Visualizing and Modifying Difficult Pixels in Cell
Image Segmentation
Daisuke Matsuzuki and Kazuhiro Hotta
Meijo University, 1-501 Shiogamaguchi, Tempaku-ku, Nagoya 468-8502, Japan
Keywords: Cell Image Segmentation, Difficult Pixels, Visualization, Modification, U-net.
Abstract: In this paper, we visualize and modify difficult pixels to recognize for deep learning. In general, an image
includes pixels that are easy or difficult to recognize. At the final layer, many deep learning methods use a
softmax function to convert the outputs of network to probabilities. Pixels with small maximum probability
are often difficult to recognize. We visualize those difficult pixels in a test image using the relationship
between confidence and pixel-wise difficulty. By visualizing difficult pixels, we confirm the connection of
cell membrane that could not be recognized by conventional method. We can connect the cell membrane by
modifying difficult pixels. In experiments, we use cell image of mouse liver dataset including three classes;
“cell membrane”, “cell nucleus” and “cytoplasm”. Our proposed method shows high recall score for “cell
membrane”. We also confirmed the connection of cell membrane in qualitative evaluation.
1 INTRODUCTION
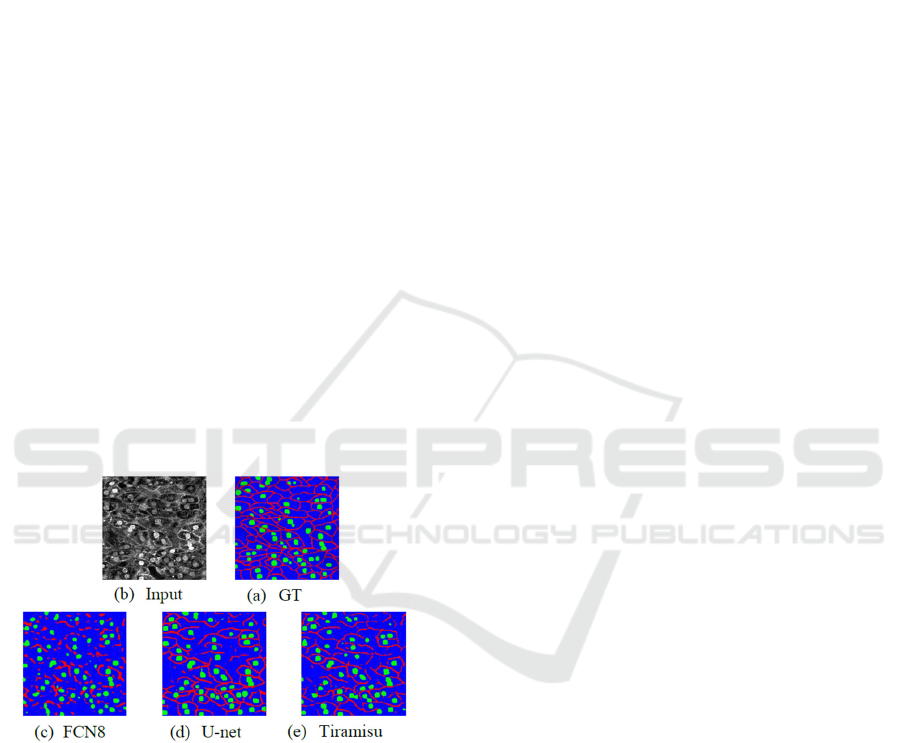
Figure 1: Segmentation results in cell image. (a) represent
input image in mouse of cell image dataset, (b) is ground
truth and (c), (d), (e) results of segmentation by FCN, U-net
and FC-DenseNet (Tiramisu). Red area represents cell
membrane, green area is nucleus and blue area show
cytoplasm.
In recent years, the amount of usable data has
increased in various fields. ImageNet (Russakovsky,
2015) is a big dataset includes a lot of images and
classes. Most of segmentation methods use the
ImageNet dataset as pre-training, and the accuracy of
semantic segmentation was improved. However, the
effect of pre-training with ImageNet is small in cell
image segmentation because ImageNet does not
include cell images. It is still difficult to prepare a
large amount of cell data and ground truth. Therefore,
cell image segmentation is difficult task yet.
Figure 1 shows the segmentation result by
conventional methods. From Figure 1, the most of
methods could not connect cell membrane well.
Especially, FCN8 (Long, 2015) recognized cell
membrane discontinuously. U-net (Ronneberge,
2015) is a famous method in medical image
segmentation, and the accuracy is higher than FCN.
Tiramisu (Simon, 2017) includes 103 convolution
layers, and it is able to extract features effectively.
However, it is difficult to connect cell membrane
well. It is very important to recognize cell membrane
in cell image segmentation. To address this problem,
we propose to visualize and modify difficult pixels in
segmentation result of a test image. When we predict
segmentation result, various methods use a softmax
function to convert the network outputs to
probabilities. Each pixel in a test image is classified
to the class with the maximum probability. Pixels
with small maximum probability are often difficult to
recognize. We use the relationship between the
maximum probability among all classes and pixel-
wise difficulty. We visualized difficult pixels
according to the maximum probability in test phase.
We confirmed that many difficult pixels are
misclassified. Thus, we modified the segmentation
result
for a test image using the difficult pixels, and
314
Matsuzuki, D. and Hotta, K.
Visualizing and Modifying Difficult Pixels in Cell Image Segmentation.
DOI: 10.5220/0009164203140318
In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2020) - Volume 4: BIOSIGNALS, pages 314-318
ISBN: 978-989-758-398-8; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

Figure 2: Overview of the proposed methods.
the segmentation result is improved. In experiments
on cell dataset, we compared our method with
conventional methods. We confirm that the proposed
method can connect cell membrane that could not be
recognized by conventional methods. Our proposed
method can reduce false negative and achieved high
recall scores for cell membrane.
This paper is organized as follows. In section 2, we
introduce related works. In section 3, we explain the
details of the proposed method. Section 4 shows
experimental results. We also compare our method
with conventional methods. In section 5, we describe
conclusion and future works.
2 RELATED WORKS
Semantic Segmentation.
Famous semantic segmentation methods are Fully
Convolutional Networks (FCN) (Long, 2015) and
encoder- decoder CNN. FCN consists of convolution
layers and upsampling layers to recover the spatial
information. Famous encoder-decoder CNN is the
SegNet (Badrinarayanan, 2017). However, small
objects and correct location are vanished in encoder
part. Thus, U-net (Ronneberge, 2015) used skip
connections between encoder and decoder to
compensate for the information.
Cell Image Segmentation.
In the field of cell image segmentation, almost of all
methods used the U-net (Ronneberge, 2015). U-net++
(Zhou, 2018) shows high accuracy that introduce
deep supervision and Resnet (He, 2016) architecture
in backbone network. Those methods show the
effectiveness of U-net architecture.
Murata et al. (Murata, 2018) proposed a
segmentation method of cell membranes and nucleus
by integrating different branches in U-net. Hiramatsu
et al. (Hiramatsu, 2018) used a Mixture-of-Experts
(Jacobs, 1991) structure with multiple U-nets. Tsuda
et al. (Tsuda, 2019) used multiple pix2pix (Isola,
2017) for each class. Those methods improved the
accuracy on Intersection over Union but could not get
connection of cell membrane in difficult cell image
dataset.
In this paper, we visualize the difficult pixels in a
test image and improve the accuracy of cell membrane
that conventional methods cannot segment well.
3 PROPOSED METHODS
Our goal is to get connection of cell membrane well.
To achieve the objective, we modify difficult pixels.
Figure 2 shows the overall architecture of our method.
First, we predict results using CNN in test phase. This
process is shown as a black arrow in Figure 2. We use
U-net in this paper. The predicted result is used as a
segmentation result in conventional methods.
However, as shown in Figure 1, it could not get the
connection of cell membrane well.
We would like to modify difficult pixels in the
segmentation result. When we predict segmentation
result, various methods based on CNN apply a
softmax function to convert the outputs of network to
probabilities. Then, each pixel is classified to the
class with the highest probability. Pixels with small
maximum probability among all classes are often
difficult to recognize. Therefore, small maximum
probability shows low confidence of prediction.
Modify
white area
Input: Image
N
etwor
k
Prediction
Visualize
difficult pixel
Modify resul
t
Visualizing and Modifying Difficult Pixels in Cell Image Segmentation
315
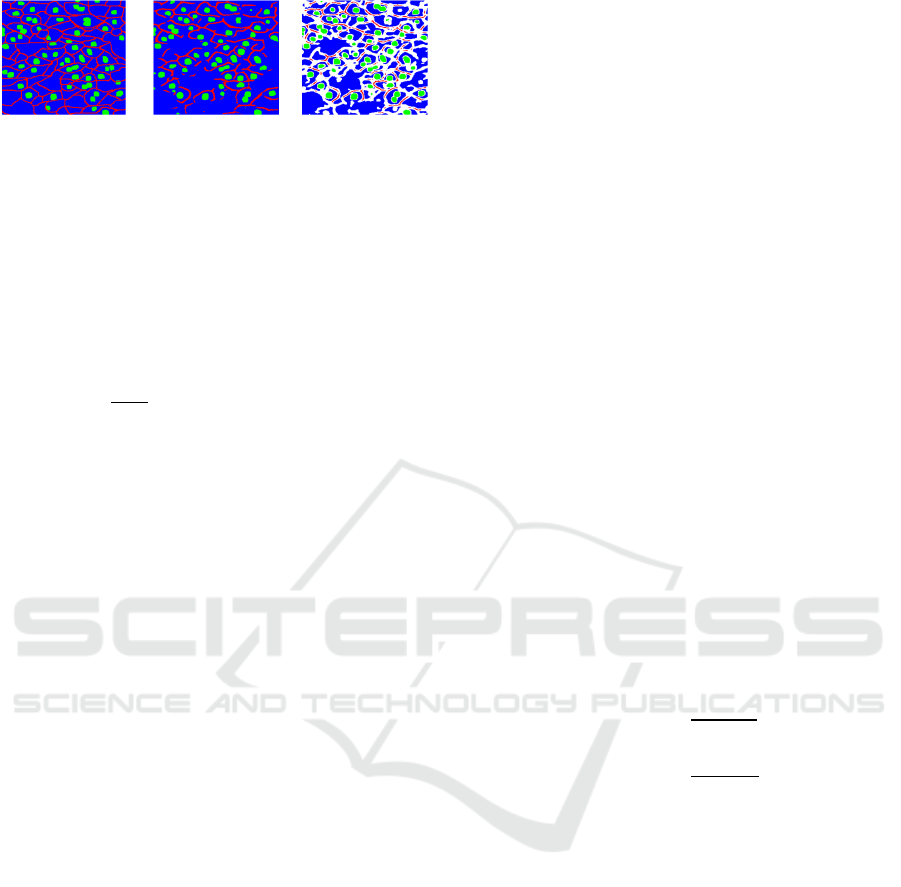
Figure 3: Visualization of difficult pixels. In visualized
difficult pixels, white area represents difficult pixels
according to confidence.
We visualized difficult pixels according to the
confidence. Figure 3 shows difficult pixels in
prediction result. White pixels represent the pixels
below the threshold α. α is defined as
1
∗
,
(1)
where y*x is image size and f(i,j) is the confidence
map. Each pixel in the confidence map represents the
maximum probability among all classes. By
calculating the threshold value based on the average
value, it is possible to define relatively difficult pixels
in a test image. From Figure 3, we confirmed that
white pixels on cell membrane are connected well.
Therefore, if the white area can be modified, it is
possible to get connection of cell membrane.
3.1 Modification of Prediction Results
Figure 4 shows how to modify difficult pixels in a test
image by our proposed method. First, we visualized
difficult pixels according to the confidence of the
network. White pixels represent difficult pixels in the
Figure. We use relationship in a cell image. The
relationships are as follows.
・Most of cell membranes are connected each other.
・Cell nucleus are not represented by one pixel.
After we visualize difficult pixels in a test image,
we modify cell membrane. Most of cell membranes
are connected to each other. If white area is adjacent
cell membrane, the pixel is defined as cell membrane.
The same flow repeats multiple times. In this way, it
is possible to connect cell membranes each other.
Next, we modify cell nucleus. The cell nucleus is not
represented by a little pixel like one pixel. However,
the network sometimes recognizes cell nucleus with
very small area. From Figure 1, it can be confirmed
that conventional methods recognize only part of cell
nucleus. To address this problem, if white area is
adjacent cell nucleus, we defined that the white pixels
are as cell nucleus. In this way, it is possible to obtain
the result of the cell nucleus more accurately. Finally,
the remaining white pixels are defined as cytoplasm.
4 EXPERIMENTS
4.1 Dataset
We use cell images of mouse liver dataset (Imanishi,
2018). The dataset is fluorescence images of the liver
of transgenic mice that expressed fluorescent markers
on the cell membrane and nucleus. The size of image
is 512×512 and include three classes; cell membrane,
cell nucleus and cell cytoplasm. It contains 35/5/10
images for training, validation and test.
4.2 Implementation Details
In this experiment, we use Adam (Kingma, 2014) as
optimizer and learning rate is set to 1e-3. We used
batch renormalization (Ioffe, 2017) with batchsize 2,
and single GPU with GeForce GTX 1080 TITAN.
Since various cell image segmentation methods are
based on U-net, we also used U-net. We used early
stopping according to the mIoU for validation dataset.
We use precision score and recall score as an
evaluation measure. Calculation of precision and
recall are followed as
(2)
(3)
where TP represent true positive, FP and FN are false
positive and false negative.
4.3 Evaluation
Figure 5 shows segmentation results by our proposed
method and conventional methods. From Figure 5, we
confirmed that our proposed method can connect
membrane well with small number of false positives.
However, our method tends to recognize cell
membrane with thicker than ground truth. This is
because we reconstruct preferentially cell membrane
when we modified difficult pixels.
Next, we evaluate precision and recall score.
Table 1 shows the recall score of the proposed method
and conventional U-net. From Table 1, we confirmed
that the proposed method shows very high recall
score for cell membrane. This result shows small false
negatives.
Table
2
shows
the
precision
score
of
the
GT Prediction
Visualize
difficult pixel
BIOSIGNALS 2020 - 13th International Conference on Bio-inspired Systems and Signal Processing
316
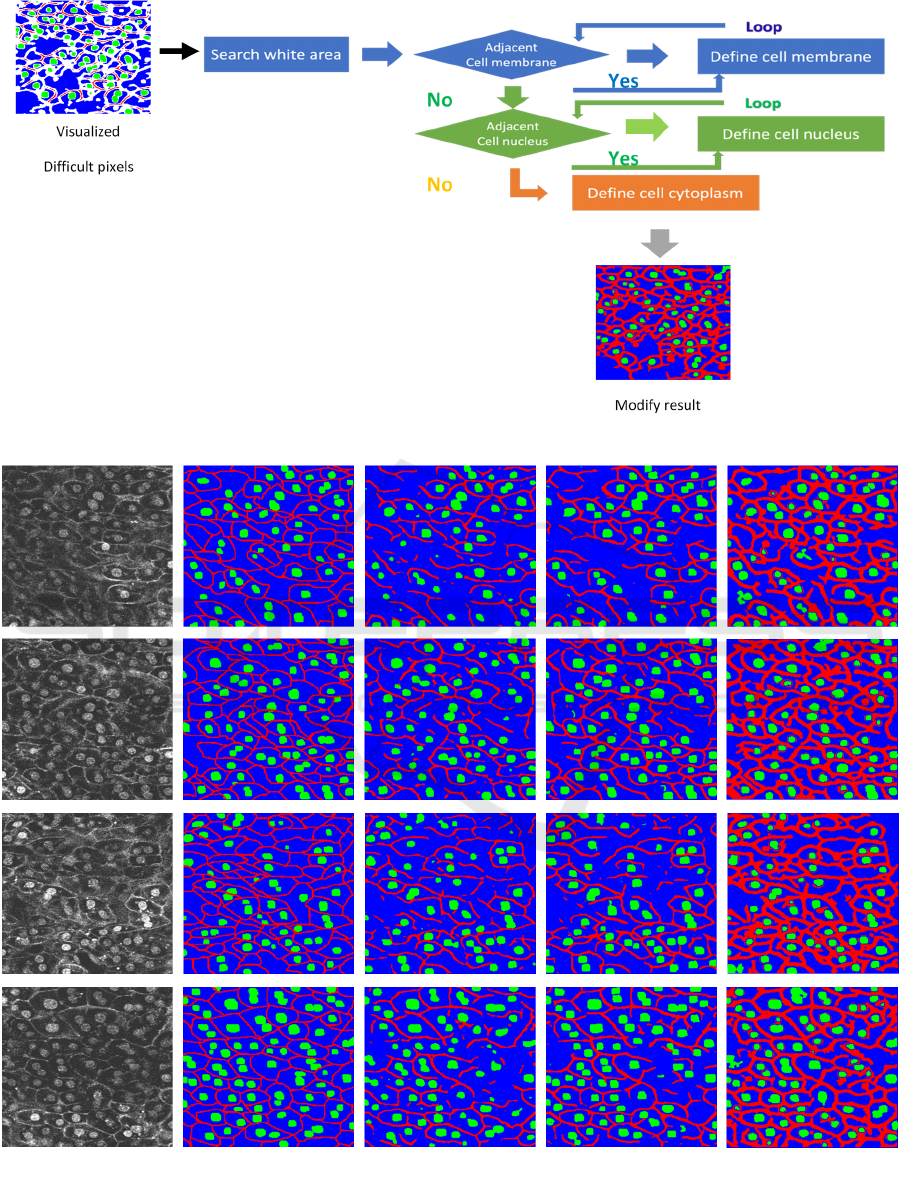
Figure 4: Flow of proposed method.
Figure 5: segmentation results. (a) represent input images and (b) are ground truth images. (c) and (d) are the results by
conventional Tiramisu103 and U-net and (e) shows the results by our proposed method.
(b) GT
(c) Tiramisu
(
d
)
U-ne
t
(e) Ou
r
(a) Inpu
t
Visualizing and Modifying Difficult Pixels in Cell Image Segmentation
317
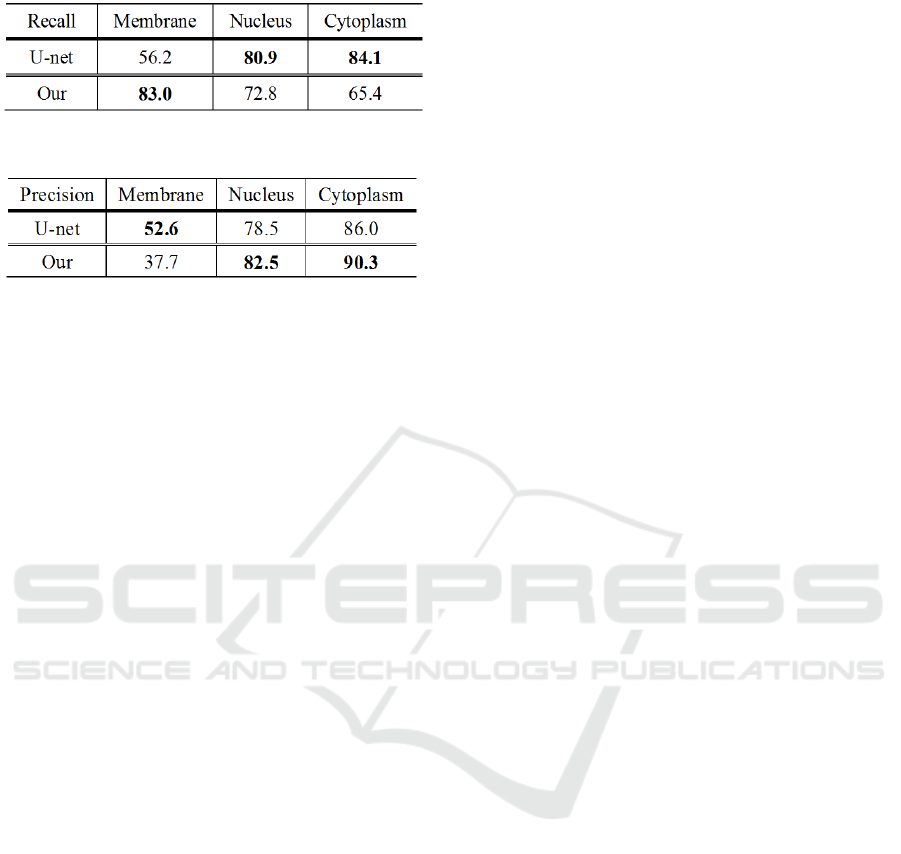
Table 1: Result of recall score.
Table 2: Result of precision score.
proposed method and conventional U-net. From
Table 2, we confirmed that cell nucleus is also
improved. This result shows that our proposed
method can reduce false positive like a noise. On the
other hand, U-net shows high precision score for cell
membrane. The reason is that our method recognizes
the cell membrane thickly. Therefore, our proposed
method is high recall score but precision score is low
score. However, our goal is to get connection of cell
membrane. High recall score is the result we
expected.
5 CONCLUSIONS
In this paper, we proposed to visualize and modify
difficult pixels according to the confidence of
network output. Difficult pixels often show the
connection between cell membrane, and we can
modify those pixels. We confirmed that the proposed
method can get connection of cell membrane well.
However, our method tends to that cell membrane is
recognized thicker than ground truth. Thus, we would
like to study another method for modifying prediction
results more accurately.
ACKNOWLEDGEMENTS
This work is partially supported by MEXT/JSPS
KAKENHI Grant Number 18H04746 "Resonance
Bio" and 18K111382.
REFERENCES
O. Russakovsky, J. Deng, H. Su, J. Krause, S. Satheesh, S.
Ma, Z. Huang, A. Karpathy, A. Khosla, M. Bernstein,
A. C. Berg, and L. Fei-Fei. Imagenet large scale visual
recognition challenge. IJCV, 2015, pp211- 252.
J. Long, E. Shelhamer, and T. Darrell. Fully convolutional
networks for semantic segmentation. In CVPR, 2015,
pp3431-3440.
O. Ronneberge, F. Philipp, and B. Thomas. U-net:
Convolutional networks for biomedical image
segmentation. In MICCAI, 2015, 234-241.
J. Simon, et al. The one hundred layers tiramisu: Fully
convolutional densenets for semantic segmentation. In
CVPR Workshop, 2017, pp11-19.
V. Badrinarayanan, A. Kendall, and R. Cipolla. Segnet: A
deep convolutional encoder-decoder architecture for
image segmentation. IEEE transactions on pattern
analysis and machine intelligence 39.12: 2481-2495,
2017.
Z. Zhou, MMR. Siddiquee, N. Tajbakhsh, et al. Unet++: A
nested u-net architecture for medical image
segmentation. In Deep Learning in Medical Image
Analysis and Multimodal Learning for Clinical
Decision Support, Cham, 2018, pp3-11.
K. He, X. Zhang, S. Ren, and J. Sun. Deep residual learning
for image recognition. In CVPR, 2016, pp770-778.
T. Murata, K, Hotta., et al. Segmentation of cell membrane
and nucleus using branches with different roles in deep
neural network. In BIOSIGNAL, 2018, pp256-261.
Y. Hiramatsu, K. Hotta, et al. Cell image segmentation by
integrating multiple CNNs. In CVPR Workshops, 2018,
pp2205-2211.
R. A. Jacobs, et al. Adaptive mixtures of local experts,
Neural Computation, vol.3, Issue.1, pp.79–87, 1991.
H. Tsuda and K. Hotta. K. Cell Image Segmentation by
Integrating Pix2pixs for Each Class. In CVPR
Workshops, 2019, pp0-0.
P. Isola, JY. Zhu, T. Zhou, and A. A. Efros, Image-to-
Image Translation with Conditional Adversarial
Networks. In CVPR, 2017, pp. 5967-5976.
A. Imanishi, T. Murata, et al. A Novel Morphological
Marker for the Analysis of Molecular Activities at the
Single-cell Level. Cell Structure and Function, 43: 129-
140, 2018
P. D. Kingma and Jimmy Ba. Adam: A method for
stochastic optimization. arXiv preprint arXiv:
1412.6980. 2014
S. Ioffe. Batch renormalization: Towards reducing
minibatch dependence in batch-normalized models. In
NeurlPS, 2017, pp1945-1953.
BIOSIGNALS 2020 - 13th International Conference on Bio-inspired Systems and Signal Processing
318
