
3D Plant Growth Prediction via Image-to-Image Translation
Tomohiro Hamamoto, Hideaki Uchiyama
a
, Atsushi Shimada and Rin-ichiro Taniguchi
b
Kyushu University, Fukuoka, Japan
Keywords:
Plant Image Analysis, Prediction, CNN, LSTM, GAN, Image-to-Image Translation.
Abstract:
This paper presents a method to predict three-dimensional (3D) plant growth with RGB-D images. Based
on neural network based image translation and time-series prediction, we construct a system that gives the
predicted result of RGB-D images from several past RGB-D images. Since both RGB and depth images
are incorporated into our system, the plant growth can be represented in 3D space. In the evaluation, the
performance of our proposed network is investigated by focusing on clarifying the importance of each module
in the network. We have verified how the prediction accuracy changes depending on the internal structure of
the our network.
1 INTRODUCTION
In the field of agricultural research, the design of plant
growth model has been conducted to manage the cul-
tivation of crops (Prasad et al., 2006). For instance,
such technologies are useful for optimizing planned
cultivation by predicting plant growth in plant facto-
ries. Plant growth prediction is obviously a challeng-
ing issue because the degree of growth varies greatly
depending on both genetic and environmental factors.
In addition, quantitative measurement of plant condi-
tions is also an important issue. To tackle the latter
issue, the conventional measurement and analysis of
the plant growth have been based on the measurement
of body weight and dry matter (Reuter and Robinson,
1997). However, such measurements are inappropri-
ate when monitoring the plant growth for a while be-
cause they need to destruct plants. Therefore, the ap-
plications of image processing has been considered as
an attempt to measure plant growth in a non-contact
manner (Mutka and Bart, 2015).
As growth indices, leaf areas, stem diameters, leaf
lengths, etc. from plants in the image are generally
used (O’Neal et al., 2002). Especially, the leaf area is
used as a basic criterion representing the plant growth.
More concretely, specific colored pixels in the images
are first obtained via thresholding based binarization,
and then the number of pixels is used as the size of the
leaf area. Existing techniques generally focus on mea-
suring the condition of the plant growth in the images
a
https://orcid.org/0000-0002-6119-1184
b
https://orcid.org/0000-0002-2588-6894
with classical image processing methods. However,
the results do not represent physically-correct shape
in three dimensional (3D) space such that the leaf ar-
eas cannot correctly be computed in the images if the
camera is inclined against the leaves. Therefore, the
design of the plant growth model in 3D space is desir-
able for further quantitative analysis.
In this paper, we propose a method that can obtain
the 3D plant growth model from RGB-D images cap-
tured at a top view (Uchiyama et al., 2017) by using
an image-to-image translation algorithm proposed in
the field of deep neural network. In other words, we
aim at reproducing RGB-D plant images that will be
captured several hours or several days later from past
time-series images. This issue is categorized as a fu-
ture image prediction task (Srivastava et al., 2015).
Especially, we extend the network on image-to-image
translation that changes the image domain between
images to prediction tasks. As a result, our network
outputs a future 3D plant shape by simultaneously
predicting both RGB and depth images. In the evalua-
tion, the performance of several architectures derived
from our basic one is investigated to clarify the im-
portance of each module in the network.
2 RELATED WORK
Our goal is to generate future RGB-D images based
on a time-series prediction technique so that plant
growth prediction in 3D space can be achieved. In
our approach, an image prediction technology based
Hamamoto, T., Uchiyama, H., Shimada, A. and Taniguchi, R.
3D Plant Growth Prediction via Image-to-Image Translation.
DOI: 10.5220/0008989201530161
In Proceedings of the 15th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2020) - Volume 5: VISAPP, pages
153-161
ISBN: 978-989-758-402-2; ISSN: 2184-4321
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
153

on image-to-image translation is proposed to gener-
ate the plant growth model, as neural network based
image analysis. In this section, we discuss related
work from the following aspects: plant image anal-
ysis, image-to-image translation, and time-series pre-
dictions relevant to our study.
2.1 Plant Image Analysis
Plant image analysis has been studied since 1970s.
For instance, statistical analysis of plant growth has
been conducted simply from the size of leaves in im-
ages (MATSUI and EGUCHI, 1976). As time-series
image analysis, plant growth is analyzed by using
optical flow (Barron and Liptay, 1997). In recent
years, predictive analysis based on high-quality or
large-scale plant images has been performed (Hart-
mann et al., 2011; Fujita et al., 2018).
Recently, neural networks have been used for
plant image analysis with the advance of deep learn-
ing based neural networks. Research issues such as
detection of plant leaves and measurement of plant
center positions by feature extraction tasks (Aich and
Stavness, 2017; Giuffrida et al., 2016; Chen et al.,
2017), and padding of plant image data by image gen-
eration tasks (Zhu et al., 2018) have been investigated.
In addition, there is an attempt to estimate the posi-
tion of a branch from a multi-view plant image by im-
age transformation and restore it in 3D space (Isokane
et al., 2018). One work investigated future plant im-
ages from the past images in 2D (Sakurai et al., 2019).
However, the plant growth model in 3D has not been
investigated in the literature.
2.2 Image-to-Image Translation
The basic idea of image-to-image translation is that
the source image is converted to the target image
based on learning image domain features. In the
field of deep learning, the original image was first
restored by auto encoder (Hinton and Salakhutdinov,
2006), and then the image synthesis by changing the
latent space is performed by using variational auto en-
coder (Kingma and Welling, 2013). VAE-GAN syn-
thesizes an image with high visual fidelity by combin-
ing Generative Adversarial Network (GAN) structure
with encoder-decoder structure (Larsen et al., 2015).
In our work, we will use this idea in addition to ex-
isting time-series prediction methods for prediction
tasks.
2.3 Time-series Prediction
Time-series prediction represents the research issue to
predict future images from past ones so that what will
happen in the future is predicted. Long-Short Term
Memory (LSTM), which is a deep neural network
with a structure suitable for time series analysis, was
proposed to learn long-term relationships between
frames (Hochreiter and Schmidhuber, 1997). More
effective video representation can be achieved with
encoder-decoder structure that divides LSTM into an
encoder capturing the features of a video and a de-
coder generating a prediction image (Srivastava et al.,
2015). In addition, Convolutional LSTM, which
combines spatial convolution with LSTM structure,
has been devised to capture spatial features of im-
ages (Xingjian et al., 2015).
3 PROPOSED METHOD
Our method first takes a series of RGB plant images
and corresponding depth images of the plants as input,
and then generates future plant images predicted from
the input through the network.
3.1 Network Architecture
We primarily use encoder-decoder LSTM to create
images to learn time-series changes of plant growth,
as similar to (Sakurai et al., 2019). Input images are
converted from time-series representation into feature
space by passing through the encoder LSTM. The de-
coder LSTM first reads the image converted to the
feature space by the encoder, and then restores the
time-series representation from the information. It
tries to compress the amount of information by go-
ing through the transformation from image to feature
space, and allows to obtain an output image sequence
independent of the number of input images.
Further, we propose to incorporate GAN as an er-
ror function, in addition to the inter-pixel error be-
tween the output image and the correct image. We
apply GAN to the frames of the generated image se-
quence to learn the time-series characteristics. In ad-
dition, the accuracy of prediction for each image is
improved by applying GAN to one image taken at ran-
dom from the image sequence.
In summary, our network utilizes the structure of
GAN, which is divided into a generator that generates
images and a discriminator that identifies images, as
illustrated in Figure 1. The generator learns so that
the generated image will be similar to the real image
VISAPP 2020 - 15th International Conference on Computer Vision Theory and Applications
154
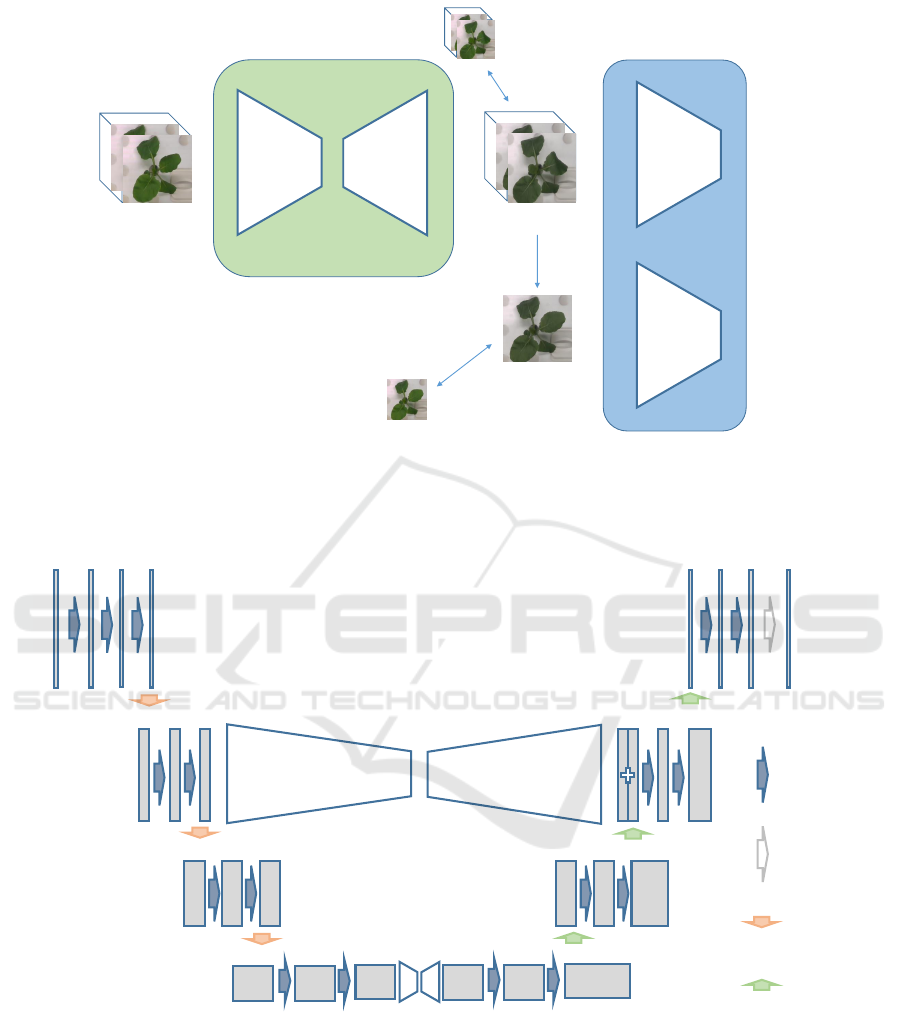
Generator
Encoder Decoder
Random
crop
real
or
fake
real
or
fake
Discriminator
Generate images
Cropped flame
MSE loss
MSE loss
Input images
Figure 1: Proposed network for 3D plant growth prediction. The network converts the RGB-D plant image input through the
generator into a predicted one several hours later. In the GAN process, the generated image is discriminated from the correct
image by discriminator.
2 × 2 conv
Stride = 2
Pixel shuffler
1 × 1 conv
Stride = 1
3 × 3 conv
Stride = 1
LSTM1
LSTM2
4 16 16 16 4
32 32 32 32+32 64 64
64 64 64 64
64
128
128
128
128
128
128
256
Encoder LSTM
Decoder LSTM
16 16 16
Input images Generate images
1/2 size
1/4 size
1/8 size
Figure 2: Generator. Each image is spatially convoluted through the CNN, and transformed with time information through
the LSTM. The converted image is restored by CNN and pixel shuffler.
while the discriminator learns to distinguish the ac-
tual image from the image synthesized by the genera-
tor. We try to refine more realistic images by learning
these iteratively. In the learning process, the mean
squared error (MSE) between pixels of the images is
considered as a loss to stabilize the learning, in addi-
tion to the loss of GAN.
3.2 Generator
The generator has an encoder-decoder structure, and
uses LSTM between each of encoder and decoder, as
illustrated in Figure 2. The encoder plays the role of
dimension compression while the decoder plays the
role of restoring the image from the compressed in-
3D Plant Growth Prediction via Image-to-Image Translation
155
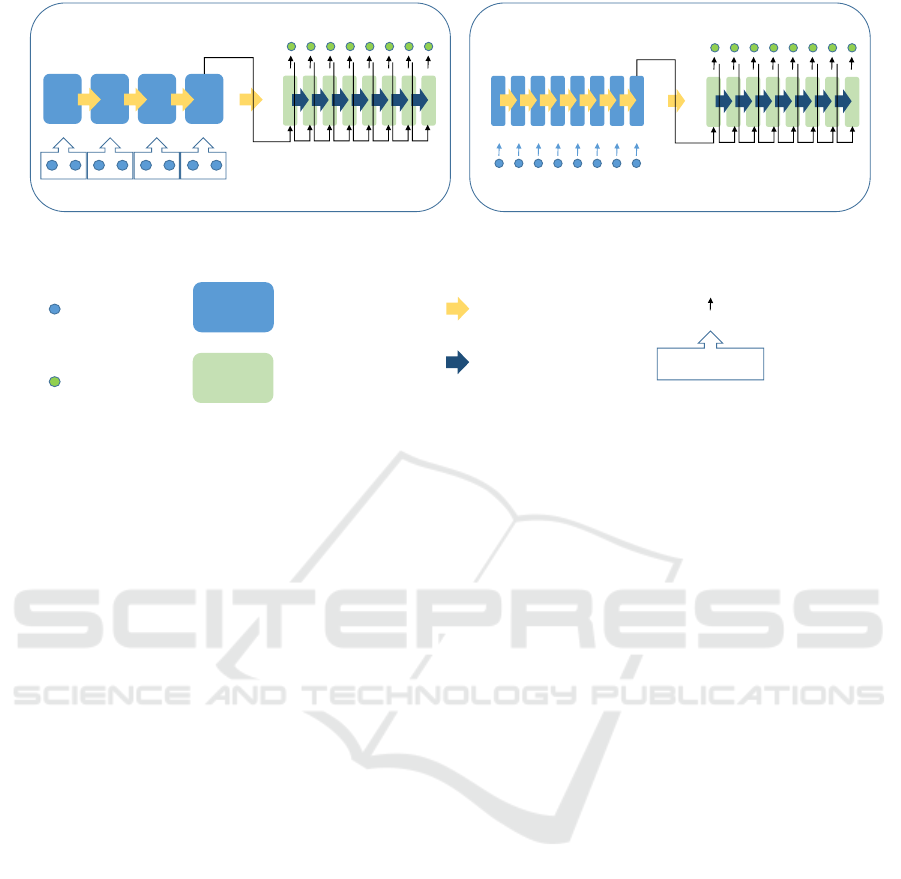
LSTM1 (1/2 size input) LSTM2 (1/8 size input)
Encoder LSTM
Decoder LSTM
Encoder LSTM
cell state and output
Decoder LSTM
cell state and output
mean
LSTM output
Input
Output
Figure 3: Encoder-decoder LSTM. The encoder of LSTM1 has an input that averages two elements of 1/2 size of the original
image, and LSTM2 takes 1/8 element of the original image as input. The decoder receives the output and cell state of the final
layer of the encoder.
formation. In the encoder, the image information is
compressed by CNN, and then the time information
is compressed by encoder LSTM. In the decoder, the
number of image representations obtained as output is
generated from the time information compressed by
the encoder LSTM, and then the original image size
is restored by CNN. Regarding CNN computation,
spatial features are obtained by calculating weighted
sums with surrounding pixels in multiple layers for
each image. The image size can be compressed by
changing the kernel stride. Pixel shuffler is a tech-
nique to increase the size of an image by applying
array transformation to the channel, and is used for
tasks such as generating super-resolution images.
3.3 Encoder-decoder LSTM in
Generator
Images extracted by CNN are converted into output
images through two LSTM structures with different
sizes, as illustrated in Figure 3. The upper LSTM
reads local changes between images while the lower
LSTM does global changes of images. In the encoder,
the input arranged in time series is put into the LSTM
in order. The cell state and output are updated by the
spatio-temporal weight calculation with the input of
the previous time. The output at the encoder is used
only for updating the next state, and then the output at
the final layer is passed to the decoder along with the
cell state. In the decoder, the output of the encoder is
given as the input of the first layer, and then the in-
put of the subsequent layer is given the output of the
previous layer of the decoder itself. The decoder can
generate the output in a self-recursive manner, which
allows to generate output independent of the number
of inputs. Both encoder and decoder are implemented
by using convolutional LSTM, which is used for time-
series image analysis.
3.4 Discriminator
The discriminator performs discrimination for single
image and time-series change between images in par-
allel, as illustrated in Figure 4. The time-series dis-
criminator performs the dimensional compression of
the image in the 2D convolution part, and then iden-
tifies the time series features by 3D convolution. The
single image discriminator takes out one random im-
age from the generated time-series images, and per-
forms feature extraction by two-dimensional convo-
lution.
The discriminator classifies the output of the final
layer into a binary class of 0 to 1 by activating it with
a sigmoid. When learning the generator, it learns so
that the output is 1 for the generated image with the
weight of the discriminator fixed. When learning the
discriminator, the generator weight is fixed so that the
correct image output is 1 and the generated image out-
put is 0. Based on this GAN property, the loss func-
tion of the generator G and the loss function of the
discriminator D are set. Since our network has two
discriminators, the loss function of a time-series dis-
criminator is denoted as G
t
, D
t
, and the loss function
of a single image discriminator is denoted as G
s
, D
s
.
In order to stabilize the learning process, MSE loss
VISAPP 2020 - 15th International Conference on Computer Vision Theory and Applications
156
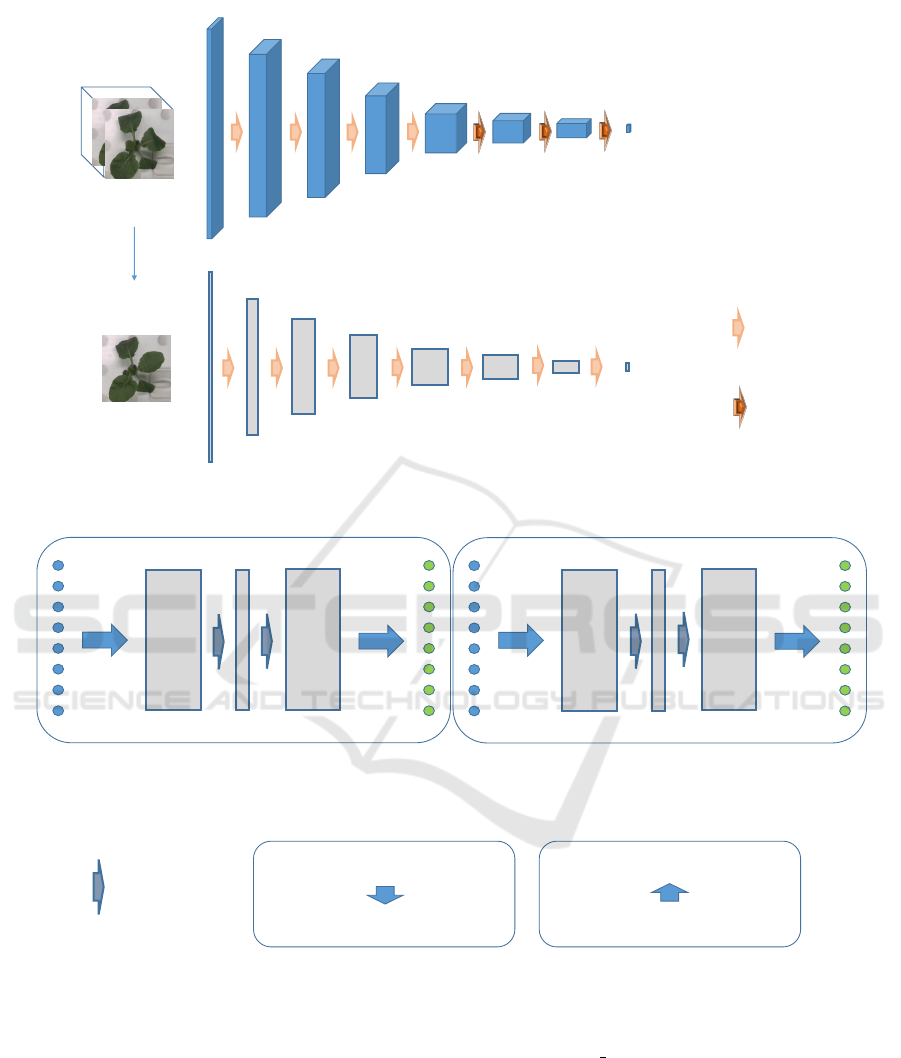
Random
crop
Generate images
Cropped flame
4
16 32
64 128
128
128
1
4
16 32
64 128
128
128
1
[8,8,8] [4,4,4] [2,2,2]
[1,1,1]
2 × 2
2Dconv
2 × 2 × 2
3Dconv
0~1
Real or fake
0~1
Real or fake
(channel)
(channel)
Figure 4: Discriminator. The discriminator has a single image identification function and a time-series image identification
function. 3D convolution is used for the time-series image discrimination.
Encoder_Decoder1 (1/2 size input)
Channel
concat
Channel
divide
32m
32n
m input
n output
Channel
concat
Channel
divide
128m
128n
m input
n output
32
128
Encoder_Decoder2 (1/8 size input)
32
32
128
128
3 × 3 conv
Stride = 1
Channel concat
[time, batch, height, width, channel]
[batch, height, width, time*channel]
Channel divide
[time, batch, height, width, channel]
[batch, height, width, time*channel]
Figure 5: Encoder-decoder with time-series inter-channel concatenation. The time information is compressed and decom-
pressed by combining the time axes with channels.
L
MSE
is added during generator learning. The loss
function of the generator L
G
and the loss function of
the discriminator L
D
in the whole network are as fol-
lows. α = 10
−3
, β = 10
−3
is given as the experimental
value.
L
G
= L
MSE
+ αG
t
+ βG
s
(1)
L
D
= αD
t
+ βD
s
(2)
As the learning activation function, relu is used for
the final layer of the generator, sigmoid is used for the
final layer of the classifier, tanh is used for the LSTM
layer, and leaky relu is used for the other layers.
4 EVALUATION
To investigate the performance of our proposed
method, the evaluation with both RGB and depth im-
ages in the KOMATSUNA dataset (Uchiyama et al.,
2017) was performed. Especially, the effectiveness of
3D Plant Growth Prediction via Image-to-Image Translation
157

each module is investigated by removing LSTM and
GAN from our network for the comparison. In addi-
tion, we present the result of the plant growth predic-
tion in 3D.
4.1 Dataset
In our study, we used the KOMATSUNA dataset in
this evaluation. This dataset is composed of time-
series image sequences of 5 Komatuna vegetables.
For each plant, there are 60 frames taken every 4
hours from a bird’s-eye view. In this experiment,
both RGB and depth images included in the dataset
are used. This dataset assumes an environment where
data can be obtained uniformly by plant factories.
In the experiment, plant data is divided into a
training set and a test one. During the network train-
ing, four plants were used as a training set, and the
remaining one was used as a test set. The resolution
of the image is [128, 128]. For each plant, images are
reversed left and right, and rotated by 90, 180, and
270 degrees, as data augmentation.
In addition, the change of the generated image due
to the multiple layers of LSTM was verified. The ex-
periment uses a structure in which three layers of dif-
ferent LSTMs are superimposed on each of the en-
coder and decoder.
4.2 Comparison Network
To investigate the effectiveness of our network archi-
tecture, we made comparison networks by changing
our structure at the structure of the encoder-decoder
part of the network described in Section 3.
In order to investigate the change of the generated
image with and without LSTM, we used the encoder-
decoder structure based on the time series operation
by the sequence conversion between channels, which
is not based on LSTM. Dimensional compression is
performed by concatenating input images into chan-
nels and convolving them at once. In the subsequent
convolution, the amount of information in the im-
age sequence is restored by increasing the number of
channels.
Furthermore, the output of the network with and
without GAN is displayed to examine changes to
GAN. As a loss function without GAN, the network
only calculates the MSE loss for the generator.
4.3 Result
The train trained an image sequence of four strains of
plants through the network, and predicted the growth
of the remaining one plant strain from the images
through the trained network. The input is a time-
series image of 8 plants. Based on the input, the future
growth of the plant is predicted and output as an im-
age. Figure 7 shows the results of learning for each
network.
Images generated by channel-concatenation
encoder-decoder are high reproducible as individual
images, and there is little blurring of the image
for the later prediction. On the other hand, the
image generated by encoder-decoder LSTM tends to
maintain the continuity of motion over time, although
the image is less sharp due to the prediction in the
back. Increasing the layer of LSTM improves the
sharpness of the image, but overreacts to movement.
As a result of applying a discriminator by GAN,
a clearer image was generated, but a real image could
not be generated. The reason is that the network be-
comes more sensitive to changes, and the response to
minute differences becomes too high.
Table 1 shows the MSE loss values calculated for
each predicted time and the average value for the im-
ages output in the network. The loss values are higher
in the later prediction, and this trend is particularly
strong when LSTM is not used. In a network with
three layers of LSTM, the MSE loss is higher than
when only one layer of LSTM is used.
4.4 3D Reconstruction
By predicting the depth image, the prediction result
can be displayed in 3D. The depth image prediction
can be performed simultaneously with RGB predic-
tion by adding a channel for depth image prediction.
In order to generate a more natural 3D model, it is
recommended to replace missing values in the depth
image with standard values such as the median of the
image as a preliminary preparation.
The 3D model of plants predicted using RGB-D
images is displayed, as shown in Figure 8. The sur-
face of the point cloud was applied using Meshlab’s
ball pivoting.
5 CONCLUSION
We proposed a method for generating 3D plant
growth model by applying a machine learning frame-
work for plant growth prediction. We applied an im-
age sequence prediction network based on time-series
changes to plant images, and verified how the gener-
ated images change in response to changes in the net-
work structure. It was shown that 3D growth predic-
tion is performed using RGB-D images for prediction.
VISAPP 2020 - 15th International Conference on Computer Vision Theory and Applications
158
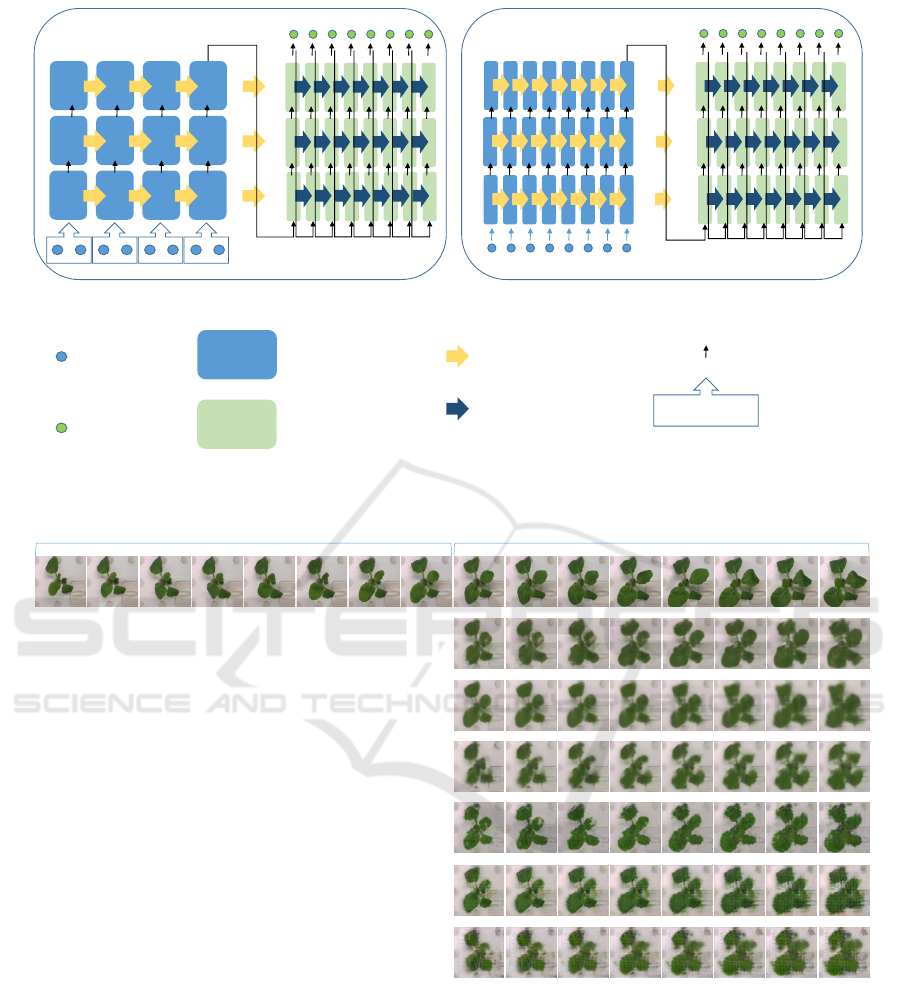
1 1 1 1
1 1 1 1 1 1 1 1
3 3 3 3 3 3 3 3
3 3 3 3 3 3 3 3
2 2 2 2 2 2 2 2
3 3 3 3 3 3 3 3
2 2 2 2 2 2 2 2
1 1 1 1 1 1 1 1
2 2 2 2
3 3 3 3
2 2 2 2 2 2 2 2
1 1 1 1 1 1 1 1
LSTM1 (1/2 size input) LSTM2 (1/8 size input)
Encoder LSTM
Decoder LSTM
Encoder LSTM
cell state and output
Decoder LSTM
cell state and output
mean
LSTM output
Input
Output
Figure 6: 3-layer encoder-decoder LSTM. The LSTM output is passed to the next LSTM. The output of the last layer of the
decoder LSTM is regarded as the output of the entire encoder-decoder.
Input images Ground truth
Channel concat
1-layer LSTM
3-layer LSTM
Channel concat +
GAN
1-layer LSTM +
GAN
3-layer LSTM +
GAN
Figure 7: Image sequence generated through network based on given input image sequence. The time-series images of 8
plants are input, (left) and the subsequent growth is output as 8 images (right).
As a future work, we will perform quantitative
analysis of the growth factors by the network by
adding to the prediction network data that is an in-
dicator of plant growth such as leaf area in addition to
the image. In addition, by inputting data such as the
amount of water given together with images, it is nec-
essary to develop a responsive system that presents
the possibility of poor growth for growing plants.
ACKNOWLEDGMENT
A part of this work is supported by JSPS KAKENHI
Grant Number JP17H01768 and JP18H04117.
3D Plant Growth Prediction via Image-to-Image Translation
159

Channel concat
1-layer LSTM
3-layer LSTM
Channel concat + GAN
1-layer LSTM + GAN
3-layer LSTM + GAN
Figure 8: 3D restoration result of predicted image. 3D point cloud (left) and faceted image by Ball Pivoting (right) are
visualized.
VISAPP 2020 - 15th International Conference on Computer Vision Theory and Applications
160
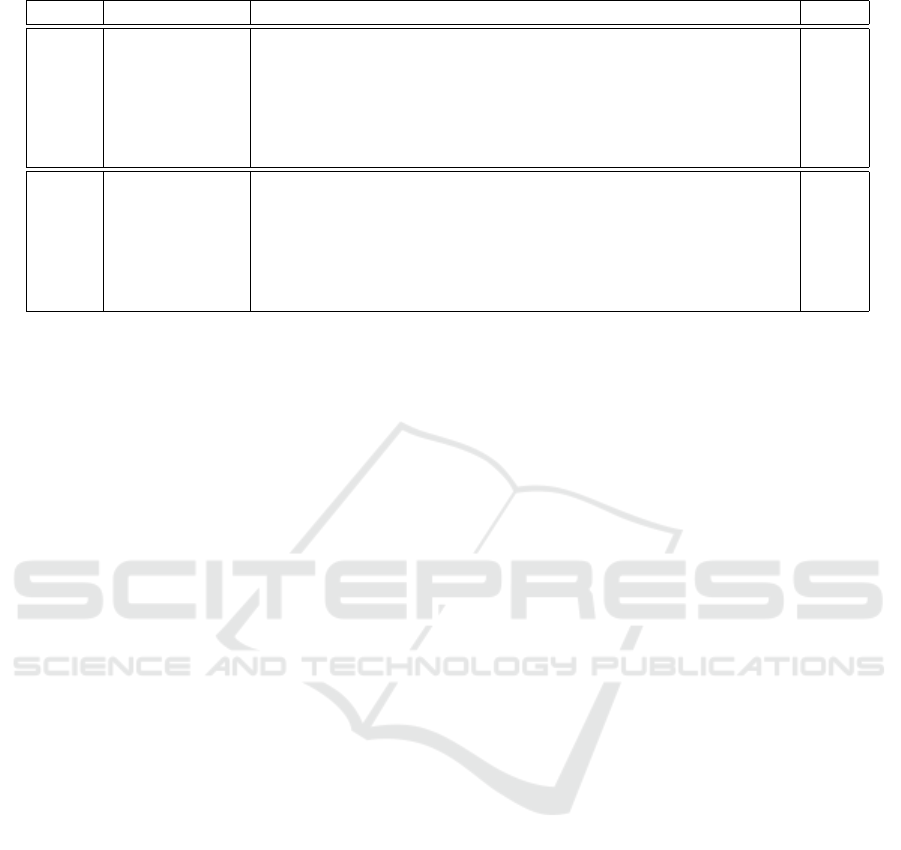
Table 1: MSE loss of the generated images: output per hour and its mean.
model leaf1 leaf2 leaf3 leaf4 leaf5 leaf6 leaf7 leaf8 mean
RGB Channel concat 0.707 1.006 1.009 1.413 1.800 1.867 2.324 2.754 1.610
+ GAN 0.975 1.364 1.371 1.636 2.060 2.348 2.587 2.949 1.911
(10
−2
) 1-layer LSTM 0.866 1.284 1.127 1.759 2.446 2.286 2.228 2.282 1.785
+ GAN 0.913 1.368 1.200 1.692 2.107 2.043 2.256 2.453 1.754
3-layer LSTM 1.629 1.454 1.463 2.182 2.745 2.679 2.912 3.029 2.262
+ GAN 1.564 1.377 1.305 2.053 2.560 2.685 2.993 3.100 2.205
Depth Channel concat 1.846 2.328 2.619 2.998 3.264 3.200 2.751 3.107 2.764
+ GAN 2.994 3.100 3.249 3.292 3.477 3.239 2.577 2.623 3.069
(10
−4
) 1-layer LSTM 1.730 1.998 2.424 2.735 3.305 3.236 2.690 2.567 2.586
+ GAN 1.916 2.072 2.457 2.719 3.344 3.359 3.142 3.408 2.802
3-layer LSTM 1.856 2.108 2.495 2.781 3.331 3.311 2.826 2.817 2.690
+ GAN 2.558 2.665 3.050 3.239 3.687 3.614 3.328 3.495 3.204
REFERENCES
Aich, S. and Stavness, I. (2017). Leaf counting with deep
convolutional and deconvolutional networks. In Pro-
ceedings of the IEEE International Conference on
Computer Vision, pages 2080–2089.
Barron, J. and Liptay, A. (1997). Measuring 3-d plant
growth using optical flow. Bioimaging, 5(2):82–86.
Chen, Y., Ribera, J., Boomsma, C., and Delp, E. (2017). Lo-
cating crop plant centers from uav-based rgb imagery.
In Proceedings of the IEEE International Conference
on Computer Vision, pages 2030–2037.
Fujita, M., Tanabata, T., Urano, K., Kikuchi, S., and Shi-
nozaki, K. (2018). Ripps: a plant phenotyping system
for quantitative evaluation of growth under controlled
environmental stress conditions. Plant and Cell Phys-
iology, 59(10):2030–2038.
Giuffrida, M. V., Minervini, M., and Tsaftaris, S. A. (2016).
Learning to count leaves in rosette plants.
Hartmann, A., Czauderna, T., Hoffmann, R., Stein, N., and
Schreiber, F. (2011). Htpheno: an image analysis
pipeline for high-throughput plant phenotyping. BMC
bioinformatics, 12(1):148.
Hinton, G. E. and Salakhutdinov, R. R. (2006). Reducing
the dimensionality of data with neural networks. sci-
ence, 313(5786):504–507.
Hochreiter, S. and Schmidhuber, J. (1997). Long short-term
memory. Neural computation, 9(8):1735–1780.
Isokane, T., Okura, F., Ide, A., Matsushita, Y., and Yagi,
Y. (2018). Probabilistic plant modeling via multi-
view image-to-image translation. In Proceedings of
the IEEE Conference on Computer Vision and Pattern
Recognition, pages 2906–2915.
Kingma, D. P. and Welling, M. (2013). Auto-encoding vari-
ational bayes. arXiv preprint arXiv:1312.6114.
Larsen, A. B. L., Sønderby, S. K., Larochelle, H., and
Winther, O. (2015). Autoencoding beyond pixels
using a learned similarity metric. arXiv preprint
arXiv:1512.09300.
MATSUI, T. and EGUCHI, H. (1976). Computer control
of plant growth by image processing. Environment
Control in Biology, 14(1):1–7.
Mutka, A. M. and Bart, R. S. (2015). Image-based pheno-
typing of plant disease symptoms. Frontiers in plant
science, 5:734.
O’Neal, M. E., Landis, D. A., and Isaacs, R. (2002). An
inexpensive, accurate method for measuring leaf area
and defoliation through digital image analysis. Jour-
nal of Economic Entomology, 95(6):1190–1194.
Prasad, A. K., Chai, L., Singh, R. P., and Kafatos, M.
(2006). Crop yield estimation model for iowa using
remote sensing and surface parameters. International
Journal of Applied Earth Observation and Geoinfor-
mation, 8(1):26–33.
Reuter, D. and Robinson, J. B. (1997). Plant analysis: an
interpretation manual. CSIRO publishing.
Sakurai, S., Uchiyama, H., Shimada, A., and Taniguchi, R.-
I. (2019). Plant growth prediction using convolutional
lstm. In 14th International Conference on Computer
Vision Theory and Applications, VISAPP 2019-Part
of the 14th International Joint Conference on Com-
puter Vision, Imaging and Computer Graphics Theory
and Applications, VISIGRAPP 2019, pages 105–113.
SciTePress.
Srivastava, N., Mansimov, E., and Salakhudinov, R. (2015).
Unsupervised learning of video representations using
lstms. In International conference on machine learn-
ing, pages 843–852.
Uchiyama, H., Sakurai, S., Mishima, M., Arita, D.,
Okayasu, T., Shimada, A., and Taniguchi, R.-i.
(2017). An easy-to-setup 3d phenotyping platform
for komatsuna dataset. In Proceedings of the IEEE
International Conference on Computer Vision, pages
2038–2045.
Xingjian, S., Chen, Z., Wang, H., Yeung, D.-Y., Wong, W.-
K., and Woo, W.-c. (2015). Convolutional lstm net-
work: A machine learning approach for precipitation
nowcasting. In Advances in neural information pro-
cessing systems, pages 802–810.
Zhu, Y., Aoun, M., Krijn, M., Vanschoren, J., and Campus,
H. T. (2018). Data augmentation using conditional
generative adversarial networks for leaf counting in
arabidopsis plants. In BMVC, page 324.
3D Plant Growth Prediction via Image-to-Image Translation
161
