
Channel-wise Aggregation with Self-correction Mechanism for
Multi-center Multi-Organ Nuclei Segmentation in Whole Slide Imaging
Mohamed Abdel-Nasser
1,3 a
, Adel Saleh
2
and Domenec Puig
1 b
1
Computer Engineering and Mathematics Department, University Rovira i Virgili, Tarragona, Spain
2
Gaist Solutions Ltd., U.K.
3
Electrical Engineering Department, Aswan University, Aswan, Egypt
Keywords:
Computational Pathology, Nuclei Segmentation, Whole Slide Imaging, Deep Learning.
Abstract:
In the field of computational pathology, there is an essential need for accurate nuclei segmentation methods for
performing different studies, such as cancer grading and cancer subtype classification. The ambiguous bound-
ary between different cell nuclei and the other objects that have a similar appearance beside the overlapping
and clumped nuclei may yield noise in the ground truth masks. To improve the segmentation results of cell
nuclei in histopathological images, in this paper, we propose a new technique for aggregating the channel maps
of semantic segmentation models. This technique is integrated with a self-correction learning mechanism that
can handle noisy ground truth. We show that the proposed nuclei segmentation method gives promising results
with images of different organs (e.g., breast, bladder, and colon)collected from medical centers that use de-
vices of different manufacturers and stains. Our method reaches the new state-of-the-art. Mainly, we achieve
the AJI score of 0.735 on the Multi-Organ Nuclei Segmentation benchmark, which outperforms the previous
closest approaches.
1 INTRODUCTION
Currently, digital pathology has an essential role in
clinics and laboratories. Thousands of tissue biop-
sies are taken from cancer patients yearly, and in turn,
the whole-slide imaging (WSI) technique permits the
acquisition of high-resolution images of slides. Cell
segmentation indicates the segmentation of the cell
nuclei. Notably, there is a necessity for precise nuclei
segmentation techniques in the context of computa-
tional pathology for facilitating the extraction of de-
scriptors for the morphometrics of cell nuclei. Several
descriptors, such as cell nuclei shape and number of
cell nuclei in WSI images, can be used to conduct var-
ious studies such as the determination of cancer types,
cancer grading, and prognosis (Moen et al., 2019).
Indeed, there is a diverse tissue type, variations in
staining and cell type leads to different visual char-
acteristics of WSI images. This makes the segmenta-
tion of nuclei segmentation a challenging task. Nuclei
segmentation task necessitates a vast effort to manu-
ally create the pixel-wise annotations that can be used
for training machine learning techniques. There are
nuclei segmentation toolboxes available in Cell Pro-
a
https://orcid.org/0000-0002-1074-2441
b
https://orcid.org/0000-0002-0562-4205
filer (Carpenter et al., 2006) and ImageJ-Fiji (Schin-
delin et al., 2012). However, the visual characteris-
tics of WSI images makes it very difficult to develop
traditional image processing based segmentation al-
gorithms that give acceptable nuclei segmentation re-
sults with WSI images taken from several cancer pa-
tients and collected at different medical centers for
various organs, such as breast, colon, lung, and stom-
ach (Niazi et al., 2019).
Recently, a review and comprehensive compar-
ison are presented in (Vicar et al., 2019) for cell
segmentation methods for label-free contrast mi-
croscopy. These segmentation methods studied are
categorized as follows: 1) single-cell segmentation
methods, 2) foreground segmentation methods, such
as thresholding, feature-extraction, level-set, graph-
cut, machine learning-based, and 3) seed-point ex-
traction methods, namely Laplacian of Gaussians, ra-
dial symmetry and distance transform, iterative radial
voting, maximally stable extremal region. The study
concluded that the machine learning-based methods
give accurate segmentation results.
In the last years, deep learning models have
been employed for performing different segmentation
tasks in biology (Niazi et al., 2019). Naylor et al.
(Naylor et al., 2017) introduced a fully automated
method for cell nuclei segmenting in histopathology
466
Abdel-Nasser, M., Saleh, A. and Puig, D.
Channel-wise Aggregation with Self-correction Mechanism for Multi-center Multi-Organ Nuclei Segmentation in Whole Slide Imaging.
DOI: 10.5220/0009156604660473
In Proceedings of the 15th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2020) - Volume 4: VISAPP, pages
466-473
ISBN: 978-989-758-402-2; ISSN: 2184-4321
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
images based on three semantic segmentation mod-
els: PangNet,a fully convolutional network (FCN)
and DeconvNet. They ensembled the three seman-
tic segmentation models and obtained an F1-score of
0.80. In (Al-Kofahi et al., 2018), a three-step cell nu-
clei segmentation approach is proposed: 1) the detec-
tion of the cells using a deep learning-based model
to obtain pixel probabilities for nuclei, cytoplasm,
as well as background, 2) the separation of touching
cells based on blob detection and shape-based water-
shed techniques that can distinguish between the indi-
vidual nuclei from the nucleus prediction map and 3)
the segmentation of the nucleus and cytoplasm). With
four different datasets, they obtained an accuracy of
0.84.
In (Qu et al., 2019), a weakly supervised
deep nuclei segmentation using points annotation in
histopathology images is proposed. In this study, the
original WSI images and the shape prior of nuclei are
employed to obtain two types of coarse labels from
the points annotation using the Voronoi diagram and
the k-means clustering algorithm. These coarse la-
bels are used to train a deep learning model, and then
the dense conditional random field is utilized in the
loss function to fine-tune the trained model. With
the multi-organ WSI dataset, a dice score of 0.73 is
achieved. In (Mahmood et al., 2019), a conditional
generative adversarial network (cGAN) model is pro-
posed for nuclei segmentation. A large dataset of syn-
thetic WSI images with perfect nuclei segmentation
labels is generated using an unpaired GAN model.
Both synthetic and real data with spectral normaliza-
tion and gradient penalty for nuclei segmentation are
used to train the cGAN model.
In (Zhou et al., 2019), a deep learning-based
model called contour-aware informative aggregation
network (CIA-Net) with a multilevel information ag-
gregation module between two task-specific decoders.
Instead of using independent decoders, this model ex-
ploits bi-directionally aggregating task-specific fea-
tures to merge the advantages of spatial and texture
dependencies between nuclei and contour. Besides, a
smooth truncated loss that modulates losses is utilized
to mitigate the perturbation from outliers. As a result,
the CIA-Net model is almost built using informative
samples, and so its generalization capability could be
enhanced (i.e., with multi-organ multi-center nuclei
segmentation tasks). With the 2018 MICCAI chal-
lenge of the multi-organ nuclei segmentation dataset,
they produced a Jaccard score of 0.63.
In addition, in (Wang et al., 2019), a multi-path di-
lated residual network is proposed for nuclei segmen-
tation and detection. The network includes the fol-
lowing: 1) multi-scale feature extraction based on D-
Figure 1: WSI images.
ResNet and feature pyramid network (FPN), 2) can-
didate region network, and 3) a final network for de-
tection and segmentation. The detection and segmen-
tation network involves three parts: segmentation, re-
gression, and classification sub-networks. With the
MonuSeg dataset, an aggregated Jaccard index (AJI)
of 0.46 is obtained.
Although methods above achieved promising cell
nuclei segmentation results, the ambiguous bound-
ary between different cell nuclei and the other ob-
jects that have a similar appearance beside the over-
lapping and clumped nuclei may yield noise in the
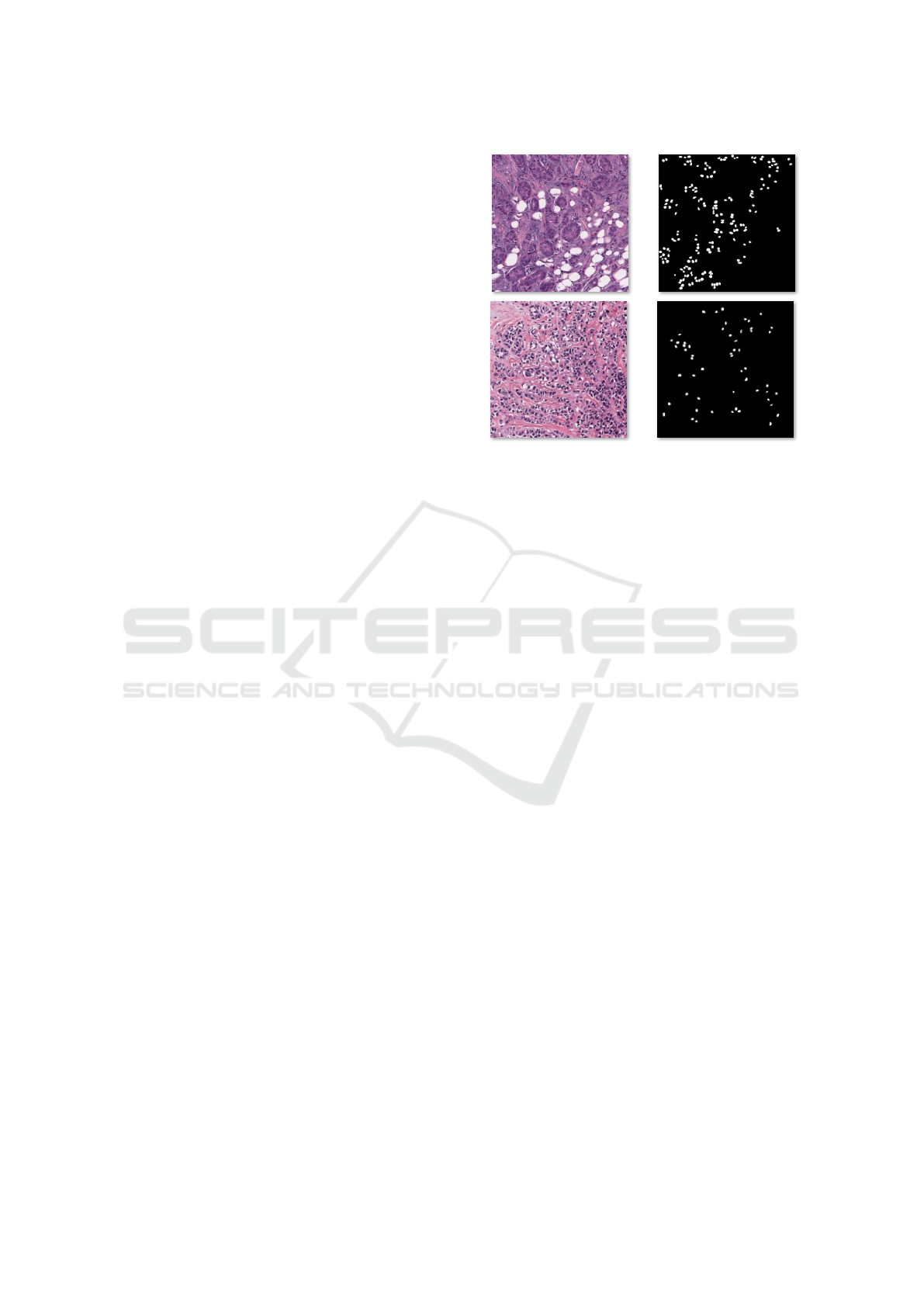
ground truth masks (see Fig. 1). To cope with these
issues, in this paper, we propose a new technique for
aggregating the channel maps of semantic segmenta-
tion models. This technique is integrated with a self-
correction learning mechanism that can handle noisy
ground truth. Notably, we do not claim any novelty
of the self-correction mechanism or the channel-wise
aggregation mechanism, but only the superiority of
the performance of the proposed method with the nu-
clei segmentation task.
The rest of this paper is organized as follows. Sec-
tion 2 presents the proposed methods. Section 3 pro-
vides the results and discussion. Section 4 concludes
the paper and gives some points of future work.
2 METHODOLOGY
The ambiguous boundary between different cell nu-
clei and the other objects that have a similar appear-
ance beside the overlapping and clumped nuclei may
yield noise in the ground truth masks. To improve
the segmentation results of cell nuclei in histopatho-
logical images, we propose a new technique for ag-
Channel-wise Aggregation with Self-correction Mechanism for Multi-center Multi-Organ Nuclei Segmentation in Whole Slide Imaging
467

gregating the channel maps of semantic segmenta-
tion models. This technique is integrated with a self-
correction learning mechanism that can handle noisy
ground truth.
2.1 Channel-wise Aggregation
Mechanism
The aggregation operators reduces a set of numbers
(x
1
, x
2
, . . . , x
n
) into a single representative number y.
This operation can be expressed as follows (Lucca
et al., 2017):
y = A(x
1
, x
2
, . . . , x
n
) (1)
here A : [0, 1]
n
×[0, 1] → [0, 1] is said to be aggregation
function Iff it has the following conditions:
• Identity in the Case of Unary: A(x) = x
• Boundary Conditions: A(0
1
, ..., 0
n
) = 0 and
agg(1
1
, ..., 1
n
) = 1
• Non Decreasing: A(x
1
, ..., x
n
) ≤ A(y
1
, ..., y
n
)
when (x
1
, ..., x
n
) ≤ (y
1
, ..., y
n
)
For the sake of simplicity, in this paper, we use the
maximum operator (max{x
1
, . . . , x
n
}). In the decoder
part of the segmentation model, we apply a element-
wise max. aggregation function on the channel maps.
If we have channel maps {C1, C2, . . . , C
n
}, the aggre-
gation function will produce one channel map (C
a
gg).
Then, we concatenate the C
a
gg with the original fea-
ture maps and feed all together to the next layers as
folows {C
a
gg, C1, C2, . . . , C
n
}.
2.2 Self Correction Training
Mechanism
The self-correction training strategy (Li et al., 2019)
can be employed to aggregate a segmentation model
and labels, which can enhance the performance of
the segmentation model and the ground-truth labels
in an iterative manner. The improvement that can be
achieved by the self-correction training strategy de-
pends on the initial segmentation results of the basic
segmentation model. It is important to note that if the
initial segmentation results are not accurate, they may
worsen the self-correction training strategy. Thus,
the self-correction strategy should be begun after the
training loss begins to take a flat shape.
After we get good segmentation results with
the basic segmentation model, a cyclically learning
scheduler with warm restarts is used. Here, a cosine
annealing learning rate scheduler with cyclical restart
(Loshchilov and Hutter, 2016) that can be mathemat-
ically formulated as follows:
φ = φ
min
+
1
2
(φ
max
− φ
min
)
1 + cos
EP
res
EP
π
(2)
where EP is the number of epochs in each cycle, EP
res
indicates the number of epochs gone since the previ-
ous restart, φ
max
is the initial learning rate, and φ
min
is
the final learning rate.
After each cycle of the self-correction mecha-
nism, we obtain a set of weights (models) θ =
ˆ
θ
0
,
ˆ
θ
1
, . . . ,
ˆ
θ
T
, and the corresponding predicted la-
bels Y =
{
ˆy
0
, ˆy
1
, . . . , ˆy
T
}
, where T is the number of
training cycles. After each training cycle, the current
model weights
ˆ
θ are aggregated with the weights of
previous cycle
ˆ
θ
t−1
in order to obtain new weights
ˆ
θ
t
as follows:
ˆ
θ
t
=
t
t + 1
ˆ
θ
t−1
+
1
t + 1
ˆ
θ (3)
Similarly, the the ground-truth labels are be aggre-
gated as follows:
ˆy
t
=
t
t + 1
ˆy
t−1
+
1
t + 1
ˆy (4)
where t refers to the number of the current cycle (0 ≤
t ≤ T ), and ˆy is the generated pseudo-labels (pseudo-
masks) with the model
ˆ
θ
t
.
2.3 Basic Framework for Conducting
Cell Nuclei Segmentation
The self-correction training used in his paper utilizes
the A-CE2P model (Ruan et al., 2019) as the basic
framework for conducting cell nuclei segmentation.
As shown in Fig. 2, the CE2P model comprises three
branches: segmentation branch (top), fusion branch
(middle), and edge branch (bottom). The operation
of A-CE2P can be mathematically formulated as fol-
lows:
E = α
1
E
edge
+ α
2
E
parsing
+ α
3
E
consistent
(5)
where α
1
, α
2
and α
3
are hyper-parameters to control
the contribution among these three losses. The CE2P
model is jointly trained in an end-to-end manner by
minimizing E .
For an input WSI image img, assume that the cell
nuclei ground truth label is ˆy
cn
i
and the predicted mask
is y
cn
i
, where cn refers to the number of pixels for class
i. The pixel-level supervised nuclei segmentation task
can be expressed using the cross-entropy loss as fol-
lows:
E
e
= −
1
N
∑
i
∑
cn
ˆy
cn
i
log p (y
cn
i
) (6)
VISAPP 2020 - 15th International Conference on Computer Vision Theory and Applications
468
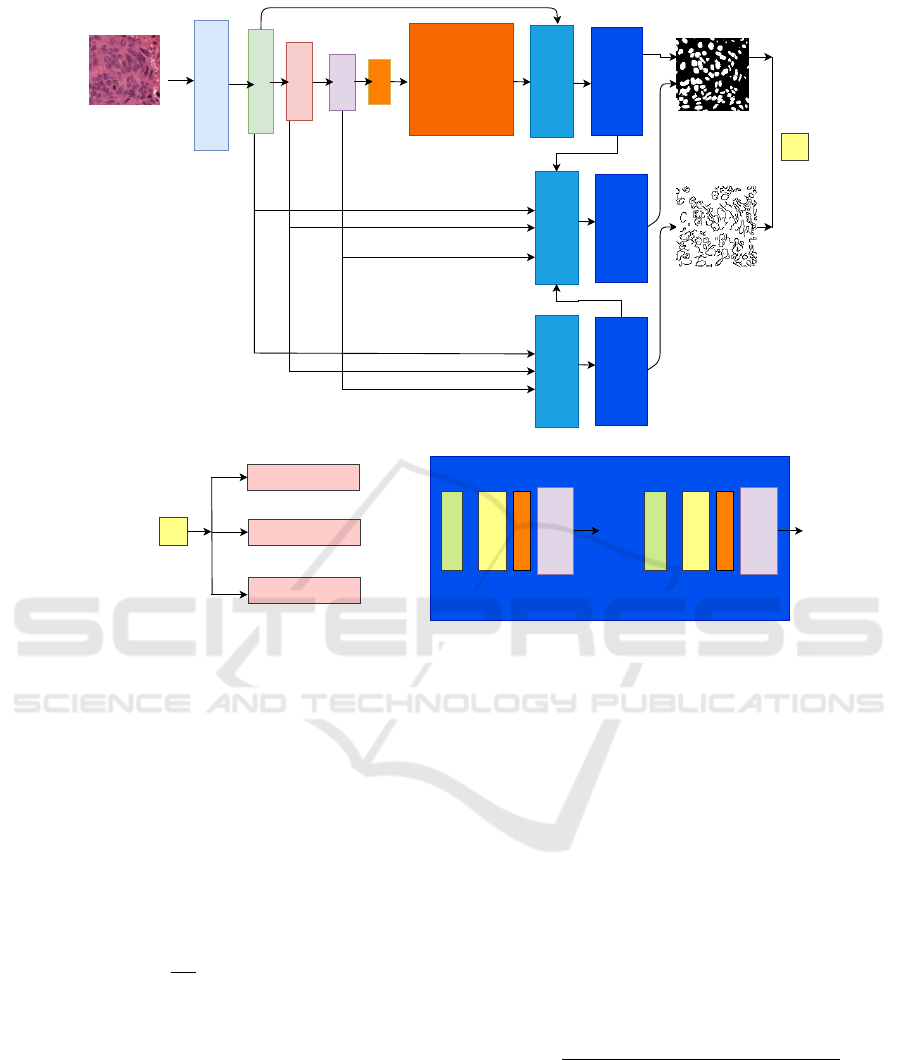
conv1
conv2
conv3
conv4
conv5
Context
Encoding
UpsampleConcat
Decoder Module
UpsampleConcat
Decoder Module
UpsampleConcat
Decoder Module
Loss
Edge
Mask
Loss
Segmentation Loss
EdgeLoss
ConsistencyConstrain
Decoder Module
conv
max channel
concat
Upsampling
conv
max channel
concat
Upsampling
* * *
a)
b)
c)
Figure 2: Self correction training mechanism.
where N is the number of pixels, K is the num-
ber of classes. To let the model facilitate the
mean intersection-over-union (mIoU) directly, the the
cross-entropy loss and the mIoU loss are combined as
follows:
E
cells
= E
e
+ L
mIoU
(7)
To preserve the consistency between the predicted nu-
clei segmentation masks and the boundary prediction,
the following constraint term is exploited:
E
consistent
=
1
|N|
∑
n∈N
pos
˜
edg
n
− edg
n
(8)
In this expression, N is the number of positive edge
pixels, edg
cn
refers to the edge maps produced by the
edge branch and
˜
edg
cn
is the edge maps produced by
the nuclei segmentation branch y
cn
i
. When computing
the loss, we should avoid that non-edge pixels domi-
nate the loss. To do so, the non-edge pixels are sup-
pressed, and only positive edge pixels cn ∈ N
pos
are
allowed to contribute to the consistency term. Here,
the ResNet-101 (He et al., 2016) is used as the back-
bone of the feature extractor with the ImageNet pre-
trained weights.
3 RESULTS AND DISCUSSION
3.1 Evaluation Metrics
Aggregated Jaccard Index (AJI) is proposed in (Ku-
mar et al., 2017) assessing the performance of nuclei
segmentation methods. If AJI equals 1, it means that
we obtain perfect nuclei segmentation results. AJI is
an modified version of the Jaccard index that divides
the aggregated intersection cardinality by the aggre-
gated union cardinality in the ground truth and seg-
mented masks. AJI can be expressed as follows:
AJI =
∑
L
i=1
GT
i
∩ NP
∗
j
(i)
∑
K
i=1
GT
i
∪ NP
∗
j
(i)
+
∑
k∈Ind
|
NP
k
|
(9)
In this expression, GT =
S
i=1,2..K
GT
i
is the ground-
truth of the nuclei pixels, NP =
S
j=1,2...L
NP
j
are the
prediction nuclei segmentation results, NP
∗
j
(i) is the
connected component from the prediction result that
maximize the Jaccard index, and Ind is the list of in-
dices of pixels that do not belong to any component
in the GT.
Channel-wise Aggregation with Self-correction Mechanism for Multi-center Multi-Organ Nuclei Segmentation in Whole Slide Imaging
469

The F1-score is the harmonic mean between pre-
cision and recall, which an be formulated as follows:
F1 =
2.T P
2.T P + FP + FN
(10)
where TP, FP, FN are the true positive, false pos-
itive, and false negative rates, respectively. The true
negative (TP) rate is defined as T N = GT ∩NP, which
is the area not belonging to any of the two masks GT
and NP.
3.2 Experimental Results and
Discussion
The dataset used in this study has been obtained from
(Kumar et al., 2017). This dataset includes 30 WSI
images with annotations from 7 organs (breast, kid-
ney, colon, stomach, prostate, liver, and bladder) col-
lected at different medical centers. The size of each
image is 1000 × 1000.
The test data includes one image from every organ
that was not exposed to the network. The rest of im-
ages is used for training. Every image in the training
and testing data was scaled to 1024x1024 and divided
into four non-overlapping patches of size 512x512.
Further random cropping of 512x512 from every im-
age was applied, as well. The overall training data
had 4906 of 512x512 patches. A batch size of one im-
age was used due the limitation in resources, namely
the GPU memory. Proposed model was trained for
50 epoch. The stochastic gradient descent (SGD) was
used as an optimizer with an initial learning rate as
1e−1, momentum as 0.99 and weight decay as1e−8.
A Titan X GPU was used to run the experiments Table
1 shows the results of the proposed method and ones
of five state-of-the-art semantic segmentation based
on deep learning models nuclei segmentation. The
five models are Fully Convolutional Network (FCN),
U-Net, Mask R-CNN, and conditional GAN (cGAN).
As shown the proposed method achieves an F1-score
of 0.876 and AJI score of 0.735. These results are bet-
ter than the ones of the previous approach (Mahmood
et al., 2019). We also show that the addition of the
channel wise aggregation improves the performance
of the baseline framework (self attention mechanism
with CE2P).
Figure 3 shows the segmentation results of the
proposed method with different organs: breast, kid-
ney, liver, prostate, bladder, colon, and stomach. As
shown, our method produces good segmentation re-
sults with bladder and stomach histopathological im-
ages with AJI scores of 0.85 and 0.83, respectively.
The proposed method gives a segmentation results
lower than 0.67 with the liver image because of the
apparent overlap between several cell nuclei.
Table 1: Comparison between the proposed model and the
related methods: FCN, U-Net, Mask R-CNN, and cGAN.
Method F1−score AJI
FCN (Long et al., 2015) 0.35 0.35
U-Net (Ronneberger et al., 2015) 0.41 0.41
Mask R-CNN (He et al., 2017) 0.50 0.50
cGAN (Mahmood et al., 2019) 0.87 0.72
Baseline 0.88 0.73
Proposed 0.89 0.74
Figure 4 shows a comparison between the number
of cell nuclei in the predicted masks and the corre-
sponding ground truth. As shown, the number of cell
nuclei obtained by the proposed method is a bit higher
than the ones of the ground-truth.
The proposed model gives promising segmenta-
tion results when we have noisy ground truth masks
because of the ambiguous boundary between differ-
ent cell nuclei and the other objects that have a simi-
lar appearance beside the overlapping and clumped.
If the cell nuclei ground-truth is almost clean, the
channel-wise aggregation, label refinement, and the
self-correction training mechanisms can be seen as an
ensembling of clones of the basic segmentation model
(i.e, CE2P), which would improve the cell nuclei seg-
mentation results and produce a generalized model
that can be used with images of different organs ac-
quired at different medical centers.
4 CONCLUSION
In this paper, we propose a new technique for ag-
gregating the channel maps of semantic segmenta-
tion models in order to improve the segmentation re-
sults of cell nuclei in histopathological images. This
technique is integrated with a self-correction learning
mechanism that can handle noisy ground truth. We
show that the proposed nuclei segmentation method
gives promising results with images of different or-
gans (e.g., breast, bladder, and colon)collected from
medical centres that use devices of different manu-
facturers and stains. Our method achieves the new
state-of-the-art. Particularly, we achieve the AJI
score of 0.735 on the Multi-Organ Nuclei Segmen-
tation benchmark, which outperforms the previous
closest approaches. In the future work, we will ex-
plore the use of different aggregation functions to
improve the segmenting cell nuclei results. We will
also use the proposed segmentation model to segment
breast masses in other modalities, such as thermogra-
phy (Abdel-Nasser et al., 2016a; Abdel-Nasser et al.,
2016b).
VISAPP 2020 - 15th International Conference on Computer Vision Theory and Applications
470

Organ: Breast
Clinic:ChristianaHealthcare
AJI=0.72
Ground truth Segmentation
Organ: Kidney
Clinic:University ofPittsburgh
AJI=0.70
Organ: Liver
Clinic:Princess Margaret Hospital
AJI=0.67
Organ: Prostate
Clinic:Roswell Park
AJI=0.72
Organ: Bladder
Clinic:Memorial Sloan
AJI=0.85
Organ: Colon
Clinic:NorthCarolina
AJI=0.73
Organ: Stomach
Clinic:HealthNetwork
AJI=0.83
Figure 3: Segmentation results of the proposed model with different organs: breast, kidney, liver, prostate, bladder, colon, and
stomach.
Channel-wise Aggregation with Self-correction Mechanism for Multi-center Multi-Organ Nuclei Segmentation in Whole Slide Imaging
471
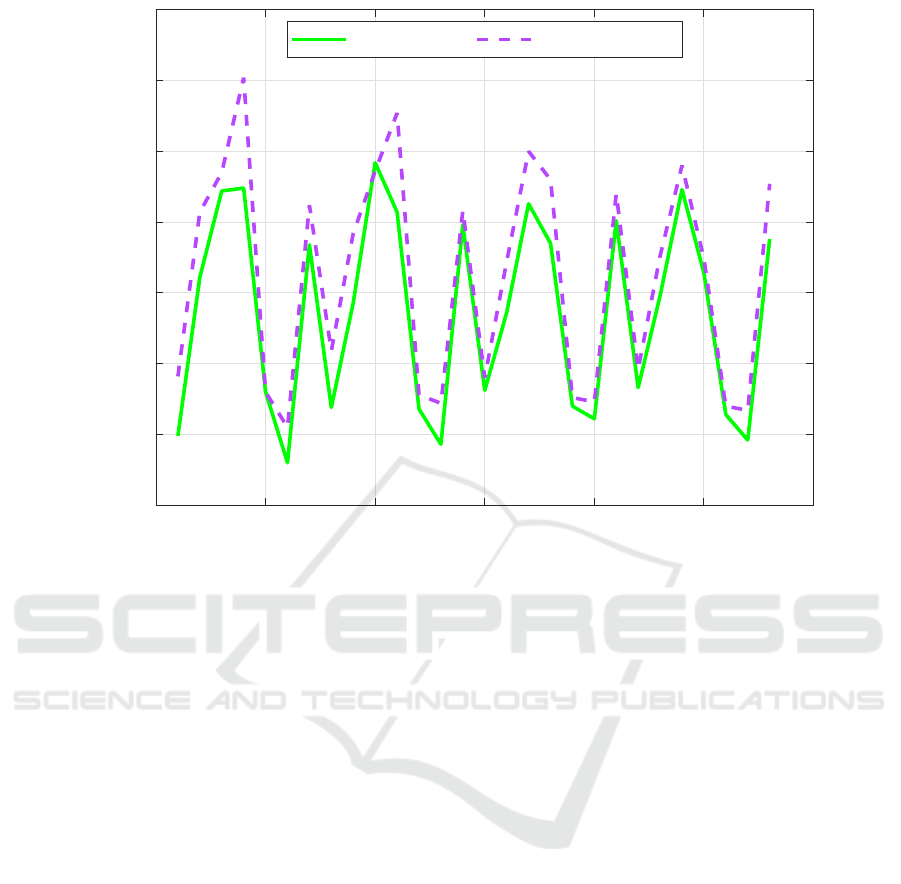
0 5 10 15 20 25 30
Patch number
0
50
100
150
200
250
300
350
Number of Cells
Ground Truth Proposed Model
Figure 4: Number of cell nuclei in the predicted masks and the corresponding ground truth.
ACKNOWLEDGEMENTS
This research was partly supported by the Spanish
Govern-ment through project DPI2016-77415-R.
REFERENCES
Abdel-Nasser, M., Moreno, A., and Puig, D. (2016a). Tem-
poral mammogram image registration using optimized
curvilinear coordinates. Computer methods and pro-
grams in biomedicine, 127:1–14.
Abdel-Nasser, M., Saleh, A., Moreno, A., and Puig, D.
(2016b). Automatic nipple detection in breast ther-
mograms. Expert Systems with Applications, 64:365–
374.
Al-Kofahi, Y., Zaltsman, A., Graves, R., Marshall, W., and
Rusu, M. (2018). A deep learning-based algorithm
for 2-d cell segmentation in microscopy images. BMC
bioinformatics, 19(1):365.
Carpenter, A. E., Jones, T. R., Lamprecht, M. R., Clarke, C.,
Kang, I. H., Friman, O., Guertin, D. A., Chang, J. H.,
Lindquist, R. A., Moffat, J., et al. (2006). Cellprofiler:
image analysis software for identifying and quantify-
ing cell phenotypes. Genome biology, 7(10):R100.
He, K., Gkioxari, G., Doll
´
ar, P., and Girshick, R. (2017).
Mask r-cnn. In Proceedings of the IEEE international
conference on computer vision, pages 2961–2969.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep resid-
ual learning for image recognition. In Proceedings of
the IEEE conference on computer vision and pattern
recognition, pages 770–778.
Kumar, N., Verma, R., Sharma, S., Bhargava, S., Vahadane,
A., and Sethi, A. (2017). A dataset and a technique for
generalized nuclear segmentation for computational
pathology. IEEE transactions on medical imaging,
36(7):1550–1560.
Li, P., Xu, Y., Wei, Y., and Yang, Y. (2019). Self-correction
for human parsing. arXiv preprint arXiv:1910.09777.
Long, J., Shelhamer, E., and Darrell, T. (2015). Fully con-
volutional networks for semantic segmentation. In
Proceedings of the IEEE conference on computer vi-
sion and pattern recognition, pages 3431–3440.
Loshchilov, I. and Hutter, F. (2016). Sgdr: Stochastic
gradient descent with warm restarts. arXiv preprint
arXiv:1608.03983.
Lucca, G., Sanz, J. A., Dimuro, G. P., Bedregal, B., Asi-
ain, M. J., Elkano, M., and Bustince, H. (2017).
Cc-integrals: Choquet-like copula-based aggregation
functions and its application in fuzzy rule-based
classification systems. Knowledge-Based Systems,
119:32–43.
Mahmood, F., Borders, D., Chen, R., McKay, G. N., Sal-
imian, K. J., Baras, A., and Durr, N. J. (2019). Deep
adversarial training for multi-organ nuclei segmenta-
tion in histopathology images. IEEE transactions on
medical imaging.
VISAPP 2020 - 15th International Conference on Computer Vision Theory and Applications
472

Moen, E., Bannon, D., Kudo, T., Graf, W., Covert, M., and
Van Valen, D. (2019). Deep learning for cellular im-
age analysis. Nature methods, page 1.
Naylor, P., La
´
e, M., Reyal, F., and Walter, T. (2017).
Nuclei segmentation in histopathology images using
deep neural networks. In 2017 IEEE 14th Inter-
national Symposium on Biomedical Imaging (ISBI
2017), pages 933–936. IEEE.
Niazi, M. K. K., Parwani, A. V., and Gurcan, M. N.
(2019). Digital pathology and artificial intelligence.
The Lancet Oncology, 20(5):e253–e261.
Qu, H., Wu, P., Huang, Q., Yi, J., Riedlinger, G. M., De,
S., and Metaxas, D. N. (2019). Weakly supervised
deep nuclei segmentation using points annotation in
histopathology images. In International Conference
on Medical Imaging with Deep Learning, pages 390–
400.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-net:
Convolutional networks for biomedical image seg-
mentation. In International Conference on Medical
image computing and computer-assisted intervention,
pages 234–241. Springer.
Ruan, T., Liu, T., Huang, Z., Wei, Y., Wei, S., and Zhao,
Y. (2019). Devil in the details: Towards accurate sin-
gle and multiple human parsing. In Proceedings of
the AAAI Conference on Artificial Intelligence, vol-
ume 33, pages 4814–4821.
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V.,
Longair, M., Pietzsch, T., Preibisch, S., Rueden, C.,
Saalfeld, S., Schmid, B., et al. (2012). Fiji: an open-
source platform for biological-image analysis. Nature
methods, 9(7):676.
Vicar, T., Balvan, J., Jaros, J., Jug, F., Kolar, R., Masarik,
M., and Gumulec, J. (2019). Cell segmentation meth-
ods for label-free contrast microscopy: review and
comprehensive comparison. BMC bioinformatics,
20(1):360.
Wang, E. K., Zhang, X., Pan, L., Cheng, C.,
Dimitrakopoulou-Strauss, A., Li, Y., and Zhe, N.
(2019). Multi-path dilated residual network for nuclei
segmentation and detection. Cells, 8(5):499.
Zhou, Y., Onder, O. F., Dou, Q., Tsougenis, E., Chen, H.,
and Heng, P.-A. (2019). Cia-net: Robust nuclei in-
stance segmentation with contour-aware information
aggregation. In International Conference on Informa-
tion Processing in Medical Imaging, pages 682–693.
Springer.
Channel-wise Aggregation with Self-correction Mechanism for Multi-center Multi-Organ Nuclei Segmentation in Whole Slide Imaging
473
