
A Manifold Learning Framework for the Detection of Cardiac Disorders
in Acoustic Signals
Keren Hochman
1
, Amir Averbuch
1
, Alon Schclar
2
and Raid Saabni
2
1
School of Computer Science, Tel Aviv University, POB 39040, Tel Aviv 69978, Israel
2
School of Computer Science, The Academic College of Tel-Aviv Yaffo, POB 8401, Tel Aviv 61083, Israel
Keywords:
Cardiac Disorder Detection, Manifold learning, Dimensionality Reduction, Acoustic Signal.
Abstract:
Cardiac disorders are clinical situations in which the heart does not function properly. These disorders may
be fatal to patients if they are not detected. Detecting such disorders often involves special and in some cases
very expensive medical devices such as Computer Tomography (CT), Magnetic Resonance Imaging (MRI),
Ultrasound imaging or Electrocardiograms. Acoustic detection of these disorders by simply listening to the
heart using a stethoscope - although being the cheapest detection method - requires a highly skilled doctor. We
propose a method that detects cardiac disorders from simple acoustic recordings of the heart. Acquiring such
recording is in most cases cheaper than the above mentioned devices. The proposed algorithm is composed
of two steps: an offline training step which constructs a classifier based on labeled recordings; and an online
classification step which detects cardiac disorders given a recording of the heart. Given the online nature
of the algorithm, the proposed algorithm can be implemented as a smartphone application. One of the key
elements of oth the training and detection steps is the concise and informative representation of the acoustic
signal. This representation is obtained using the application of the spline wavelet packet transform followed
by the application of the Diffusion Maps (DM) dimensionality reduction algorithm. The proposed approach is
generic and can be applied to various signal types for solving different classification problems.
1 INTRODUCTION
Classification of acoustic signals is a contemporary
problem whose application is found in many domains
e.g. Biology (Mac et al., 2018), Surveillance (Mu-
nich, 2004; Yaan Li and Zhe Chen, 2017; Schclar
et al., 2010; Averbuch et al., 2001; Averbuch et al.,
2004) and oceanic sciences (D.A.Abraham, 2019),
to name a few. In this paper we focus on detec-
tion of cardiac disorders using acoustic recordings of
the heart. The underlying assumption is that record-
ings of cardiac disorders have distinctive acoustic sig-
natures which differ from the acoustic signatures of
healthy hearts. In order to detect cardiac disorders us-
ing acoustic recordings, one must extract and recog-
nize these definitive signatures. These signatures can
be found in small intervals in the recording. There-
fore, we decompose the signal into short overlapping
windows that are used by the proposed algorithm.
Each window is treated as a high dimensional data
point.
Using the raw signal is inefficient and produces
poor results since the signal contains redundant in-
formation and noise. The redundant information is
partly due to the quasi-periodic structure of the signal
which contains only a small number of dominant fre-
quencies. Accordingly, we apply the Spline Wavelet
Packet Transform (SWPT) (Daubechies, 1992) to
each window since we assume that acoustic sig-
natures are inherent in the energy of the wavelet
packet coefficients. Furthermore, SWPT sparsifies
the smooth parts of the signal, providing better energy
compactization of the signal in a few wavelet packet
coefficients and provides better frequency coverage of
the signal.
In order to remove noise that is still present after
the application of the SWPT and derive a more con-
cise representation (using the high dimensional result
of the SWPT is impractical due to the curse of dimen-
sionality) we apply the Diffusion Maps (DM) (Coif-
man and Lafon, 2006; Schclar, 2008) dimensionality
reduction algorithm to the result of the SWPT. This
embeds the high dimensional result of the SWPT into
a lower dimension space. We chose the DM algorithm
since it was successfully applied in various algorithms
(Lafon and Lee, 2006; Lafon et al., 2006; Rabin and
Coifman, 2012; Deng and Han, 2016; Sulam et al.,
2017).
In order to derive the concise representation of a
new signal s
new
, we use the Nystr
¨
om out-of-sample
192
Hochman, K., Averbuch, A., Schclar, A. and Saabni, R.
A Manifold Learning Framework for the Detection of Cardiac Disorders in Acoustic Signals.
DOI: 10.5220/0009094401920197
In Proceedings of the 9th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2020), pages 192-197
ISBN: 978-989-758-397-1; ISSN: 2184-4313
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

extension (Nystr
¨
om, 1928) algorithm instead of ap-
plying the DM algorithm. We do so since the com-
plexity of the Nystr
¨
om algorithm is linear while the
complexity of the DM algorithm is quadratic.
The proposed algorithm is composed of two
stages:
• An offline training stage in which data with a-
priory knowledge is analyzed and features, which
characterize it, are extracted and stored.
• An online detection phase in which a new signal
that was part of the training stage undergoes pro-
cessing stages that are similar to the training stage.
The processing outcome is compared using a sim-
ple k-nearest neighbor scheme to the database that
was constructed in the training stage.
The rest of the paper is organized as follows. In Sec-
tion 2 we briefly describe the SWPT, DM and the
Nystr
¨
om algorithms. In section 3 the proposed algo-
rithm is presented. Experimental results are presented
in section 4. In section 5 we summarize the results
and outline the next steps in this research.
2 MATHEMATICAL
BACKGROUND
2.1 Spline Wavelet Packets
There are many wavelet packet libraries which dif-
fer from each other by their generating low-pass and
high-pass filters, the shape of their basic waveforms
and their frequency contents. In principle, the trans-
form of a signal of length n = 2
j
can be implemented
up to the j
th
decomposition level (scale). At this
level, there exist n different waveforms, which are
close to sine and cosine waves with multiple frequen-
cies. There is a duality in the nature of the wavelet
packet. As the decomposition level increases, a better
frequency resolution at the expense of time domain
resolution is achieved and vice versa.
Figure 1 displays a wavelet packet after decompo-
sition into three levels by a spline of sixth order (Bat-
tle -Lemarie (Daubechies, 1992)). The splines do not
have a compact support in the time domain. However,
they produce an excellent splitting of the frequency
domain (see Fig. 1-b).
The advantage of spline wavelet is that it produces
a good split of the frequency domain, however, it is
not localized as well as other wavelet packets. In this
paper we chose the sixth order spline wavelet packets
since it reduces the overlap between frequency bands
associated with different decomposition blocks, while
Figure 1: Spline of the sixth order wavelet packet in the
third scale‘.
providing a variety of waveforms with a fair time do-
main localization.
2.2 The Diffusion Maps Algorithm
Diffusion maps (Coifman and Lafon, 2006; Schclar,
2008), is a nonlinear dimensionality reduction
method. The eigenfunctions of a Markov matrix,
which define a random walk on the data, are used to
construct coordinates that provide concise representa-
tions of the underlying data sets where the geometry
of the original data set is preserved.
Consider a data set Ω = {x
1
,...x
n
}, x
i
∈ R
m
, m ∈
N. An undirected weighted graph in which each node
corresponds the a data point is constructed. Every pair
of nodes x
i
,x
j
∈ Ω, i 6= j, 1 ≤ i < j ≤ n is connected
by an edge whose weight is given by a symmet-
ric, point-wise non-negative weight function w(x
i
,x
j
),
which reflects the similarity between x
i
and x
j
. The
DM algorithm embeds Ω into a low-dimensional L
2
space in which the Euclidean distance between points
i and j approximates the connectivity between these
points in the graph. The connectivity is referred to as
the diffusion distance.
The choice of the weight function is dependents
on the application as long as the conditions of sym-
metry and non-negativity are kept. A common choice
for the kernel function, w(x
i
,x
j
), is the Gaussian ker-
nel
w(x
i
,x
j
) = exp
−
||x
i
− x
j
||
2
ε
(1)
where ε > 0 is the scale parameter. Discussion on how
to choose ε can be found in (Schclar and Averbuch,
2015).
We now create a random walk on the data set
Ω by forming the kernel p(x,y) =
w(x,y)
d(x)
where
d(x) =
∑
z∈Ω
w(x,z) is the degree of x. Since p(x,y) ≥
0 and
∑
y∈Ω
p(x,y) = 1, this defines a Markov chain
A Manifold Learning Framework for the Detection of Cardiac Disorders in Acoustic Signals
193

where p(x, y) is the probability to jump from x to y
in a single step. Let P be the N × N transition ma-
trix of this Markov chain. Let p
t
(x,y) be the ker-
nel corresponding to the t
th
power of matrix P, that
is, p
t
(x,y) is the transition probability matrix in t
time steps. When t →+∞, this Markov chain is gov-
erned by a unique stationary distribution φ
0
, such that
lim
t→∞
p
t
(x,y) = φ
0
(y) for all x and y. φ
0
is the top
left eigenvector of P, φ
T
0
P = φ
T
0
, and φ
0
(y) =
d(y)
∑
∈Ω
d(z)
holds. Let {φ
l
} and {ψ
l
} be the corresponding bi-
orthogonal left and right eigenvectors of P. The fol-
lowing eigendecomposition exists
p
t
(x,y) =
n−1
∑
l=0
λ
t
l
ψ
l
(x)φ
l
(y) (2)
where {λ
l
} is the sequence of eigenvalues of P (with
|λ
0
| ≥ |λ
1
|...).The diffusion distance between two
points x and z was introduced in (Coifman and Lafon,
2006) as
D
2
t
(x,z) =
∑
y∈Ω
((p
t
(x,y) − (p
t
(z,y))
2
φ
0
(y)
. (3)
This distance measures the connectivity between x
and z since it involves an integration along all paths
of length t starting from x or z. Unlike the shortest
path or geodesic distance, this metric is robust to noise
(perturbation) due to this integration.
The connection between the diffusion distance
and the eigenvectors is given by
D
2
t
(x,z) =
∑
l≥1
λ
2t
(ψ
l
(x) − ψ
l
(z))
2
. (4)
Note that ψ
0
does not appear in the sum because it is
a constant.
Due to the spectrum decay, only a few number of
terms are needed to achieve a given relative accuracy
δ > 0. The number of needed terms is denoted by
m(t). From Eq. 4 it follows that the right eigenvector
can be used to compute the diffusion distance and thus
the diffusion map is defined as
Ψ
t
: x → (λ
t
1
ψ
1
(x),λ
t
2
ψ
2
(x)...,λ
t
m(t)
ψ
m(t)
(x))
T
. (5)
This mapping provides coordinates for the data set
Ω and embeds the n data points into the Euclidean
space R
m(t)
. The dimensionality is reduced due to the
fast decay of {λ
l
} that ensures that m(t) << m.
2.2.1 The Nystr
¨
om Out-of-sample Extension
The Nystr
¨
om extension (NE) (Nystr
¨
om, 1928;
Williams and Seeger, 2000; Fowlkes et al., 2004) ex-
tends a known function on a given data set to include
a new data point which is not int the date set. The
NE algorithm uses both the target function and the
geometry of the training set. We use NE to embed
a new signal into the low-dimensional representation
of the training set. The Nystr
¨
om extension has been
successfully used numerous problems in the past e.g.
to accelerate kernel machines (Williams and Seeger,
2000) and spectral clustering (Fowlkes et al., 2004),
to name a few.
Let Ω be a data set and let Ψ
t
be its diffusion em-
bedding map. µ
l
and φ
l
are the eigenvalues and eigen-
vectors, respectively, of the Gaussian kernel with
width σ on the training data Ω. Denote by Ω the new
data set. σ > 0 defines the scale of the extension.
Then, µ
l
φ
l
(x) =
∑
y∈Ω
e
−||x−y||
2
/σ
2
φ
l
(y), x ∈ Ω.
Since the kernel can be evaluated in the entire space,
it is possible to use any x ∈ R
d
on the right side of the
identity.
The Nystr
¨
om extension (Nystr
¨
om, 1928) from
Ωto R
d
of the eigenfunctions is defined as:
¯
φ
l
(x) =
1
µ
l
∑
y∈Ω
e
−||x−y||
2
/σ
2
φ
l
(y), x ∈ R
d
. (6)
Any function on the training set can be decom-
posed into
f (x) =
∑
l
h
φ
l
, f
i
φ
l
(x), x ∈ Ω. (7)
The Nystr
¨
om extension of f on the rest of R
d
is
given by
¯
f (x) =
∑
l
h
φ
l
, f
i
¯
φ
l
(x), x ∈ R
d
. (8)
In particular, f can be every coordinate in the embed-
ding that the DM algorithm produces.
3 THE PROPOSED ALGORITHM
The classification algorithms for processing acoustic
signals is split into two stages, training and detection.
The input to the training stage is a data set Ω =
{s
i
}
n
i=1
that is composed of recordings signals in
a pulse mode modulation (PCM) format of healthy
hearts and hearts that suffer from disorders. The
signals may vary in their length and their class/type
are known a-priory. The training stage constructs a
concise representation of the training signals. This
is achieved by embedding the signals into a low-
dimensional space. The detection phase embeds new
signals into the low-dimensional space that was con-
structed during the training phase. A new signal
is classified into health/unhealthy heart via a simple
nearest neighbor scheme.
Both stages share common data preparation steps
which are described in Algorithm 1 and are detailed
below.
ICPRAM 2020 - 9th International Conference on Pattern Recognition Applications and Methods
194

Algorithm 1: Preprocessing of a signal s.
1. Decomposition of the signal s into overlapping
windows.
2. Application of the spline wavelet packet trans-
form to each window.
3. Summing the wavelet coefficients in each fre-
quency band.
4. Averaging every µ consecutive windows obtained
in step 2 in order to reduce the noise where µ > 0
is a parameter that indicates the number of win-
dows to average.
5. Dimensionality reduction of each averaged win-
dow using DM during training and using the
Nystr
¨
om extension during testing.
The stages differ in a final preprocessing stage that
they apply following Algorithm 1. Namely, the train-
ing stage uses the DM algorithm while the detection
phase uses the Nystr
¨
om extension.
Below is a detailed description of each step in Al-
gorithm 1.
Step 1: Decomposition into Windows. Let s
i
∈ Ω,
be a signal and let s
i
(t), t = 0,..., |s
i
| − 1 denote the
modulation value at time t where |s
i
| is the size of sig-
nal s
i
. Each signal s
i
is decomposed into a set of win-
dows W
i
- each of size l = 2
r
, r,l ∈ N, with overlap-
ping of ν% between every two consecutive windows.
The set of all windows is given by Ω
w
= ∪
n
i=1
W
i
=
{w
j
}
n
w
j=1
, w
j
∈ R
l
where n
w
is the total number of win-
dows of the signals in Ω.
Step 2: Application of the Spline Wavelet Pack-
ets. We use the sixth order spline wavelet packet.
A spline wavelet is applied to scale D ∈ N to each
window w
j
∈ Ω
w
. Typically, if l = 2
10
= 1024, then
D = 6 and if l = 2
9
= 512 then D = 5. The coef-
ficients are taken from the last scale D. This scale
contains l = 2
r
coefficients that are arranged into 2
D
blocks of length 2
r−D
. Each block is associated with
a certain frequency band. These bands form a near
uniform partition of the Nyquist frequency domain
into 2
D
parts. The outcome is Ω
wave
= {wp
j
}
n
w
j=1
,
where wp
j
∈ R
l
. At the end of this step, each win-
dow wp
j
∈ Ω
wave
is substituted for the set of its spline
wavelet coefficients.
Step 3: Calculation of the Energy. We construct
the acoustic signature using the distribution of en-
ergy among blocks which consist of wavelet packet
coefficients. The energy is calculated by summing
the coefficients in each block. The outcome is Ω
e
=
we
j
n
w
j=1
where each we
j
∈ Ω
e
is of dimension R
2
D
.
This operation reduces the dimension by a factor of
2
r−D
.
Step 4: Averaging. This step is applied in order
to reduce perturbations and noise. Given the energy,
Ω
e
= {we
j
}
n
w
j=1
, of the signals as calculated in step 3,
we calculate the average of every µ consecutive win-
dows which belong to the same signal in order to re-
ceive a more robust signature. Given a training signal
s, let Ω
e
(s) =
we
j
(s)
n
w
(s)
j=1
be the set of segments
of wavelet energy coefficients that were calculated in
step 3 for s where n
w
(s) is the number of segments
that s was decomposed to in step 1. For each segment
we
j
(s) we calculate
wa
j
(s) =
1
µ
j−µ+1
∑
k= j
we
k
(s)
The classification of wa
j
(s) is the same as s. The out-
put of this step is Ω
a
=
wa
j
n
w
j=1
, wa
j
∈ R
2
D
.
Step 5: Dimensionality Reduction. The dimen-
sionality of each segment in the output of step 4 if
further reduced by applying dimensionality reduction.
However, the training and detection stages implement
this step differently. The training stage applies the
DM algorithm to Ω
a
and produces
e
Ω
a
=
f
wa
j
n
w
j=1
,
f
wa
j
∈ R
q
where q is the reduced dimension. The de-
tection step, on the other hand, employs the Nystr
¨
om
extension algorithm in the following manner.
Let α be a test signal that is input to the detection
stage. In order to classify the k-th segment, α
k
, where
k ≥ µ, the, steps 1-4 are applied to α
k
. We denote the
averaged energy of the µ windows that precede α
k
by
b
α
k
(note that there is no need to wait for the entire
signal to be received). The Nystr
¨
om extension algo-
rithm is applied to
b
α
k
and we denote the result by
e
α
k
.
This embeds it into the reduced space that contains
Ω
a
. The classification of
e
α
k
is determined according
to the type of training window that is the nearest to
e
α
k
.
The classification phase is done online. In order to
classify a signal at time t, the algorithm only needs the
µ consecutive overlapping windows that immediately
precede time t.
4 EXPERIMENTAL RESULTS
We denote the parameters of the algorithms by: L is
the window size, ν is the overlapping percent, µ is the
number of windows to average, D is the the scale of
A Manifold Learning Framework for the Detection of Cardiac Disorders in Acoustic Signals
195
spline wavelet and q is the the target dimensionality
of the DM algorithm. The number of neighbors in
the k-nn classifier was set to 15. The values of these
parameters were determined empirically. The classi-
fication phase was tested on recordings that were not
a part of the training set.
The signals were taken from different people in
different occasions. The recording sample rates (SR)
were 22050 samples per second (SPS) and 11025
SPS. They were all down-sampled to 2205 SPS. The
classes in this experiment are: (a) normal heart beats;
and (b) a cardio vascular disorder. The data were
collected from different adults in different occasions.
The training sample set consisted from 7 recordings,
4 of them represent normal cardio behavior and 3
represent a cardio disorder. The detection set con-
tains 2 recordings that did not participate in the train-
ing phase. The following parameters were used in
the training and classification phases: L = 1024, ν =
75%, µ = 3, D = 6, q = 3. These parameters were
determined empirically.
Figure 2 depicts the clusters that are obtained
when the dimension reduced space is R
2
. It can be
seen that the first two eigenvectors provide a complete
separation into two disjoint clusters.
Figure 2: Clusters generated by the application of DM. The
plot is the embedded data onto the space spanned by the first
two eigenvectors.
Figure 3 contains the classification results of a
recording that contains a cardiac disorder.
Figure 3: Classification from a recording that contains a
cardiac disorder. Top: Original recording. Bottom: The
probability for a cardio disorder using the DM algorithm.
Figure 4 contains the classification results of a
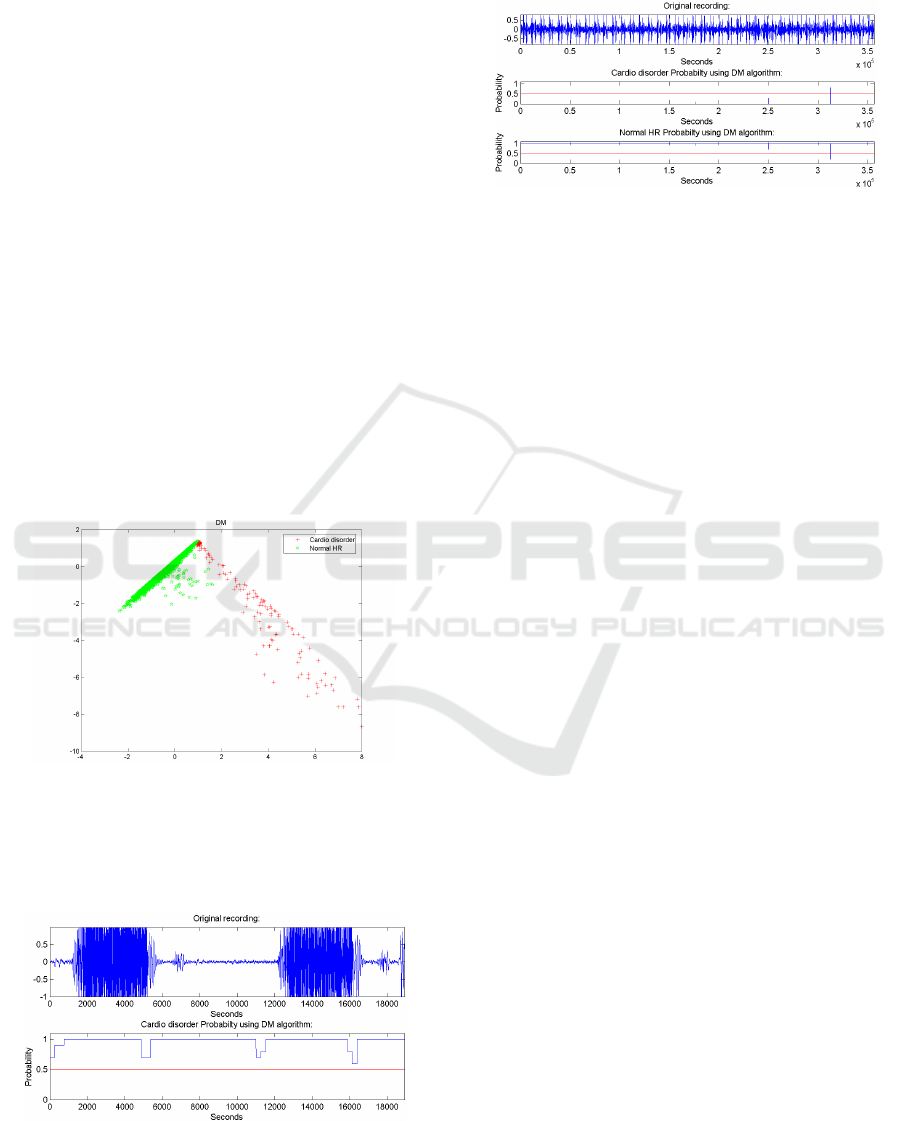
normal heart beat signal.
Figure 4: Classification from a recording that contains a
normal cardio system. Top: Original recording. Middle:
The probability for a cardio disorder using the DM algo-
rithm. Bottom: The probability for a normal cardio behav-
ior using the DM algorithm.
It can be seen that classification results are very
good.
5 CONCLUSIONS
In this work, we presented a manifold learning
scheme for the detection of cardiac disorders using
acoustic recordings. The algorithm is composed of a
training phase and a detection phase. In both phases
the signals are decomposed to overlapping windows
and each window undergoes a spline wavelet packet
transform followed by coefficients compaction and
temporal smoothing. The dimensionality of each win-
dow is further reduced via the DM dimensionality re-
duction algorithm. The set of all windows form a
training set for a nearest-neighbor classifier. During
the detection, the examined signal undergoes simi-
lar steps to those employed during the training, how-
ever, the dimension is reduced via the NE algorithm.
Each classification of each window in the examined
signal is determined according to its nearest in the
dimension-reduced training set.
The preliminary results of the proposed scheme
are very promising. However, the scheme is further
investigate in as follows. Additional experiments are
needed to corroborate the accuracy of the scheme.
A data-driven method to automatically determine the
optimal values of the algorithm’s parameters is sought
after. Other classifiers and other manifold learning al-
gorithms should be examined. The proposed scheme
is general and can be successfully applied to other do-
mains. Finally, the training set can be online extended
to include the new signals.
ICPRAM 2020 - 9th International Conference on Pattern Recognition Applications and Methods
196

REFERENCES
Averbuch, A., Hulata, E., and Zheludev, V. (2004). Identifi-
cation of acoustic signatures for vehicles via reduction
of dimensionality. International Journal of Wavelets,
Multiresolution and Information Processing, 2(1).
Averbuch, A., Hulata, E., Zheludev, V., and Kozlov, I.
(2001). A wavelet packet algorithm for classification
and detection of moving vehicles. Multidimensional
Systems and Signal Processing, 12(1).
Coifman, R. R. and Lafon, S. (2006). Diffusion maps. Ap-
plied and Computational Harmonic Analysis: special
issue on Diffusion Maps and Wavelets, 21:5–30.
D.A.Abraham (2019). Underwater Acoustic Signal Pro-
cessing: Modeling, Detection, and Estimation (Mod-
ern Acoustics and Signal Processing). Springer.
Daubechies, I. (1992). Ten Lectures on Wavelets. Society
for Industrial and Applied Mathematics, Philadelphia,
PA, USA.
Deng, S.-W. and Han, J.-Q. (2016). Towards heart sound
classification without segmentation via autocorrela-
tion feature and diffusion maps. Future Generation
Computer Systems, 60:13 – 21.
Fowlkes, C., Belongie, S., Chung, F., and Malik, J. (2004).
Spectral grouping using the nystrom method. IEEE
Transactions on Pattern Analysis and Machine Intel-
ligence, 26(2):214–225.
Lafon, S., Keller, Y., and Coifman, R. R. (2006). Data fu-
sion and multicue data matching by diffusion maps.
IEEE Transactions on Pattern Analyss and Machine
Intelligence, 28:1784–1797.
Lafon, S. and Lee, A. (2006). Diffusion maps and coarse-
graining: A unified framework for dimensionality re-
duction, graph partitioning, and data set parameteri-
zation. IEEE Transactions on Pattern Analysis and
Machine Intelligence, 28(9):1393–1403.
Mac, A. O., Gibb, R., Barlow, K. E., Browning, E., Fir-
man, M., and Freeman, R. (2018). Bat detective - deep
learning tools for bat acoustic signal detection. PLoS
Computational Biology, 14(3).
Munich, M. E. (2004). Bayesian subspace methods for
acoustic signature recognition of vehicles. 12th Eu-
ropean Signal Processing Conference, pages 2107–
2110.
Nystr
¨
om, E. J. (1928).
¨
Uber die praktische aufl
¨
osung
von linearen integralgleichungen mit anwendungen
auf randwertaufgaben der potentialtheorie. Commen-
tationes Physico-Mathematicae, 4(15):1–52.
Rabin, N. and Coifman, R. R. (2012). Heterogeneous
datasets representation and learning using diffusion
maps and laplacian pyramids. In Proceedings of the
2012 SIAM International Conference on Data Mining,
pages 189–199.
Schclar, A. (2008). A Diffusion Framework for Dimension-
ality Reduction, pages 315–325. Springer US, Boston,
MA.
Schclar, A. and Averbuch, A. (2015). Diffusion bases di-
mensionality reduction. In Proceedings of the 7th In-
ternational Joint Conference on Computational Intel-
ligence, IJCCI 2015, Lisbon, Portugal, November 12-
14, 2015., pages 151–156.
Schclar, A., Averbuch, A., Hochman, K., Rabin, N., and
Zheludev, V. (2010). A diffusion framework for de-
tection of moving vehicles. Digital Signal Process-
ing,, 20(1):111–122.
Sulam, J., Romano, Y., and Talmon, R. (2017). Dynami-
cal system classification with diffusion embedding for
ecg-based person identification. Signal Processing,
130:403 – 411.
Williams, C. K. I. and Seeger, M. (2000). Using the nystr
¨
om
method to speed up kernel machines. In Proceedings
of the 13th International Conference on Neural Infor-
mation Processing Systems, NIPS’00, pages 661–667,
Cambridge, MA, USA. MIT Press.
Yaan Li and Zhe Chen (2017). Entropy based underwater
acoustic signal detection. In 2017 14th International
Bhurban Conference on Applied Sciences and Tech-
nology (IBCAST), pages 656–660.
A Manifold Learning Framework for the Detection of Cardiac Disorders in Acoustic Signals
197
