 371–379. https://doi.org/10.1109/
TNB.2016.2564299
Kluyver, T., Ragan-kelley, B., Pérez, F., Granger, B.,
Bussonnier, M., Frederic, J., … Willing, C. (2016).
Jupyter Notebooks—a publishing format for
reproducible computational workflows. In Positioning
and Power in Academic Publishing: Players, Agents
and Agendas. https://doi.org/10.3233/978-1-61499-
649-1-87
Kurniati, A. P., Johnson, O., Hogg, D., & Hall, G. (2016).
Process Mining in Oncology: a Literature Review.
Information Communication and Management
(ICICM). https://doi.org/10.1109/INFOCOMAN.
2016.7784260
Kusuma, G., Bennett, B., & Johnson, O. (2017). Process
analysis in cardiovascular disease using process
mining. Abstract 621 in Scott P.J. et Al. Journal of
Innovation in Health Informatics, 24(1), 171.
https://doi.org/10.14236/jhi.v24i1.939
Process Mining of Disease Trajectories: A Feasibility Study
711

Kusuma, G., Sykes, S., McInerney, C., & Johnson, O.
(2019). Resource of Process Mining for Disease
Trajectory Mining. Retrieved from https://github.com/
gpkusuma/dtm_processmining
Mannhardt, F., & Blinde, D. (2017). Analyzing the
trajectories of patients with sepsis using process mining
(Vol. 1859). Retrieved from APA website: www.
tue.nl/taverne
Mans, R. S., Reijers, H. A., Van Genuchten, M., &
Wismeijer, D. (2012). Mining processes in dentistry.
IHI’12 - Proceedings of the 2nd ACM SIGHIT
International Health Informatics Symposium, 379–388.
https://doi.org/10.1145/2110363.2110407
Mans, R. S., van der Aalst, W. M. P., & Vanwersch, R. J.
B. (2015). Process Mining in Healthcare Evaluating
and Exploiting Operational Healthcare Processes.
https://doi.org/10.1007/978-3-319-16071-9
Mans, R. S., van der Aalst, W. M. P., Vanwersch, R. J. B.,
& Moleman, A. J. (2013). Process Mining in
Healthcare: Data Challenges when Answering
Frequently Posed Questions. In Lenz R., Miksch S.,
Peleg M., Reichert M., Riaño D., & ten Teije A. (Eds.),
Lecture Notes in Computer Science (Vol. 7738, pp.
140–153). Retrieved from http://wwwis.win.
tue.nl/~wvdaalst/publications/p707.pdf
Muhrer, J. C. (2014). The importance of the history and
physical in diagnosis. The Nurse Practitioner, 39(4),
30–35.
https://doi.org/10.1097/01.NPR.0000444648.20444.e6
Murray, S. A., Kendall, M., Boyd, K., & Sheikh, A. (2005).
Illness trajectories and palliative care. BMJ, 330(7498),
1007–1011. https://doi.org/10.1136/bmj.330.7498.100
7
Pavalko, E. K. (1997). Beyond Trajectories: Multiple
Concepts for Analyzing LongTerm Process. In M. A.
Hardy (Ed.), Studying Aging and Social Change:
Conceptual and Methodological Issues (pp. 129–147).
Thousand Oaks, CA: SAGE Publications Ltd.
Pescosolido, B. A. (2013). Patient Trajectories. In The
Wiley Blackwell Encyclopedia of Health, Illness,
Behavior, and Society (pp. 1770–1777).
https://doi.org/10.1002/9781118410868.wbehibs282
Pham, T., Tran, T., Phung, D., & Venkatesh, S. (2017).
Predicting healthcare trajectories from medical records:
A deep learning approach. Journal of Biomedical
Informatics, 69, 218–229. https://doi.org/10.1016/
j.jbi.2017.04.001
PODS4H. (2019). Retrieved from http://pods4h.com
Process Mining Group. (2010). ProM - Process Mining
Toolkit. Retrieved from www.promtools.org
Rojas, E., Munoz-Gama, J., Sepulveda, M., & Capurro, D.
(2016). Process mining in healthcare: A literature
review. Journal of Biomedical Informatics, 61(April),
224–236. https://doi.org/10.1016/j.jbi.2016.04.007
Rothman, K. J., & Greenland, S. (2005). Causation and
causal inference in epidemiology. American Journal of
Public Health, 95(SUPPL. 1), S144-50.
https://doi.org/10.2105/AJPH.2004.059204
Steinhaeuser, K., & Chawla, N. V. (2009). A network-
based approach to understanding and predicting
diseases. Social Computing and Behavioral Modeling,
209–216. https://doi.org/10.1007/978-1-4419-0056-2-
26
Sumida, K., & Kovesdy, C. P. (2017). Disease Trajectories
Before ESRD: Implications for Clinical Management.
Seminars in Nephrology, 37(2), 132–143. https://doi.
org/10.1016/j.semnephrol.2016.12.003
van der Aalst, W. M. P. (2011). Process Mining: Discovery,
Conformance and Enhancement of Business Processes.
https://doi.org/10.1007/978-3-642-19345-3
van Eck, M. L., Lu, X., Leemans, S. J. J., & van Der Aalst,
W. M. P. (2015). PM2: A process mining project
methodology. In J. Zdravkovic, M. Kirikova, & P.
Johannesson (Eds.), Lecture Notes in Computer Science
(including subseries Lecture Notes in Artificial
Intelligence and Lecture Notes in Bioinformatics) (Vol.
9097, pp. 297–313). https://doi.org/10.1007/978-3-
319-19069-3_19
Wang, X., Sontag, D., & Wang, F. (2014). Unsupervised
Learning of Disease Progression Models.
https://doi.org/10.1145/2623330.2623754
WHO. (2019). Classification of Diseases. Retrieved from
http://www.who.int/classifications/icd/en/
Williams, R., Rojas, E., Peek, N., & Johnson, O. A. (2018).
Process mining in primary care: A literature review.
Studies in Health Technology and Informatics, 247,
376–380. https://doi.org/10.3233/978-1-61499-852-5-
376
World Health Organization. (2002). Medical Records
Manual : A Guide for Developing Countries. Retrieved
from https://apps.who.int/iris/bitstream/handle/10665/
208125/9290610050_rev_eng.pdf?sequence=1&isAllo
wed=y
World Health Organization. (2016). Diagnostic errors:
technical series on safer primary care. Retrieved from
http://apps.who.int/bookorders.
HEALTHINF 2020 - 13th International Conference on Health Informatics
712
