
Federated Learning on Distributed Medical Records for
Detection of Lung Nodules
Pragati Baheti, Mukul Sikka, K. V. Arya and R. Rajesh
ABV-Indian Institute of Information Technology and Management Gwalior, Gwalior, 474015, India
Keywords:
Federated Learning, Distributed Database, Decentralized training, Electronic Medical Records, Blockchain.
Abstract:
In this work, the concept of federated Learning is applied on medical records of CT scans images for detection
of pulmonary lung nodules. Instead of using the naive ways, the authors have come up with decentralizing
the training technique by bringing the model to the data rather than accumulating the data at a central place
and thus maintaining differential privacy of the records. The training on distributed electronic medical records
includes two models: detection of location of nodules and its confirmation. The experiments have been carried
out on CT scan images from LIDC dataset and the results shows that the proposed method outperformed the
existing methods in terms of detection accuracy.
1 INTRODUCTION
(Sheller et al., 2018) showed that the amount of health
data generated increases by 48% annually and may
reach 2314 exabytes by 2020. If this ’big data’ is
available for training, the model would be much ac-
curate. Training data in a central place becomes re-
source consuming but it is hard to obtain due to pri-
vacy and ownership concerns.
Federated learning(Brisimi et al., 2018) is a ma-
chine learning technique involving training of algo-
rithm across multiple decentralized servers holding
local data samples, without exchanging their data.
This approach of collaboratively learning a shared
prediction model while keeping all the training data
on device hence decoupling the ability to do machine
learning from the need to store the data at a central
place. It is a concept where AI model can be gov-
erned by multiple owners and trained securely on an
unseen, distributed dataset. Instead of bringing all the
data in one place, federated learning bring the model
to the data. This allows a data owner to maintain the
only copy of their information and solve the problem
of security. In this learning model trusted hospitals
are considered as federated severs and data-holders
where the sensitive information of their patients are
secured, yet available when needed for training. All
such hospitals acting as nodes are supposed to update
the model by further training.
In this work the concept of federated learning is
applied for detection of pulmonary lung nodules. The
lung nodules are too small to be detected manually
and is often conflicted with blood vessels and other
small underlying biological structures due to similar-
ity in shape and size. If detected, it may be wrongly
confused to be tuberculosis. Further tests are required
for surety which delays the confirmation of lung can-
cer and its treatment. This delays the survival rate of
patients by 67%(Sivakumar and Chandrasekar, 2013).
In all the traditional models, the data was accu-
mulated in a central place and trained together. But
to get a better accuracy for detection of nodules, the
data required for training should be very large. Pa-
tients do not want to share their medical records even
for research purposes. Our focus was to improve the
accuracy by increasing the dataset while working for
the privacy as well. Medical records should contribute
to training explicitly without actually being shared.
To deal with patient’s differential privacy many
theories were proposed. Applying the model on dis-
tributed databases and accumulating the updates for
further improving the initial model from different
nodes forms the basis of federated learning.
In case of sketched update(Kone
ˇ
cn
`
y et al.,
2016),the individual data centres apply the model on
their data and send complete the updated model to
the central trusted server and replace the initial model
with the sent model. Another approach highlighted
was that of structured approach in which instead of
sending the entire model, they send the updates in
the form of model parameters changes. The sketched
method of updation forms the bottleneck of federated
Baheti, P., Sikka, M., Arya, K. and Rajesh, R.
Federated Learning on Distributed Medical Records for Detection of Lung Nodules.
DOI: 10.5220/0009144704450451
In Proceedings of the 15th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2020), pages 445-451
ISBN: 978-989-758-402-2
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
445

learning due to bandwidth limitation. If the model
stored at the Central server is completely replaced by
new model, it would require good link connection and
would increase the number of bits being uploaded. In
the structured way of updation, it may lead to devia-
tion from accuracy. (Kim et al., 2019) has shown that
assigning the equal weightage to a model update with
less no of data samples and the model update with
more number of data samples may lead to misleading
results.
In literature many methods have been proposed on
detection of lung nodules. One such method proposed
by (Murphy et al., 2009) forms clusters of closely oc-
curring volumes and hence, forming one large vol-
ume by application of KNN in order to reduce false
cases. The input to such model were shape index,
maximum and minimum dimensions of structures to
be considered as nodules, number of voxels to be con-
sidered in the cluster to be classified into nodular or
non-nodular region the drawback of this method of
supervised classification using KNN/SVM is the in-
crease in false positives as two or more non-nodular
regions of smaller volumes in cluster may be portrait
as a bigger voxel giving the false essence of a nodule.
The intensity based genetic method of detection
(Dehmeshki et al., 2007) of cancer initiating lung
nodules based on intensity in which the intensity in-
side lung nodules is higher than the surrounding vol-
ume. The shape based detection that takes the index
of sphere as 1 and of blood vessels as 0.75 whereas
threshold of nodules is taken to be 0.95. But this
method leads to difficulty in identifying nodules of
irregular shapes and density patterns. The partly solid
and non-solid nodules are not detected as they fail to
cross the threshold value of the shape index. The nod-
ules close to the pleural surface and those attached
to blood vessels are also not detected leading to false
cases.
2 PROCEDURE OF WORK
This work of decentralizing the ML model in dis-
tributed databases for detection of lung nodules and
predict its severity followed a series of work. It started
with 1. Designing the initial ML model 2. Distribut-
ing the model to all nodes while ensuring the security
3. Updating the model.
2.1 Initial Model
The basic model deployed in this work is an integra-
tion of two sequential models. The first model detects
the occurrence of nodules while the second model
confirms it’s presence. The different stages of the pro-
posed model are discussed below:
2.1.1 Dataset Acquisition
The LIDC dataset used for detection of pulmonary
nodules contains 1010 CT scans from 1010 different
patients(Armato III et al., 2011). Seven cases where
the scans were incomplete are excluded. Each scan
was checked and annotated by four radiologists man-
ually and a total of 2632 nodules were found in the
dataset. ’annotations’ is a csv file included in the
dataset that contains the information which are used
as a standard reference for nodule detection. An-
other csv file under the name ’candidates’ used for the
LUNA16 workshop contains a set of candidate loca-
tions for checking the correctness and completeness
of the nodule location thereby ensuring reduction in
false cases.
2.1.2 Preprocessing
Segmentation of lungs from the surrounding region
was the first step involved where lungs from the CT
scan images are segmented by using predefined edge
detection techniques so having the focus is within the
pulmonary region.
The next step in pre-processing is masking of the
segmented lung images to highlight the region of in-
terest based on the annotations file in LIDC dataset.
Based on the coordinate values and the radius of the
nodules specifies in the annotations.
Both the segmented and masked images are sliced
layer by layer. The segmented images serve as input
images and masked images as corresponding labels.
The masked and segmented images to be fed as input
to the model are concentrated to the desired region of
interest where the nodules are present. The size of the
largest nodule found in the LIDC dataset upon traver-
sal through 2632 nodules is not more than 64× 64 pix-
els. Therefore, the setting of the ROI to this size will
sufficiently enclose all nodules in the LIDC dataset
and remove any chances of missing out any nodule.
For this, the segmented images and masked images
are converted to 64 × 64 pixels images and stacked
into 16 layers.
For the second model, cubes of size of the nodules
are generated based on the candidates file that con-
tains the location as coordinates and radius and the
corresponding label as 1/0 denoting nodular or non-
nodular region respectively. The number of dataset
examples depicting nodular regions labeled as 1 were
much less than non-nodular region. So, balancing
of dataset is required. Hence, augmentation is used
which is a technique that can be used to artificially
VISAPP 2020 - 15th International Conference on Computer Vision Theory and Applications
446
expand the size of a training dataset by creating mod-
ified versions of images in the dataset. Image data
augmentation is used to expand the training dataset in
order to improve the performance and ability of the
model to generalize. Here, the number of cases with
diseases is relatively smaller than the number of nor-
mal cases So each nodule is rotated in x, y, z direc-
tion. After augmentation, the class 1 examples were
increased by 40 times to balanced the other class.
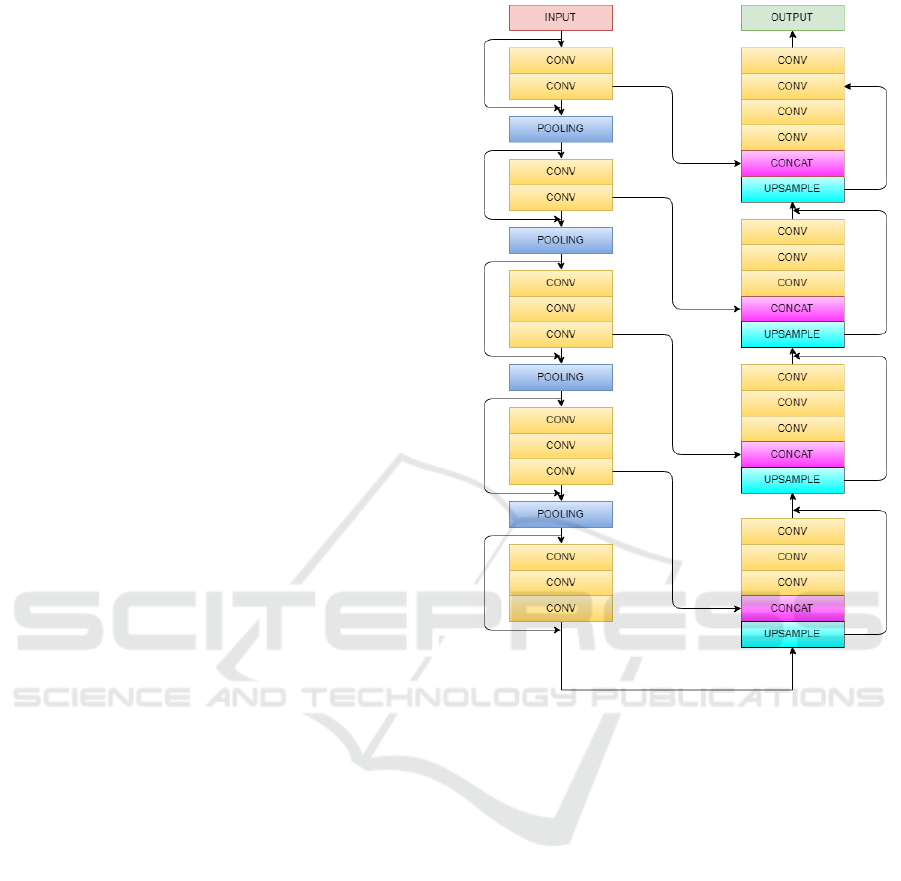
2.1.3 Architecture of Models
A feed forward network with a single massive layer
is prone to over fitting the data. Therefore, there is
a common trend that the network architecture should
go deeper not wider as long and wider network tend
to have too many weights and is not good in terms of
memory usage and computational speed. A good way
to make the model learn interesting features deeper
and by working on kernel surface. However, it has
been noticed that after some depth, the performance
degrades.
The core idea is that stacking the layers or making
the model deeper should not degrade the performance
and the loss from the current model should be less
than the residual layer. Stacking the layers is not a so-
lution due to vanishing gradient problem. The authors
have used skip connections for the better functioning
of model, the stack of layers over which each skip
connections passes are called residual blocks which
helps to calculate the true output and also makes it
easier to calculate identity function i.e. setting the
output of residual block to zero if it doesn’t contribute
for the betterment of model.
A combination of downsampling and upsampling
is done in our model. Downsampling is done to teach
the network to focus on fewer activation points than
all of it to reduce the redundancy in the feature map.
Reducing the number of parameters ensures higher
computational speeds and makes the output tolerant
to small translational changes in input. Downsam-
pling might cause information loss due to its role of
extracting only the useful information.
Upsampling is applied after downsampling when
the features are reduced to a one dimensional array.
It is done to enlarge the sparse feature maps and im-
prove the resolution. This layer densifies these feature
maps through convolution-like operations with mul-
tiple trainable filters and has nothing to do with re-
construction of lost information. The sole purpoe of
applying down/up sampling layrs is to reduce compu-
tations in each layer which keeping the dimension of
input/output as before.
Convolution layer is the first layer applied that
makes the relationship between pixels of learning im-
Figure 1: Schematic representation of Vnet model for ex-
traction of feature maps.
age using small squares of 96 × 96 × 16 of input im-
age. Convolving the image with different filters or
kernels helps to extract better features. The stride is
2 × 2 × 2 as our kernel moves by 2 pixels at a time.
We drop the part of the image where the filter did not
fit and keep only the valid part of the image. ReLu is
non linear action function which consist of two linear
equation can be written as
f (x) = max(0, x)
which helps to bring non-linearities in the network.
Stacking convolution layers vertically and keep-
ing the kernel size 3× 3× 3 which helps to makes the
model lighter and tends to improve the results. Each
input image will pass through a series a convolution
layers with kernals(filters) and pooling the image and
downsampling it to a particular value. The gradients
can flow directly through the skip connections back-
wards from later layers to initial filters.
Pooling is done after a sequence of convolution
layers to reduce the number of parameters and down
Federated Learning on Distributed Medical Records for Detection of Lung Nodules
447

sample the image while retaining the important prop-
erties as well. We apply max pooling which takes the
maximum of the pixel value from the feature map.
The first model is an implementation of Vnet 3D
architecture(Milletari et al., 2016) in which we gave
input of 16 images stacked upon each other which
contains the nodules. First the image is downsam-
pled to extract important features at each step, at ev-
ery step the feature map calculated are two times more
interesting than that of previous layer. Output of ev-
ery layer is connected to output of previous layer to
know if this layer plays a role in the proposed model.
After this Upsampling is done to further extract im-
portant features and to expand low resolution feature
map generated after down sampling. Down sampling
helps in better extraction of features whereas up sam-
pling gives overall presence of features, in our case as
lung nodules.
In second model Resnet architecture(He et al.,
2016) is used that comprises of convolution layers for
downsampling until the feature map is flattened. This
is fed to a fully connected layer with an activation
function of Softmax to give the probabilistic value to
classify into 1/0 classes.
2.1.4 Integration of Models
The output of the first model that gives the predicted
region of nodules in the CT scans as 384 × 384 pix-
els image which is a collection of 16 layers each of
96 × 96 in the form of a 4 × 4 grid structure which is
fed to our second model that cross validates the pre-
dicted ROI. This provides confidence that the nodules
are present in this region by classifying into nodular
and non-nodular with a certain probability.
2.2 Network
The checkpoints that include meta graph, index and
data files are saved after training. The meta graph de-
scribes the structure of the models which are formed
by various layers described above. Index file is an im-
mutable object that contains the name of the variables
used in each layer and data file is a collection that
saves the value of all such variables used in the graph.
These files are added to the IPFS(Benet, 2014) server
which acts as a protocol and creates a network for
a content-addressable, peer-to-peer method of storing
and sharing resources in a distributed file system.The
IPFS provides an accessible hash link given to all the
nodes that want to use the model. Once a node gets all
these files it creates the structure of the graph and ini-
tializes the values of the variables from the data files.
The structure of learning process used in the pro-
posed model is depicted in Fig2, where all the hos-
Figure 2: Federated Learning Architecture.
pitals acting as nodes can use this link generated
to download the latest model/global updates. The
nodes can further train this model on their accessi-
ble EMR’s. In this the differential privacy of the
records are maintained still being used for training the
model explicitly and come up with updates in form of
changes in model parameters as local updates.
The structure of the model is not changed in any of
the further iterations and for further updates only the
data files consisting of the values of the variables are
sent through the network. This file contains array of
numbers which consumes very small space and hence,
requires a small bandwidth. These changes are stored
in IPFS and to maintain the security. The IPFS hash
can only be accessed by the central accumulator by
generating a pair of key pairs for every node. The
generated IPFS link after storing the weight changes
is first encrypted with the public key of the central
accumulator and stored in blockchain. To access this
encrypted hash it is decrypted by the owner using its
private key.
2.3 Updation
The aggregation should be such that the local updates
i.e. the updates which are sent from every node af-
ter training model on the nodes with minimum loss
with maximum records should be given more weigh-
tage as compared to the others and not the standard
techniques.If the aggregation was only on the basis
of number of files(Kim et al., 2019), we came to no-
tice that it was biased as the nodes that processed less
number of files but with negligible loss would not be
given importance. Also, if only loss was considered
as the deciding factor it would be wrong as the nodes
that processed very less files would naturally come
up with lower loss and be given more priority which
may lead result in wrong direction. So, here must be
a trade off between the loss and the number of files.
VISAPP 2020 - 15th International Conference on Computer Vision Theory and Applications
448
The updates from nodes are computed using eq(1).
W
t+1
= W
t
+
∑
n
i=1
h
ti
W
Ni
W
Li
∑
n
i=1
W
Ni
W
Li
(1)
W
Li
=
L
ti
∑
n
i=1
L
ti
× 100
(2)
W
Ni
=
N
ti
∑
n
i=1
N
ti
× 100
(3)
where
h
ti
= W
ti
− W
t
(4)
W
t
: Weights distributed to all the nodes
W
ti
: New Weights calculated by i
th
node on its
own dataset
n : No of nodes in distributed network
t : No of iterations/updates performed
h
ti
: Weight difference of i
th
node
L
ti
: Learning Loss of i
th
node in t
th
iteration
N
ti
: No. of reports processed by i
th
node in t
th
iteration
Eq (2). signifies the normalized loss weightage
which is the ratio of the loss of the i
th
node to the sum-
mation of the loses from all the nodes in the present
updation performed.
Eq (3). signifies the normalized files weightage
which is the ratio of the number of files processed by
the i
th
node to all the files processed till then in the
present updation.
The updated weights are the aggregation of the
weights from the previous iteration and the change in
weights obtained by the above aggregation technique.
3 TRAINING
In the training mechanism initial model was trained
for 5000 cases in batch of 16 for 50000 epochs.Then
this latest model saved to IPFS server and the cor-
responding link is added to blockchain. Verified
users access that link to download the global updates.
Three nodes acting as updation servers were taken
for testing with sample size of 2500, 1500 and 500
CT scan images. Local Updates are generated after
these nodes train the model on their local records.
These updates from all the nodes are accumulated at
the federated server and aggregated by a time based
scheduler- cron. The aggregation follows the mecha-
nism of normalization stated above which leads to the
global update and provides weightage to all the local
updates. The central model is updated according to
the global update and is ready to follow the next it-
eration of updates. These server run 4,3,5 iteration
respectively of the above steps for the better aggre-
gation of models. This batch size of 500,1500,2500
records are not accumulated at a central place and is
not shared among other nodes but it’s inference is use-
ful to all the models.
4 TESTING & RESULTS
For testing, the latest model is initialized by apply-
ing the global update that was stored in blockchain
through federated server. The input to the first model
is provided with a random image from the dataset in
a batch of 16. The output of which was sent to the
second model focusing on the nodule region .
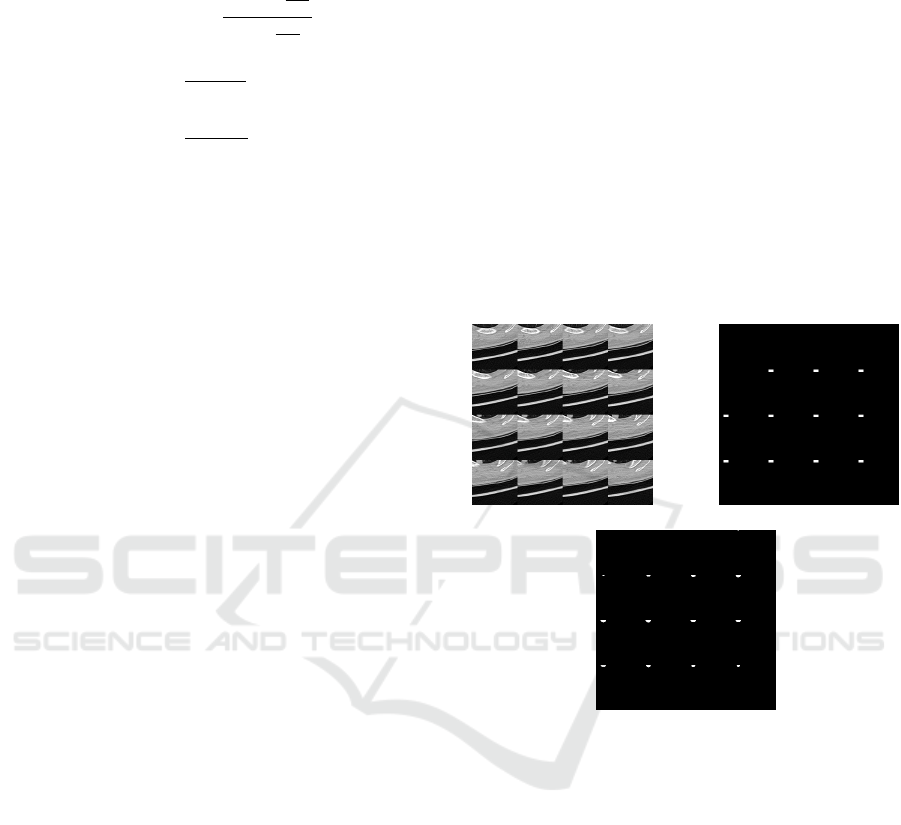
(a) (b)
(c)
Figure 3: (a) Input test image taken in a batch of 16 (b) Ex-
pected masked image highlighting the region of interest of
nodules (c) Predicted image obtained from our first model.
The set of 16 layers of one of the test image for
the first model is shown in Fig 3(a). The predicted
image for the layer by layer input of the segmented
image is shown in Fig 3(c) which is very much close
to the expected image of Fig 3(b). The white patches
in the figure signifies the presence of nodule in the
corresponding layer.
The accuracy of the first model is measured
in terms of similarity index between the predicted
masked image and the actual masked image by us-
ing cumulative color histograms(Stricker and Orengo,
1995).
The overall accuracy of first model (tested over
1305 data samples) was found to be 90.87% and that
was 97.65% for the second model (tested over 8750
samples). The loss of the first model is gauged based
Federated Learning on Distributed Medical Records for Detection of Lung Nodules
449
on the metric of dice coefficient that works as on find-
ing similarity between the masked and predicted im-
age keeping our prime focus in the white patches de-
noting the nodular regions. The Dice score is not only
a measure of how many positives pixel match found,
but it also penalizes for the false matches that the
method found. For the second model, cross-entropy is
used that measures the performance of classification
model whose output is a probability value between 0
and 1. Cross-entropy increases as the predicted prob-
ability diverges from the actual label. The training
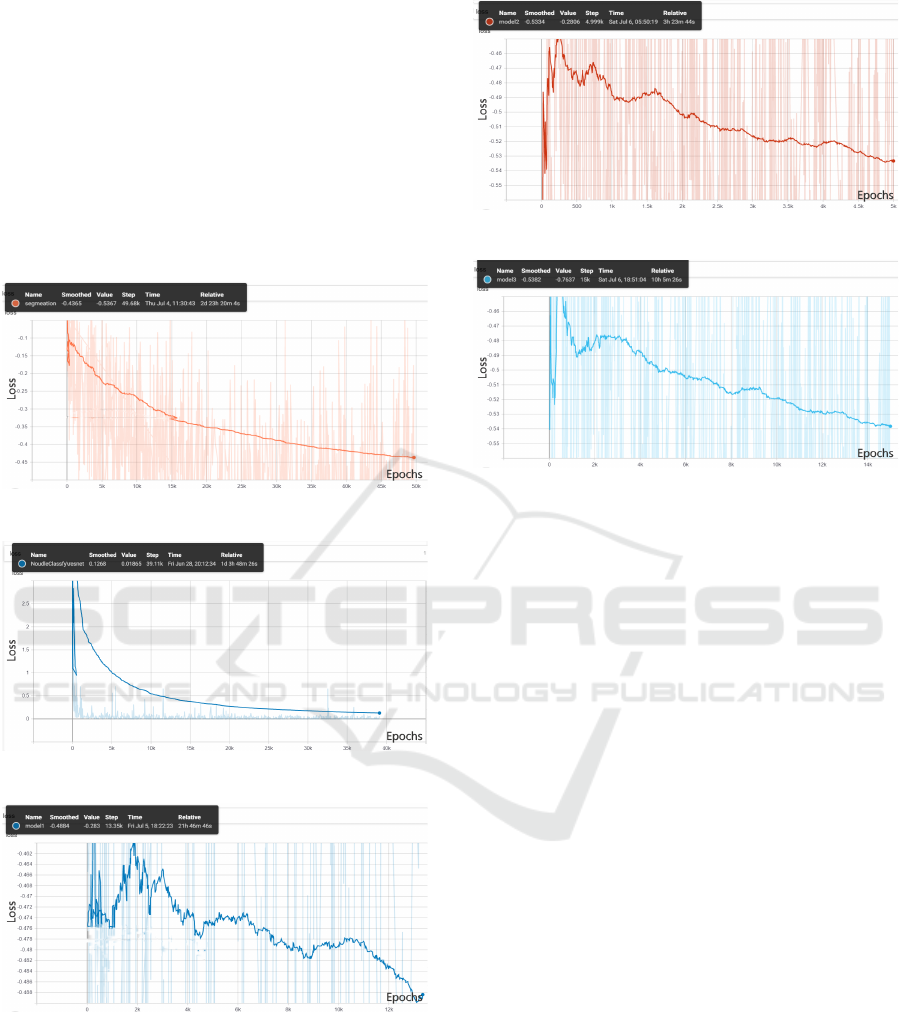
loss graphs are shown in Fig 4-Fig 8.
Figure 4: Initial first model loss vs epochs.
Figure 5: Initial second model loss vs epochs.
Figure 6: Logs of first node loss vs training epochs.
5 CONCLUSION
In this paper an approach is proposed to effectively
utilize the medical health records for disease predic-
tion using federated learning. The proposed approach
helps to maintain health records for disease predic-
tion using federated learning. The proposed approach
Figure 7: Logs of second node loss vs training epochs.
Figure 8: Logs of third node loss vs training epochs.
helps to maintain medical ethics and confidentiality of
patients record by training the system even on unseen
data in a distributed environment. This decentralized
training reduces the time of computation by not accu-
mulating the huge data at a central place. The initial
model is distributed and the updates are aggregated
by maintaining the trade off between the number of
samples and error. The proposed model evaluated on
LIDC dataset has outperformed all the existing mod-
els by exhibiting the prediction accuracy of 97.65%.
REFERENCES
Armato III, S. G., McLennan, G., Bidaut, L., McNitt-Gray,
M. F., Meyer, C. R., Reeves, A. P., Zhao, B., Aberle,
D. R., Henschke, C. I., Hoffman, E. A., et al. (2011).
The lung image database consortium (lidc) and image
database resource initiative (idri): a completed refer-
ence database of lung nodules on ct scans. Medical
Physics, 38(2):915–931.
Benet, J. (2014). Ipfs-content addressed, versioned, p2p file
system. arXiv preprint arXiv:1407.3561.
Brisimi, T. S., Chen, R., Mela, T., Olshevsky, A., Pascha-
lidis, I. C., and Shi, W. (2018). Federated learning
of predictive models from federated electronic health
records. International Journal of Medical Informatics,
112:59–67.
Dehmeshki, J., Ye, X., Lin, X., Valdivieso, M., and Amin,
H. (2007). Automated detection of lung nodules in
ct images using shape-based genetic algorithm. Com-
puterized Medical Imaging and Graphics, 31(6):408–
417.
VISAPP 2020 - 15th International Conference on Computer Vision Theory and Applications
450

He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep resid-
ual learning for image recognition. In 2016 IEEE Con-
ference on Computer Vision and Pattern Recognition
(CVPR), pages 770–778.
Kim, H., Park, J., Bennis, M., and Kim, S.-L. (2019).
Blockchained on-device federated learning. IEEE
Communications Letters.
Kone
ˇ
cn
`
y, J., McMahan, H. B., Yu, F. X., Richt
´
arik, P.,
Suresh, A. T., and Bacon, D. (2016). Federated learn-
ing: Strategies for improving communication effi-
ciency. arXiv preprint arXiv:1610.05492.
Milletari, F., Navab, N., and Ahmadi, S.-A. (2016). V-
net: Fully convolutional neural networks for volumet-
ric medical image segmentation. In 2016 Fourth Inter-
national Conference on 3D Vision (3DV), pages 565–
571. IEEE.
Murphy, K., van Ginneken, B., Schilham, A. M., De Hoop,
B., Gietema, H., and Prokop, M. (2009). A large-
scale evaluation of automatic pulmonary nodule de-
tection in chest ct using local image features and k-
nearest-neighbour classification. Medical Image Anal-
ysis, 13(5):757–770.
Sheller, M. J., Reina, G. A., Edwards, B., Martin, J., and
Bakas, S. (2018). Multi-institutional deep learning
modeling without sharing patient data: A feasibil-
ity study on brain tumor segmentation. In Interna-
tional MICCAI Brainlesion Workshop, pages 92–104.
Springer.
Sivakumar, S. and Chandrasekar, C. (2013). Lung nodule
detection using fuzzy clustering and support vector
machines. International Journal of Engineering and
Technology, 5(1):179–185.
Stricker, M. A. and Orengo, M. (1995). Similarity of color
images. In Storage and retrieval for image and video
databases III, volume 2420, pages 381–392. Interna-
tional Society for Optics and Photonics.
Federated Learning on Distributed Medical Records for Detection of Lung Nodules
451
