
SalivaPrint as a Non-invasive Diagnostic Tool
Eduardo Esteves
a
, Igor Cruz
b
, Ana Cristina Esteves
c
, Marlene Barros
d
and Nuno Rosa
e
Universidade Católica Portuguesa, Faculty of Dental Medicine,
Center for Interdisciplinary Research in Health (CIIS), Portugal
Keywords: Saliva Diagnostic, Saliva Protein Profile, Machine Learning Strategies, Health Diagnostic.
Abstract: Currently, the molecular diagnosis is based on the quantification of RNA, proteins and metabolites because
they present changes in their quantity related to clinical situations. The same molecules are not generally
suitable for early diagnosis or to follow clinical evolution, making necessary strategies to evaluate the
complete molecular scenario. There are already experimental strategies that allow the determination of total
protein profiles from saliva samples (the SalivaPrint). The goal of this work is to identify a profile of saliva
proteins (similar to a fingerprint) and, using computational methods, identify how this profiles changes with
age and gender. So far it has been possible to collect 79 samples as well as the metadata associated with each
sample using an electronic questionnaire developed by us. A total protein profile was obtained and their
association with gender was verified using statistical methods. Currently we are developing the Python scripts
for automatic data acquiring and normalization. Total protein profiles annotation on a database
(SalivaPrintDB) and their integration with the factors that affects them using machine learning strategies can
empower the use of the approach proposed on this work as a tool for monitoring the individual's health status.
1 INTRODUCTION
In the last decade, saliva has been studied as a
noninvasive, easily obtainable fluid, with diagnosis
potential (Loo et al., 2010).
Saliva reflects the secretions of the 3 largest
salivary glands (parotid, submandibular and
sublingual), smaller salivary glands, crevicular fluid
and also contains serum components, transported by
blood capillaries and subsequently transferred by
diffusion, transport and/or ultrafiltration. The
presence of serum proteins in saliva enhances its use
as a systemic health status monitoring tool
(Castagnola et al., 2017; Kaczor-Urbanowicz et al.,
2017; Kaushik and Mujawar, 2018; Wang, Kaczor-
Urbanowicz, and Wong, 2017).
We recently described an automated capillary
electrophoresis-based strategy that allows to obtain a
salivary protein profile – the SalivaPrint Toolkit
(Cruz et al., 2018).
a
https://orcid.org/0000-0001-5458-4978
b
https://orcid.org/0000-0002-7082-297X
c
https://orcid.org/0000-0003-2239-2976
d
https://orcid.org/0000-0003-0631-4062
e
https://orcid.org/0000-0003-4604-0780
Since proteins are separated according to their
molecular mass, changes in peak morphology or
fluorescence intensity (translated by changes in peak
height) correspond to changes in the amount of
proteins or in the type of proteins being expressed.
The association of clinical and personal data to saliva
protein signatures, the SalivaPrint, to alterations in
different health/disease situations, will allow to build
a powerful framework for the creation of noninvasive
diagnostic strategies.
Preliminary results of data extraction
(SalivaPrint-Toolkit, Cruz et al., 2018) from capillary
electrophoresis allowed to identify the potential for
the creation of a SalivaPrint database.
It is known that some factors such as age, sex, or
circadian rhythm (Castagnola et al., 2011, 2017; Murr
et al., 2017), contribute to altering the expression of
salivary proteins . It is essential to define the signature
of proteins correspondent to the "healthy individual",
and simultaneously consider the intra-individual
variations.
Esteves, E., Cruz, I., Esteves, A., Barros, M. and Rosa, N.
SalivaPrint as a Non-invasive Diagnostic Tool.
DOI: 10.5220/0009163506770682
In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2020) - Volume 5: HEALTHINF, pages 677-682
ISBN: 978-989-758-398-8; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
677
Additionally, will be useful to identify potential
proteins that could reflect the changes in the
SalivaPrint profiles characteristic of different
situations. Data on proteins in saliva associated with
disease and health is already extensive. Our group
compilated this information in SalivaTecDB database
(http://salivatec.viseu.ucp.pt/salivatec-db, Arrais et
al., 2013; Rosa et al., 2012), which is an important
support for the identification of proteins that may
potentially be associated with certain signatures.
SalivaTecDB has currently stored more than 4000
human salivary proteins and is constantly being
updated.
The profile of salivary proteins, expressed in
different situations, has the potential of being used for
noninvasive diagnosis. The objectives of this work
are:
1- Establish the Health-SalivaPrint(s). The factors
that influence the protein profile of saliva in healthy
individuals will be considered.
2- Build a SalivaPrint database. These profiles
will posteriorly be used for the development of a tool
for heath monitoring using machine learning
strategies.
2 MOTIVATION
Nowadays, molecular diagnosis is based on the
quantification of RNA, proteins or metabolites whose
concentration can be correlated to clinical situations.
Usually, these molecules are not suitable for early
diagnosis or to follow clinical evolution. Therefore,
strategies to evaluate the complete molecular scenario
– early diagnosis, diagnosis and clinical evolution –
are necessary.
The potential of proteins for a large-scale
diagnosis depends on cheap and preferably
noninvasive strategies for screening. Bioinformatics
strategies and solutions to work on different types of
data: from biological related data to personal and
clinical information’s, is the best approach. Data
integration by these methods is an asset to predict the
pathological status before clinical outcomes.
This work explores the potential of saliva protein
profile obtained by capillary electrophoresis – the
SalivaPrint - as a strategy to obtain signatures of
health/disease states. The application of
computational methods (machine learning strategies)
will allow the establishment of a signature that
characterizes the healthy individual and how it varies
with diverse factors.
At this time, we are only interested in establishing
the entire SalivaPrint methodology, from capillary
electrophoresis to determining which factors
influence the protein profile (of healthy individuals)
including the creation of the database that stores all
this information.
3 METHODOLOGY
This study was approved by the Ethics Committee of
the Portuguese Catholic University (UCP). In
addition, donors will be asked, prior to any collection,
the consent for samples’ collection. All work will be
carried out in accordance with the principles of the
Helsinki declaration, as reviewed in 2008.
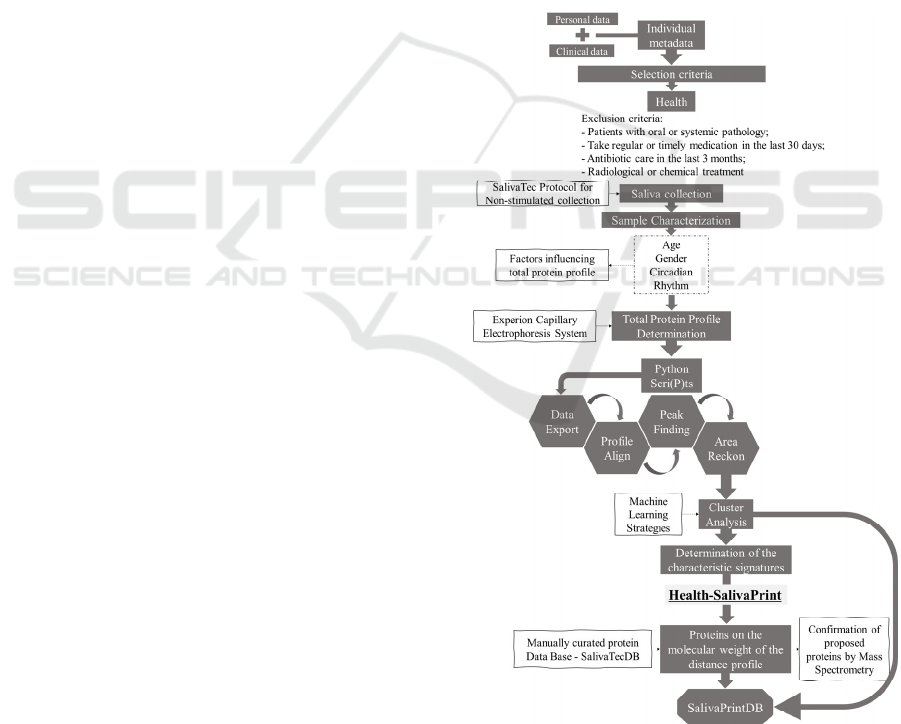
In order to achieve the proposed objectives, the
strategy presented in figure 1 will be followed:
Figure 1: Flowchart of proposed strategy plan.
HEALTHINF 2020 - 13th International Conference on Health Informatics
678

3.1 Questionnaire Preparation to
Characterize the Biological Sample
An online filling questionnaire will be created using
Qualtrics® software, for the collection of clinical and
personal information.
Questions will be defined to address the
individual's personal data and clinical status
(SalivaTec Donor Form). Each donor will be
observed by a dentist (collaboration with the Dental
Clinic of UCP), for oral health status evaluation,
taking into account the PSR index (Periodontal
Screening Recording) and DMFT (Decayed, Missing,
Filled Teeth) (Anón, 2002; Schuller and Holst, 2001).
3.2 Collection and Processing of Saliva
Samples
To define the protein profile of healthy individuals,
samples of non-stimulated saliva will be collected,
using an established protocol (Rosa et al., 2016), from
patients from the dental clinic of the Portuguese
Catholic University and from seniors in the Senior
Activity program from the municipality of Viseu. The
criteria for exclusion of healthy donors were defined
according to the literature: patients with oral or
systemic pathology; intake of regular or punctual
medication in the last 30 days; antibiotic
administration in the last 3 months; never subjected
to radiological or chemical treatment (Mozaffari et
al., 2019; Murr et al., 2017; Wang et al., 2015).
Samples will be categorized into groups of different
ages and genders.
These collections will be made under partnerships
already established with each institution.
Total protein concentration, pH and volume of
each sample will be determined.
3.3 Determination of the Total Profile
of Proteins by Capillary
Electrophoresis
Saliva protein profile (SalivaPrint) will be determined
by capillary electrophoresis, using the Experion™
Automated Electrophoresis System (BioRad) in
standard protein chips (Experion™ Pro260 Analysis
Kit). The samples will be analyzed according to the
technical specifications provided by BioRad.
Protein profile and the quantification of bands
will be obtained using the Experion™ Software,
version 3.20.
3.4 Determination of Factors
Influencing Health-SalivaPrint
The clustering of profiles will be tested according to
potentially influencing factors of salivary proteome
[age, gender and circadian rhythm (Castagnola et al.,
2017), among others].
For this, a variety of statistical analysis models
(e.g. Kruskall-Wallis) will be applied for comparing
each profile between influence groups created. Also,
data classification techniques will be implemented
with high robustness and effectiveness, such as
Random Forest, as well as approaches based on
unsupervised learning models for clustering analysis.
The performance of the different existing techniques,
the pertinence ratio of the data involved, the
effectiveness of the quality of the results obtained,
analyzing accuracy and tests of positive and negative
diagnoses, among others, will be evaluated.
Parallelly a distance profile will be determined
between the mean profile of healthy individuals’
groups formed according impact factors on protein
profile. This approach will allow the identification of
the molecular mass corresponding to the peaks that
demonstrate to be different. A threshold will be set at
half peak height to identify these intervals. This step
will allow the association of potential proteins in
these ranges using saliva protein database
SalivaTecDB (http://salivatec.viseu.ucp.pt/salivatec-
db). The area of each peak in each profile will be
compared between all individual profiles, using a t-
test, in order to identify peaks characteristic of each
factor.
From these analyses will result intervals of
molecular masses corresponding to proteins of
interest (later confirmed by mass spectrometry).
3.5 Construction of the Database for
the Storage of SalivaPrints
Protein profiles representing different conditions will
result from this work and will be gathered in
SalivaPrint database. Each SalivaPrint will be linked
to the metadata collected in the questionnaires
developed in 3.1. Tools will be developed to
categorize, edit, visualize and export the data
obtained to different formats, creating the necessary
conditions for its use in the development of
computational strategies for analysis and comparison
of saliva protein profiles for diagnostic purposes.
SalivaPrint as a Non-invasive Diagnostic Tool
679
4 PRELIMINARY RESULTS
The implementation of the proposed strategy (Figure
1) has so far yielded the results described below.
So far it has been possible to collect 79 samples
meeting the defined inclusion criteria (Table 1) and
the metadata associated with each sample using the
questionnaire developed by us.
Table 1: Number of saliva samples collected form healthy
individuals after applying the selection criteria.
Gender
Total
Female Male
Age Group
(years)
<13 12 4 24
13-24 14 10 39
25-50 22 17 16
Total 48 31 79
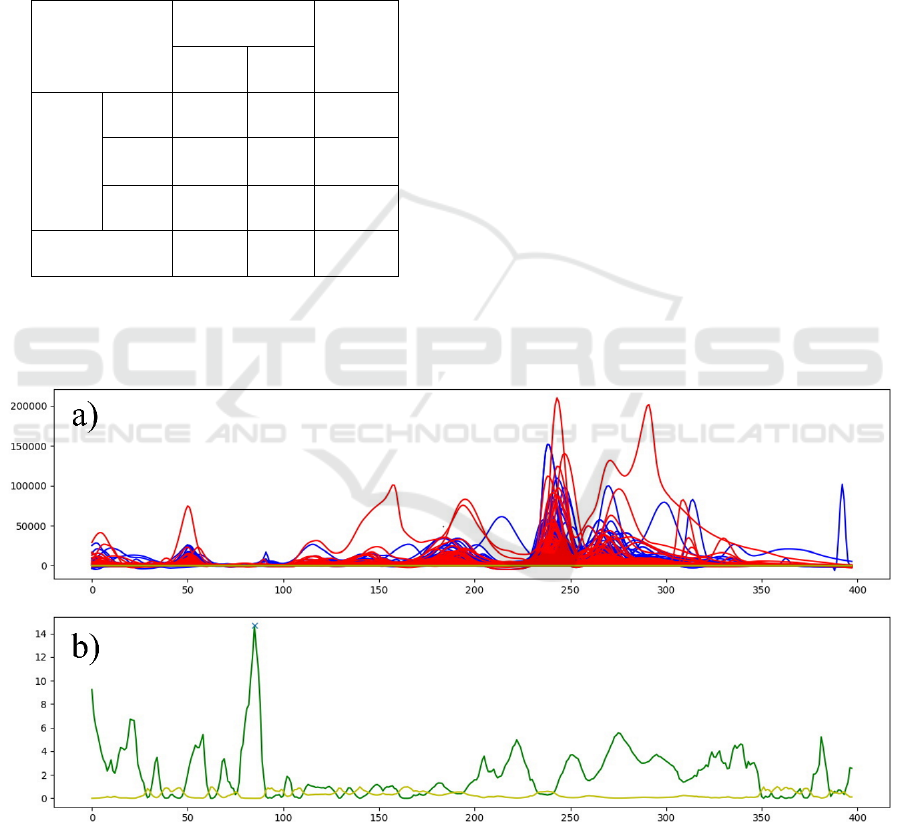
A protein profile for each gender (Female n=48;
Male n=31) was built (Figure 2 a), according to the
methodology described in the section 3.3.
In order to determine how gender impact, the
saliva total protein profile in health individuals
(Health-SalivaPrint) a Kruskal-Wallis hypothesis test
was applied (Figure 2). In top graph we could see the
protein profiles from Female (red) and Male (blue)
individuals. When Kruskal-Wallis test is applied, on
lower graph the higher peak (green) with lower p
value (yellow) corresponds to molecular weight
where it exists a significant difference between
genders, so the gender characteristic signature.
A distance profile for each gender was built
according to the methodology described earlier. The
comparison of the mean-profile obtained from saliva
of Female and Male individuals was used to establish
a distance-profile (Figure 3). This distance profile
reflects the molecular weight (MW) range
corresponding to the proteins that are altered between
genders. The creation of the distance profile allows
the identification of the specific molecular mass
according to different expressed peaks on each group
(Figure 3).
The t-test applied on half-peak areas of all profiles
showed the main differences were found in 2 of the
typical 6 peaks of the saliva protein profile. Within
these peaks, the MW ranges of
peak 5 (58.5 - 63.2
kDa) and peak 6 (66.8 - 74.2 kDa).
Figure 2: Application of the Kruskal-Wallis hypotheses test in determining profile signatures for gender identification. In a),
red lines correspond to the profiles of female individuals and blue lines to the profiles of male individuals. In b) the green line
corresponds to the test value and the yellow line to the p value. Higher hypothesis test value (green) and lower p value (yellow)
correspond to the molecular weight were exist a clear difference on fluorescence between genders.
HEALTHINF 2020 - 13th International Conference on Health Informatics
680
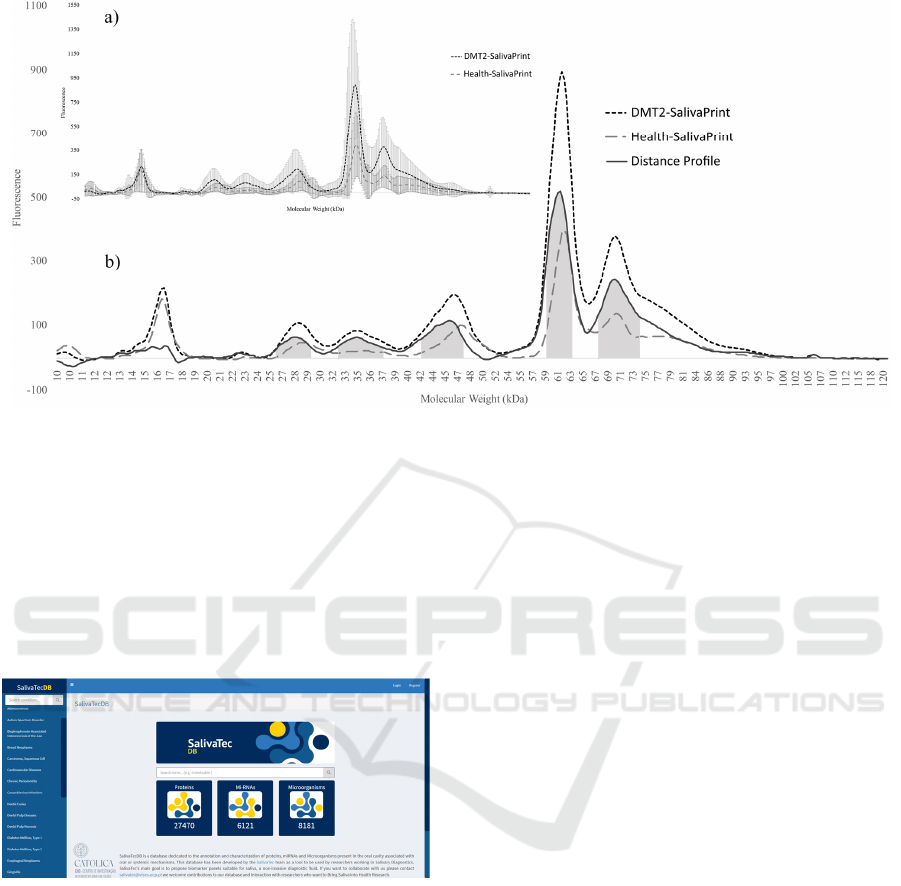
Figure 3: Protein profile distribution in Female (----) and Male (– – –) individuals. a) Female and Male mean-profile
corresponding to the average of fluorescence per molecular weight. The standard deviations are represented by the vertical
error lines. b) Protein profile for Female (----) and Male (– – –) and the distance-profile (–). Shaded areas correspond to the
MW range of the proteins most altered between genders.
Once the distance profile and the respective MW
ranges are established, it is possible, use
SalivaTecDB (Figure 4), to associate potential
proteins with molecular mass within the defined
ranges and after that confirmed by mass
spectrometry.
Figure 4: SalivaTecDB homepage. SalivaTecDB is
dedicated to the annotation and characterization of proteins,
miRNAs and Microorganisms present in the oral cavity,
associated with oral or systemic mechanisms. This database
was developed as a tool to be used by researchers working
in Salivary Diagnostics.
5 FUTURE WORK
Currently we are developing the Python scripts for:
data acquiring (from electrophoresis system) and
treatment (profile alignment, peak determination and
peak area calculation). These scripts will allow the
total protein profile comparing between individuals
and groups, with peak areas for statistical analysis, as
well as analysis with machine learning strategies. The
application of machine learning, for example Random
Forest based on unsupervised models, allow the
clusterization according to impact factors on profile
(if it’s the case) and thus define a characteristic
Health-SalivaPrint.
6 CONCLUSIONS
This paper presents the SalivaPrint strategy and
preliminary results already obtained with this
approach. This work already allowed us to develop
the methodologies that permits to obtain the total
protein profile of health individuals (in a convenience
sample). Through the analysis of these profiles it is
possible to associate the potential proteins more
representative of the differences between them using
SalivaTecDB. The application of the Kruskal-Wallis
test is a first approach on the identification of
signatures, although there still need for other analysis
to confirm the results.
Future work will be directed to python scripts
development to acquire aligned protein profiles and
determine the factors that influence the profile, as
well as establish the machine learning strategy for
cluster determination and its association to database
(SalivaPrintDB).
It is expected with this work to use the database
with SalivaPrints associated with an unsupervised
machine learning algorithm that allows protein
SalivaPrint as a Non-invasive Diagnostic Tool
681

patterns recognition through profiles and personal
data, without defined diagnosis, pathology or health
situations. This approach will consider the
morphology of the protein profile obtained with
capillary electrophoresis in an automated manner and
the influencing factors of saliva proteome, namely,
the total protein concentration, age, gender and inter-
individual and intra-individual variability. These
variables will be analyzed through the application of
supervised analysis models in a first phase to identify
the influence of each factor in the profile. Later with
a higher number of individuals we resorted to
unsupervised analysis models.
For this strategy to be used for diagnosis
purposes, it is necessary to compare Health-
SalivaPrint with a Disease-SalivaPrint. Since
differences in saliva protein profiles have been
observed in healthy people, depending on age and
gender, it is important to take this into account in
order to isolate the effect of the disease from the
effects of these parameters.
This type of work is essential to find less invasive
forms of diagnosis that take into account all the
molecular and physiological variability of the
individual.
ACKNOWLEDGEMENTS
Thanks are due to FCT/MCTES, for the financial
support of Centre for Interdisciplinary Research in
Health (UID/MULTI/4279/2019). Thanks, are also
due to FCT and UCP for the CEEC institutional
financing of AC Esteves.
REFERENCES
Anón. 2002. «Periodontal Screening and Recording (PSR)
Index: precursors, utility and limitations in a clinical
setting - Landry - 2002 - International Dental Journal -
Wiley Online Library». Obtido 24 de Março de 2019.
Arrais, Joel P., Nuno Rosa, José Melo, Edgar D. Coelho,
Diana Amaral, Maria José Correia, Marlene Barros, e
José Luís Oliveira. 2013. «OralCard: A bioinformatic
tool for the study of oral proteome». Archives of Oral
Biology 58(7):762–72.
Castagnola, M., P. M. Picciotti, I. Messana, C. Fanali, A.
Fiorita, T. Cabras, L. Calò, E. Pisano, G. C. Passali, F.
Iavarone, G. Paludetti, e E. Scarano. 2011. «Potential
applications of human saliva as diagnostic fluid». Acta
Otorhinolaryngologica Italica 31(6):347–57.
Castagnola, M., E. Scarano, G. C. Passali, I. Messana, T.
Cabras, F. Iavarone, G. Di Cintio, A. Fiorita, E. De
Corso, e G. Paludetti. 2017. «Salivary biomarkers and
proteomics: future diagnostic and clinical utilities». Acta
Otorhinolaryngologica Italica 37(2):94–101.
Cruz, Igor, Eduardo Esteves, Mónica Fernandes, Nuno Rosa,
Maria José Correia, Joel P. Arrais, e Marlene Barros.
2018. «SalivaPRINT Toolkit - Protein profile evaluation
and phenotype stratification.» Journal of proteomics
171:81–86.
Formiga, F., M. Camafort, e F. J. Carrasco Sánchez. 2019.
«Insuficiencia cardiaca y diabetes: la confrontación de
dos grandes epidemias del siglo xxi». Revista Clínica
Española.
Kaczor-Urbanowicz, Karolina Elżbieta, Carmen Martin
Carreras-Presas, Katri Aro, Michael Tu, Franklin Garcia-
Godoy, e David TW Wong. 2017. «Saliva diagnostics –
Current views and directions». Experimental Biology
and Medicine 242(5):459–72.
Kaushik, Ajeet e Mubarak A. Mujawar. 2018. «Point of Care
Sensing Devices: Better Care for Everyone». Sensors
(Basel, Switzerland) 18(12).
Loo, J. A., W. Yan, P. Ramachandran, e D. T. Wong. 2010.
«Comparative Human Salivary and Plasma Proteomes».
Journal of Dental Research 89(10):1016–23.
Mozaffari, Hamid Reza, Roohollah Sharifi, Asad Vaisi-
Raygani, Masoud Sadeghi, Samad Nikray, e Rozita
Naseri. 2019. «Salivary Profile in Adult Type 2 Diabetes
Mellitus Patients: A Case-Control Study». J Pak Med
Assoc 69(02):5.
Murr, Annette, Christiane Pink, Elke Hammer, Stephan
Michalik, Vishnu M. Dhople, Birte Holtfreter, Uwe
Völker, Thomas Kocher, e Manuela Gesell Salazar.
2017. «Cross-Sectional Association of Salivary Proteins
with Age, Sex, Body Mass Index, Smoking, and Edu-
cation». Journal of Proteome Research 16(6):2273–81.
Rosa, Nuno, Maria José Correia, Joel P. Arrais, Pedro Lopes,
José Melo, José Luís Oliveira, e Marlene Barros. 2012.
«From the Salivary Proteome to the OralOme:
Comprehensive Molecular Oral Biology». Archives of
Oral Biology 57(7):853–64.
Rosa, Nuno, Jéssica Marques, Eduardo Esteves, Mónica
Fernandes, Vera M. Mendes, Ângela Afonso, Sérgio
Dias, Joaquim Polido Pereira, Bruno Manadas, Maria
José Correia, e Marlene Barros. 2016. «Protein Quality
Assessment on Saliva Samples for Biobanking
Purposes». Biopreservation and Biobanking 14(4):289–
97.
Schuller, Annemarie A. e Dorthe Holst. 2001. «Oral Status
Indicators DMFT and FS-T: Reflections on Index
Selection». European Journal of Oral Sciences
109(3):155–59.
The American Diabetes Association. 2019. «Classification
and Diagnosis of Diabetes: Standards of Medical Care in
Diabetes—2019». Diabetes Care 42 (Supplement
1):S13–28.
Wang, Xiaoqian, Karolina Elżbieta Kaczor-Urbanowicz, e
David T. W. Wong. 2017. «Salivary Biomarkers in
Cancer Detection». Medical oncology (Northwood,
London, England) 34(1):7.
Wang, Zhihui, Yanyi Wang, Hongchen Liu, Yuwei Che,
Yingying Xu, e Lingling E. 2015. «Age-Related
Variations of Protein Carbonyls in Human Saliva and
Plasma: Is Saliva Protein Carbonyls an Alternative
Biomarker of Aging?» AGE 37(3).
HEALTHINF 2020 - 13th International Conference on Health Informatics
682
