
A Flexible Semantic Integration Framework for Fully-integrated EHR
based on FHIR Standard
Ahmed Dridi
1 a
, Salma Sassi
1
, Richard Chbeir
2
and Sami Faiz
3
1
VPNC Lab., FSJEGJ, University of Jendouba, Jendouba, Tunisia
2
Universite Pau et Pays de l’Adour, E2S/UPPA, LIUPPA, EA3000, Anglet, 64600, France
3
ISAMM, University of Manouba, La Manouba, Tunisia
Keywords:
Electronic Health Records, Health Data Integration, Semantic Interoperability, FHIR, Web Semantic, IoT
Data.
Abstract:
Despite the huge efforts focused on EHR development and massive many years of widespread availability
of this latter, health care providers and organizations are still looking for innovative solutions to bring in all
IoT data, and unstructured data into electronic health record (EHR) systems. There is a growing need to
semantically integrate health-related data from different sources to support decision-making and improve the
quality of care services provided. In this paper, we propose a flexible semantic integration framework for IoT,
unstructured and structured healthcare data in EHR systems called SF4FI-EHR. It is built on a novel approach
that applying semantic web technologies and the HL7 FHIR standard to handle integration challenges. Our
experiment results from the proof-of-concept study show that the use of such approach does enhance healthcare
data integration as well as overcome obstacles that prevent the optimal exploitation of these data.
1 INTRODUCTION
Over the years, a large amount of health-related data
is produced, captured and usually accumulated con-
tinuously in Electronic Health Records (EHRs) (Men-
achemi and Collum, 2011). EHR is the most widely
used application in the modern healthcare industry,
which has been used with the intent to improve qual-
ity of care and patient outcomes. With the advent of
Internet of Things (IoT) (Ashton et al., 2009), a new
research topic in many academic and industrial disci-
plines, especially in healthcare, has emerged. Wear-
able medical devices connected to the internet, can
help cut costs and improve patient care by collecting
invaluable data that give extra insight on the individ-
uals physical condition. These data collected from
different sources introduce new challenges related to
data integration and processing, due to their structural
characteristics, their heterogeneity, and the lack of se-
mantics. It is worthy to note that the main objec-
tives of the IoT-based healthcare systems cannot be
achieved without consolidate these data using a well-
designed and established methodology so to properly
integrate them with others clinical data into the EHR.
a
https://orcid.org/0000-0001-6374-1883
Despite the massive effort and investment in
health information systems to make clinical data in-
teroperable and integrable, the widespread use of the
IoT in EHR remains a long-term goal. The need to de-
sign appropriate solutions for seamless and effective
integration of different health data including IoT and
unstructured data into the EHR is more than before.
This paper addresses the problem of health-related
data integration, where data from multiple sources
needs to be aggregated and integrated in a unified and
rich-semantic representation. Healthcare data integra-
tion is not a trivial task, but its vital to allow clini-
cians to get the whole picture of an individuals health
related-data. Meanwhile, there are some stark chal-
lenges need to be addressed over this task.
The IoT devices continuously generate large
amounts of data that can reflect patient’s conditions.
This data is produced in a mix of data formats, mostly,
without a well-defined structure and additional se-
mantic. It has no value in its raw state, so it must be
converted into a unified representation, and semanti-
cally described to extract high-level knowledge which
can be used to make decisions, as well as to make it
easy to integrate with other data. IoT data integration
in EHR is the first challenge we will focus on in this
research.
684
Dridi, A., Sassi, S., Chbeir, R. and Faiz, S.
A Flexible Semantic Integration Framework for Fully-integrated EHR based on FHIR Standard.
DOI: 10.5220/0008981506840691
In Proceedings of the 12th International Conference on Agents and Artificial Intelligence (ICAART 2020) - Volume 2, pages 684-691
ISBN: 978-989-758-395-7; ISSN: 2184-433X
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

Another challenge that needs to be addressed is
the unstructured health data integration. Roughly
80% of clinical data is unstructured data and still
largely untapped after it is created. This data is often
recorded as free text, without standard content speci-
fications, that makes it more difficult to be handled by
EHR systems. Clinician notes, prescriptions, pathol-
ogy and radiology reports, and scanned medical docu-
ments contains meaningful data to deliver high-value
care. Therefore, this useful data needed to be effec-
tively extracted and integrated into the EHR.
In this paper, we are also motivated by enabling
semantic interoperability among structured health
data. These data are standardized by different stan-
dards, encoded with heterogeneous terminologies and
stored in various legacy systems and EHRs. For sub-
sequent seamless integration with other data, these
data need to be retrieved from their original sources
first, and then restructured into a common format and
standard terminologies.
Currently, several state of the art solutions, have
tried to solve the issue of interoperability and seman-
tic integration of healthcare data. But, to the best of
our knowledge, still there is no works are able to per-
form such integration efficiently, taking into consid-
eration all the challenges aforementioned.
Here, we propose a flexible semantic integration
framework, called SF4FI-EHR, to capture heteroge-
neous health-related data from different sources and
integrate them into an interoperable integrated EHR.
SF4FI-EHR is based on a set of semantic web
technologies and the HL7 FHIR standard (Bender and
Sartipi, 2013) to:
• Process non-standardized health related data
• Create structured and machine-readable informa-
tion and provide means to raw data of various
forms
• Seamlessly integrate data to a consistent descrip-
tion format
The rest of the paper is organized as follows. Sec-
tion 2 introduces main previous work carried-out on
health data integration. In Section 3, we present our
framework. The subsequent sections detail the main
components of the framework architecture. The ex-
perimental results are discussed in Section 4. Sec-
tion 5 describes an example use case to demonstrate
the health data integration process by the proposed
framework. Finally, Section 6 concludes the paper
with suggestions for future work.
2 BACKGROUND AND RELATED
WORKS
Semantic interoperability is still one of the primary
interests in the E-Health community. In this respect,
a lot of efforts have been devoted to achieve semantic
interoperability into EHRs. As a result, several EHR
standards, terminologies, and semantic frameworks
have been proposed in the literature. For instance,
many standards organizations have put countless ef-
forts to introduce standards and specifications such
as Health Level 7 (HL7) standards, CEN EN 13606,
ISO TC 215, and openEHR (Eichelberg et al., 2005).
These standards, which are currently in continuous
improvement, aim to structure and markup the clin-
ical content for the purpose of exchange (Maharatna
and Bonfiglio, 2013). A survey and analysis of EHR
standards are presented in (Eichelberg et al., 2005).
Similarly, others attempts (Mori et al., 1998) aims
at providing terminological standards and codes , as
structured lists of terms that give a controlled vocab-
ulary and structured medically pertinent expressions,
covering complex concepts (such as diseases, opera-
tions, treatments and medicines). Despite the signifi-
cant role played by the proposed standards and termi-
nologies in achieving semantic interoperability into
EHR, there is still no common consensus regarding
their adoption.
Consequently, semantic technologies-based inte-
gration frameworks and mediation approaches are re-
quired to deal with multiple health data represen-
tations. To improve the interoperability of EHRs
data, represented with different standards, Sun et
al. (Sun et al., 2015) proposed an approach to in-
tegrate data from heterogeneous resources. RDF
data mapping approach with semantic conversions
between different representations has been applied
to integrate health records from heterogeneous re-
sources and to generate integrated data in different
data formats/semantics to support various clinical re-
search applications. In (Mart
´
ınez-Costa et al., 2014),
Martnez-Costa et al. described a layered semantic-
driven architecture to improve EHR semantic interop-
erability based on ontologies that formalize the mean-
ing of clinical data. Similarly, a semantic-driven en-
gine called SeDIE, which built on a novel approach
using a statistical method and a Multiple-Criteria
Decision-Making model to overcome the barriers of
integrating unstructured and structured data, is de-
scribed in (Dhayne et al., 2018). A scalable and
standards-based framework for integrating structured
and unstructured EHR data centered on a clinical NLP
pipeline enhanced with an FHIR-based type system,
is described in (Hong et al., 2018). Ozgur Kilic et
A Flexible Semantic Integration Framework for Fully-integrated EHR based on FHIR Standard
685

al. (Kilic and Dogac, 2009) proposed mapping clini-
cal statements between EHRs, that resolve integration
issues by using archetypes, refined message informa-
tion model (R-MIM) derivations, and semantic tools.
The issue of EHR interoperability is also ad-
dressed by numerous international projects includ-
ing; the European project EHR4CR (De Moor et al.,
2015), eMERGE (Rasmussen-Torvik et al., 2014),
SHRINE (Weber et al., 2009), PONTE (Tagaris et al.,
2012), EURECA (Genitsaridi et al., 2015), CDISC
SHARE (El Fadly et al., 2007), and GALEN (Mori
and Consorti, 1998), where authors overcame the in-
teroperability challenges using a platform capable to
bring up semantic interoperability services based on
standard terminologies.
More recently, increasing interests have been fo-
cusing on the integration of the IoT data into EHRs.
As a result, much work has been carried out in or-
der to achieving the interoperability of the sensed and
EHR data. Kumar et al. (Kumar et al., 2016) suc-
cessfully integrated patient glucose data in the EHR
through an Apple HealthKit, which collects data via
Bluetooth, stores them locally on the mobile device
and then sends the data to the EHR system through
the Epic MyChart app running on the Apples mobile
operating system (iOS). In a similar way to Apple
HealthKit, V. Gay and P. Leijdekkers (Gay and Lei-
jdekkers, 2015) developed with third-party partners
a health and fitness Android application called my-
FitnessCompanion, which integrates a wide range of
wearable devices, EHR systems, and other applica-
tions. Vuppalapati et al. (Vuppalapati et al., 2016)
reviewed the role of big data and data analytics in
healthcare. They proposed a sensor integration frame-
work to integrate various sensors data with the elec-
tronic health records. Spark and Apache Kafka are
used for processing a large amount of data in real-
time. To achieve an effective health self-management,
the authors in (Peng and Goswami, 2019) presented
an OWL ontology-based approach aiming to integrate
the health and home environment data from heteroge-
neously built services and devices. Links with exter-
nal ontologies (such as SSN (Compton et al., 2012)
and FOA
1
), semantic annotation tools and ontology
mapping techniques are used to ensure efficient data
integration. Alamri A. (Alamri, 2018) described a se-
mantic middleware that exploits ontology to support
the semantic integration of IoT and EHR data. Se-
mantic Web technologies and clinical terminologies
are used to dene and normalize the structures and rela-
tionships of the data collected from the IoT healthcare
devices and sensors and data obtained from EHRs.
A clinical decision support system (CDSS) for dia-
1
http://www.foaf-project.org/
betes self-management is discussed in (El-Sappagh
et al., 2019). The proposed CDSS is based on the
OWL FASTO ontology
2
, which integrates the seman-
tic capabilities of the SSN (Compton et al., 2012),
BFO (Grenon et al., 2004), FHIR (Bender and Sartipi,
2013), clinical practice guideline (CPG), and medical
terminologies in a unified manner, in order to provide
an interoperable integration of IoT and EHR data.
3 SF4FI-EHR: SEMANTIC
FRAMEWORK FOR FULLY
INTEGRATED EHR
Ideally, EHRs must capture and integrate data on all
aspects of healthcare over time, with the data being
represented according to relevant controlled vocab-
ularies and standards, enabling consequently a dif-
ferent level of interoperability for the purpose of ex-
change and potential secondary uses.
From this perspective, we present in this pa-
per a flexible semantic framework, called SF4FI-
EHR standing for Semantic Framework for Fully-
Integrated HER, that systematically enables the ag-
gregation of heterogeneous health-related data from
different sources and integrates them as an interoper-
able integrated EHR. SF4FI-EHR intends to integrate
3 categories of health-related data: 1) IoT and med-
ical devices generated data, 2) Unstructured, and 3)
Structured EHR data. The overall architecture of the
proposed framework is depicted in Fig.1.
To deal with the different data categories, a partic-
ular integration process is expected for each one. IoT
raw-data must be preprocessed, modeled, and then se-
mantically annotated by a modular ontology built by
reusing existing knowledge resources. For unstruc-
tured EHR data, an advanced Natural Language Pro-
cessing (NLP) technique is required to identify ac-
tionable insights and generate structured output. For
this purpose, we chose to use MetaMap tool. The
structured EHR data are needed to be retrieved from
their heterogeneous sources and then converted into a
FHIR resources. All collected, preprocessed and stan-
dardized data are converted to FHIR resource formats,
before being mapped to the FHIR OWL Ontology to
construct a comprehensive patient health profile.
As a result, an integrated FHIR-based EHR is cre-
ated and stored within a Relational DataBase for fu-
ture purposes. We chose to use RDB since it is more
popular and more stable, and most of the current EHR
databases are in RDB format.
2
https://bioportal.bioontology.org/ontologies/FASTO
ICAART 2020 - 12th International Conference on Agents and Artificial Intelligence
686
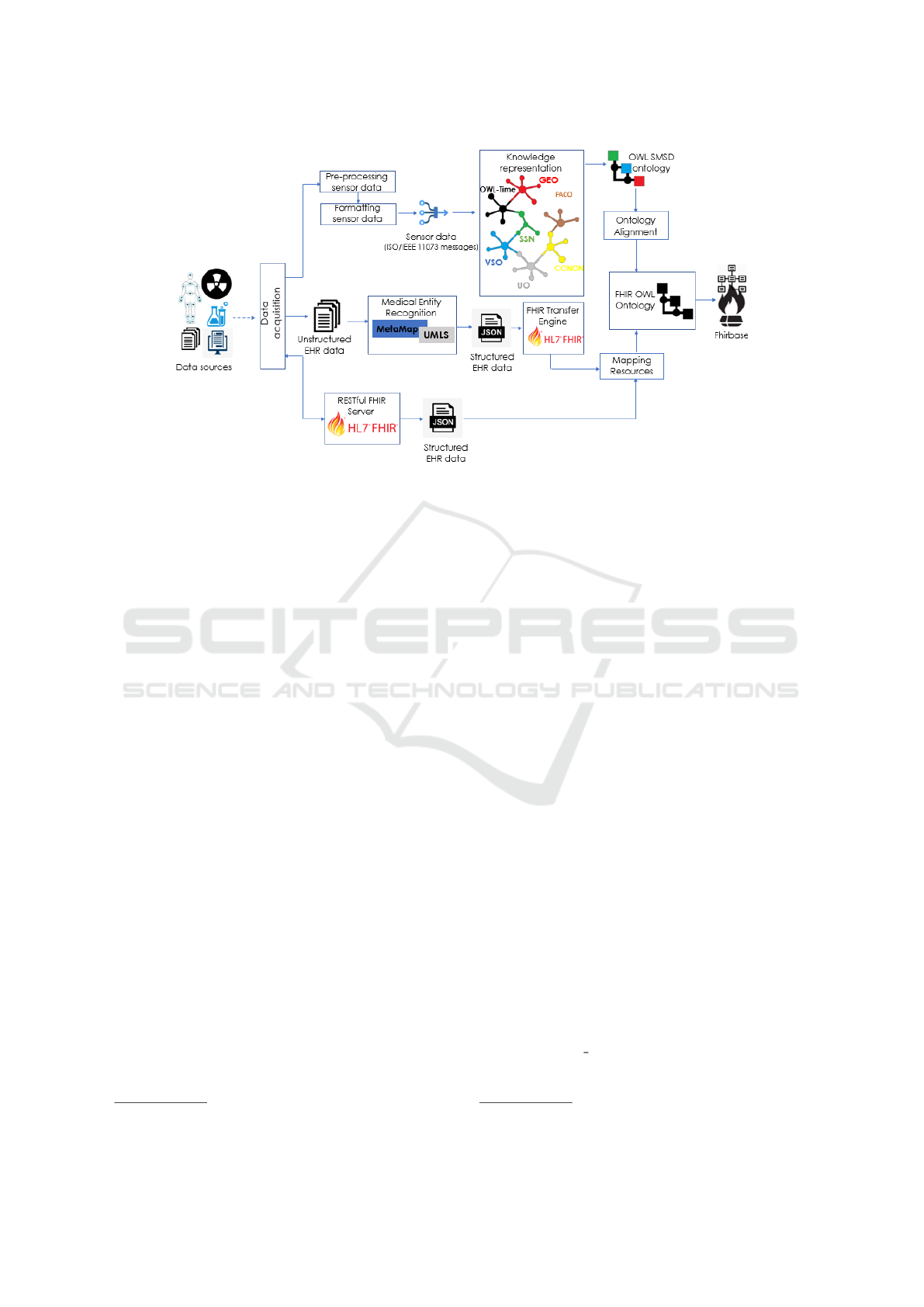
Figure 1: The architecture of the SF4FI-EHR framework.
The next subsections describe in detail the key
features of the integration process within the proposed
framework.
3.1 IoT Data Integration
IoT medical devices capable of generating data that
can be integrated into EHRs to make a significant
clinical impact and improve the way the patient is
treated by his doctor.
The problem is that IoT raw data streams, sensed
in real time, have no structure and semantics, which
makes their integration with EHR data challenging.
Thus, IoT data must undergo a preprocessing step in
order to: fill in missing values, smooth noisy data and
delete irrelevant values. Afterwards, these data are
converted into a unified format by applying ISO/IEEE
11073-104zz device specifications.
In order to use these sensor data effectively, se-
mantic annotation is applied using a modular ontol-
ogy that we called Semantic Medical Sensor Data on-
tology (SMSD). This ontology enables sensor data to
be described with semantic metadata in order to make
them machine-understandable, interoperable, as well
as to facilitate data integration. In order to ensure high
reusability, the SMSD ontology was built by extend-
ing multiple relevant ontologies. Semantic Sensor
Network ontology (SSN) (Compton et al., 2012) has
been extented in SMSD to cope with sensors networks
and data. Furthermore, We extend several concepts
from the FOAF ontology
3
, VSO ontology (Goldfain
et al., 2011), CONON ontology (Wang et al., 2004),
3
http://xmlns.com/foaf/spec/
OWL Time ontology
4
, CoDAMoS ontology (Preuve-
neers et al., 2004) and Units Ontology (Gkoutos et al.,
2012). We use distinguished colors and specific pre-
fix that indicates the ontology that each concept be-
longs to, in order to differentiate between the reused
concepts and the concepts we have proposed. Fig.2
illustrate these concepts as well as the relationships
between them. The main concepts of this ontology
are:
• SMSD:Patient: concept represents the ”who” in-
formation about the patient. A patient is described
as a person and inherits all the properties of the
class FOAF:Person which is a sub-class of the
of the FOAF:Agent class in the FOAF ontology.
It include all the demographic information neces-
sary to support the administrative, financial and
logistic procedures.
• SMSD:Device: concept describes and represents
a type of a manufactured item that is used in the
provision of healthcare. It could be a medical or
non-medical device (Smartphone, Smart Bracelet,
Wearable Glucose Level Monitoring device, etc).
The property contains is used to link between
the SMSD:Device and the SSN:Sensor classes.
SSN:Sensor concept is extended from the SSN
ontology to represent the physical objects that
observe, transforming incoming stimuli into an-
other, often digital, representation(Compton et al.,
2012).
• SMSD:Vital Signs: concept describes
the detected vital signs of patient which
are represented by the four sub-classes
4
https://www.w3.org/TR/owl-time/
A Flexible Semantic Integration Framework for Fully-integrated EHR based on FHIR Standard
687
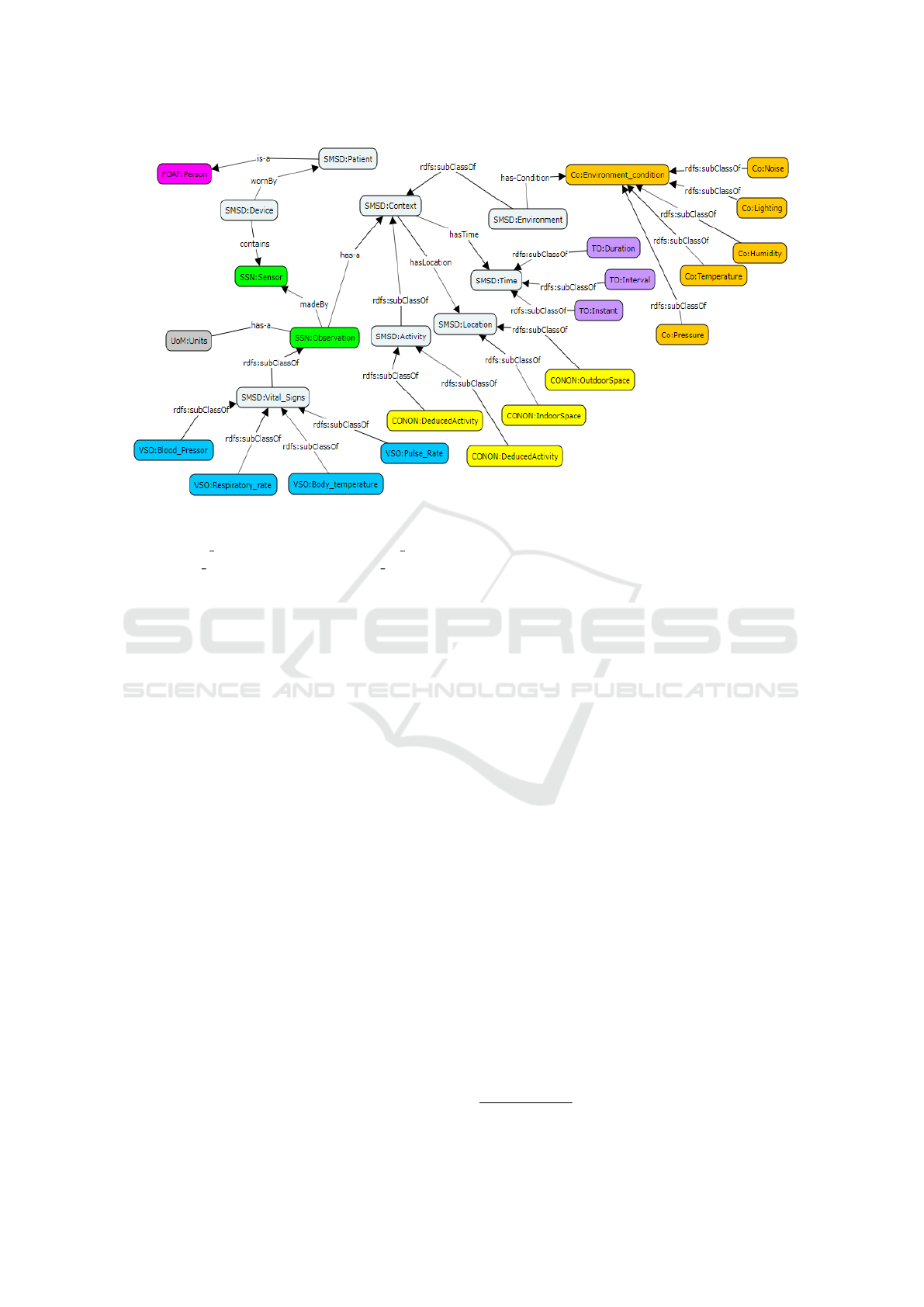
Figure 2: The SMSD ontology.
VSO:Blood Pressor, VSO:Respiratory rate,
VSO:Body temperature, VSO:Pulse Rate
which are extended by the VSO ontology that
covers the four consensus human vital signs:
blood pressure, body temperature, respiration
rate, pulse rate.
• SMSD:Context: concept represents the seman-
tics of concepts and their relationships that cap-
tured about a particular context in pervasive com-
puting environments. It provides a simple context
modeling based on four main concepts, which are:
The Activity, the Location, the Time and the En-
vironment:
– SMSD:Activity: represents based on several
sensors readings the patients activity.
– SMSD:Location: represents the physical loca-
tion of the patient.
– SMSD:Time: represents temporal concepts for
describing the temporal properties of the cap-
tured data.
– SMSD:Environment: describes the characteris-
tics of the surrounding environment condition
of the patient.
The SMSD ontology is encoded in OWL 2 (using
Protg 5.5, an open source ontology editor).
3.2 Unstructured EHR Data Integration
It is well known that a lot of relevant data for making
accurate healthcare decisions is available in unstruc-
tured form. This data needs to be converted into a
structured fashion to allow easy analysis.
Advances in artificial intelligence, such as deep
learning techniques based on Natural Language Pro-
cessing (NLP) (Mellish, 1989), enable easy and
meaningful information extraction from unstructured
data and generate a structured representation.
To make sense of unstructured data, a Named En-
tity Recognition (NER) (Chieu and Ng, 2003) ap-
proach is adopted by the proposed framework. NER
is a sub-field of information extraction (IE) that works
by searching for specific entity terms and by group-
ing them into pre-defined categories. Entity recogni-
tion has been widely developed in the context of the
medical field, usually under the name of Medical En-
tity Recognition (MER) (Abacha and Zweigenbaum,
2011) in order to identify medical entities.
MetaMap program (Aronson, 2006) developed by
the National Institute of Health (NIH) is considered a
baseline in MER. SF4FI-EHR uses MetaMap to dis-
cover and map medical text to UMLS terminologies.
MetaMap’s output generated from unstructured data
is formatted in JSON format, and then transmitted to
FHIR Transform Engine (FTE)
5
to be converted into
FHIR resources. Fig.3 illustrates the described trans-
formation process.
3.3 Structured EHR Data Integration
Structured EHR data are scattered across multiple
sources, designed by different data models, encoded
with heterogeneous terminologies and represented in
5
http://www.openmapsw.com/products/FTE.htm
ICAART 2020 - 12th International Conference on Agents and Artificial Intelligence
688

Figure 3: Unstructured EHR data integration process.
various serialization formats. These data need to be
extracted from their original sources first, and then
restructured into a common format and standard ter-
minologies, in order to make easy their subsequent
integration with other data.
A RESTful FHIR server based on FHIR Trans-
form Engine (FTE) is embedded into our framework,
to deal with structured EHR data. Since it is REST-
ful, the FHIR server can collect data from distributed
EHR systems via HTTP GET requests. The data get-
ting in response are represented as JSON-based re-
sources thanks to the FTE.
3.4 The Fully Integrated EHR
To consolidate all parts of the collected data into a
single centralized location, and maintain a compre-
hensive health patient profile, a mapping process is
required between the resulted data from the already
described processes and the W3C’s FHIR OWL On-
tology
6
.
IoT data, modeled using the SMSD OWL
ontology, are mapped to the FHIR ontol-
ogy using specific defined mapping rules.
For instance, the SSN:Obersavation and
SMSD:Activity from the SMSD ontology are
mapped to the fhir.ResourceObservation and
fhir.ResourceActivityDefinition, respectively.
For the other pieces of data (resulted from Un-
structured and structured EHR data integration pro-
cesses) which are already standardized based on
6
http://w3c.github.io/hcls-fhir-rdf/spec/ontology.html
FHIR standard, a direct map between their resources
elements and the FHIR ontology resources is per-
formed. As a result, a FHIR-based fully integrated
EHR that aggregates patient health-related data from
disparate sources, is created. Finally, a conversion
step is running to map FHIR Resources content from
the FHIR OWL ontology to their equivalent in the
FHIR Relational DataBase schema. Data obtained is
stored in FHIRbase, in order to make data available
for future purposes.
4 IMPLEMENTATIONS
As a proof-of-concept implementation, we used: 1)
Protg 5.5 to develop the OWL SMSD ontology for
modeling and semantically annotate IoT raw data, 2)
Snoggle as SWRL-based ontology mapper is used to
assist in the mapping task between the SMSD and the
FHIR ontology, 3) MetaMap tool is used to recog-
nize medical entities from unstructured data and gen-
erate a JSON-based structured output. MetaMap’s re-
sults is transformed into FHIR-based documents for-
mat by using FHIR Transform Engine, 4) RESTful
HAPI-FHIR server based on FHIR Transform Engine
(FTE) is implemented to collect data from distributed
EHR systems, and 5) JSON2OWL mapping is devel-
oped to map JSON -based data to OWL FHIR ontol-
ogy. The collected data are stored in FHIRbase, as a
standard RDB implementation provided by HL7.
5 EVALUATION RESULTS
To validate our work, a clinical data sets relating to
10 patients have been used as input to our framework,
comprising for each patient:
• An standardized electronic health record.
• Non-standardized clinical documents (e.g. bio-
logical analyzes and radiology reports)
• A vital signs data set generated from different
medical devices carried by patient (temperature,
blood pressure, heart rate and respiratory rate)
• Readings from patient smartphone multiple sen-
sors that can indicate the patients’ activities and
his surrounding environment (e.g. GPS, Ac-
celerometer, Ambient Light Sensor, etc.).
The main objective of this evaluation is to exhibit
that the health data integration process can be success-
fully conducted using SF4FI-EHR framework. Each
data set will be processed according to its category.
A Flexible Semantic Integration Framework for Fully-integrated EHR based on FHIR Standard
689

Sensor data are modeled by the modular SMSD on-
tology, unstructured data are processed by MetaMap
and then converted to FHIR resources formats, and
structured EHR data are converted to FHIR resources
formats, before being directly mapped to the FHIR
OWL Ontology. All data collected modeled by the
FHIR ontology are storing into RDB FHIRbase.
A prototype has been deployed that allowed us
to set up a first set of tests using the web browser.
SQL queries are executed to demonstrate that the dif-
ferent data resources are successfully integrated to-
gether. The SQL query shown in Fig.4 is executed to
query for a particular patient observation resources.
Figure 4: The query executed.
The query result is presented in Fig.5, in which
we can see that both the blood glucose value from
an unstructured/not standardized document (biology
analysis results) and the vital signs data from medical
devices are both retrieved in the Observation resource
section in the new Integrated EHR.
Figure 5: The result of the query executed.
To make sure that the integration was successfully
carried out, we invited domain experts to assist the
evaluation process and to assess the quality and the
validity of the integrated data. Discussions with ex-
perts led to that the integration data is well done, as
well as the correctness and the consistency of the in-
tegrated data is well ensured.
6 CONCLUSION
In this paper, we described a flexible semantic inte-
gration framework called SF4FI-EHR. Our solution
integrates its data, as well as Unstructured and Struc-
tured EHR data, coming from heterogeneous sources.
The integration of different data formats is achieved
thanks to the application of Semantic Web technolo-
gies. A modular SMSD ontology based on SSN is
proposed to describe its data. MetaMap tool is used
to deal with the unstructured data, in order to extract
meaningful information from these data and generate
a structured output. Historical patient data collected
from distributed EHR systems using a Restful FHIR
server based on FHIR Tranform Engine. All obtained
data are mapped to the OWL FHIR ontology to re-
construct a Fully integrated EHR data. The semantic
interoperability is handled based on the HL7 FHIR
standard.
Future work will focus on evaluating this frame-
work with more real-world use cases for improvement
and validation. Furthermore, We planned to improve
our approach by integrating environmental data such
as Smart Homes and Ambient Assisted Linvig envi-
ronment data.
REFERENCES
Abacha, A. B. and Zweigenbaum, P. (2011). Medical entity
recognition: A comparison of semantic and statistical
methods. In Proceedings of BioNLP 2011 Workshop,
pages 56–64. Association for Computational Linguis-
tics.
Alamri, A. (2018). Ontology middleware for integration
of iot healthcare information systems in ehr systems.
Computers, 7(4):51.
Aronson, A. R. (2006). Metamap: Mapping text to the umls
metathesaurus. Bethesda, MD: NLM, NIH, DHHS,
pages 1–26.
Ashton, K. et al. (2009). That internet of things thing. RFID
journal, 22(7):97–114.
Bender, D. and Sartipi, K. (2013). Hl7 fhir: An agile and
restful approach to healthcare information exchange.
In Proceedings of the 26th IEEE international sympo-
sium on computer-based medical systems, pages 326–
331. IEEE.
Chieu, H. L. and Ng, H. T. (2003). Named entity recogni-
tion with a maximum entropy approach. In Proceed-
ings of the seventh conference on Natural language
learning at HLT-NAACL 2003-Volume 4, pages 160–
163. Association for Computational Linguistics.
Compton, M., Barnaghi, P., Bermudez, L., Garc
´
ıA-Castro,
R., Corcho, O., Cox, S., Graybeal, J., Hauswirth, M.,
Henson, C., Herzog, A., et al. (2012). The ssn on-
tology of the w3c semantic sensor network incubator
group. Web semantics: science, services and agents
on the World Wide Web, 17:25–32.
De Moor, G., Sundgren, M., Kalra, D., Schmidt, A., Dugas,
M., Claerhout, B., Karakoyun, T., Ohmann, C., Las-
tic, P.-Y., Ammour, N., et al. (2015). Using elec-
tronic health records for clinical research: the case of
the ehr4cr project. Journal of biomedical informatics,
53:162–173.
Dhayne, H., Kilany, R., Haque, R., and Taher, Y. (2018).
Sedie: A semantic-driven engine for integration of
healthcare data. In 2018 IEEE International Con-
ference on Bioinformatics and Biomedicine (BIBM),
pages 617–622. IEEE.
ICAART 2020 - 12th International Conference on Agents and Artificial Intelligence
690

Eichelberg, M., Aden, T., Riesmeier, J., Dogac, A., and
Laleci, G. B. (2005). A survey and analysis of elec-
tronic healthcare record standards. Acm Computing
Surveys (Csur), 37(4):277–315.
El Fadly, A., Daniel, C., Bousquet, C., Dart, T., Lastic,
P.-Y., and Degoulet, P. (2007). Electronic healthcare
record and clinical research in cardiovascular radiol-
ogy. hl7 cda and cdisc odm interoperability. In AMIA
Annual Symposium Proceedings, volume 2007, page
216. American Medical Informatics Association.
El-Sappagh, S., Ali, F., Hendawi, A., Jang, J.-H., and
Kwak, K.-S. (2019). A mobile health monitoring-and-
treatment system based on integration of the ssn sen-
sor ontology and the hl7 fhir standard. BMC medical
informatics and decision making, 19(1):97.
Gay, V. and Leijdekkers, P. (2015). Bringing health and fit-
ness data together for connected health care: mobile
apps as enablers of interoperability. Journal of medi-
cal Internet research, 17(11):e260.
Genitsaridi, I., Kondylakis, H., Koumakis, L., Marias, K.,
and Tsiknakis, M. (2015). Evaluation of personal
health record systems through the lenses of ec re-
search projects. Computers in biology and medicine,
59:175–185.
Gkoutos, G. V., Schofield, P. N., and Hoehndorf, R. (2012).
The units ontology: a tool for integrating units of mea-
surement in science. Database, 2012.
Goldfain, A., Smith, B., Arabandi, S., Brochhausen, M.,
and Hogan, W. R. (2011). Vital sign ontology.
Grenon, P., Smith, B., and Goldberg, L. (2004). Biody-
namic ontology: applying bfo in the biomedical do-
main. Studies in health technology and informatics,
pages 20–38.
Hong, N., Wen, A., Shen, F., Sohn, S., Liu, S., Liu, H., and
Jiang, G. (2018). Integrating structured and unstruc-
tured ehr data using an fhir-based type system: A case
study with medication data. AMIA Summits on Trans-
lational Science Proceedings, 2018:74.
Kilic, O. and Dogac, A. (2009). Achieving clinical
statement interoperability using r-mim and archetype-
based semantic transformations. IEEE Transac-
tions on Information Technology in Biomedicine,
13(4):467–477.
Kumar, R. B., Goren, N. D., Stark, D. E., Wall, D. P.,
and Longhurst, C. A. (2016). Automated integra-
tion of continuous glucose monitor data in the elec-
tronic health record using consumer technology. Jour-
nal of the American Medical Informatics Association,
23(3):532–537.
Maharatna, K. and Bonfiglio, S. (2013). Systems Design
for Remote Healthcare. Springer Science & Business
Media.
Mart
´
ınez-Costa, C., Kalra, D., and Schulz, S. (2014). Im-
proving ehr semantic interoperability: future vision
and challenges. In MIE, pages 589–593.
Mellish, C. (1989). Natural Language Processing in Pro-
log: an introduction to computational linguistics.
Addison-Wesley.
Menachemi, N. and Collum, T. H. (2011). Benefits and
drawbacks of electronic health record systems. Risk
management and healthcare policy, 4:47.
Mori, A. R. and Consorti, F. (1998). Exploiting the ter-
minological approach from cen/tc251 and galen to
support semantic interoperability of healthcare record
systems. International journal of medical informatics,
48(1-3):111–124.
Mori, A. R., Consorti, F., and Galeazzi, E. (1998). Stan-
dards to support development of terminological sys-
tems for healthcare telematics. Methods of Informa-
tion in Medicine, 37(04/05):551–563.
Peng, C. and Goswami, P. (2019). Meaningful integration
of data from heterogeneous health services and home
environment based on ontology. Sensors, 19(8):1747.
Preuveneers, D., Van den Bergh, J., Wagelaar, D., Georges,
A., Rigole, P., Clerckx, T., Berbers, Y., Coninx, K.,
Jonckers, V., and De Bosschere, K. (2004). Towards
an extensible context ontology for ambient intelli-
gence. In European Symposium on Ambient Intelli-
gence, pages 148–159. Springer.
Rasmussen-Torvik, L. J., Stallings, S. C., Gordon, A. S.,
Almoguera, B., Basford, M. A., Bielinski, S. J., Braut-
bar, A., Brilliant, M., Carrell, D. S., Connolly, J.,
et al. (2014). Design and anticipated outcomes of the
emerge-pgx project: a multicenter pilot for preemp-
tive pharmacogenomics in electronic health record
systems. Clinical Pharmacology & Therapeutics,
96(4):482–489.
Sun, H., Depraetere, K., De Roo, J., Mels, G., De Vloed, B.,
Twagirumukiza, M., and Colaert, D. (2015). Semantic
processing of ehr data for clinical research. Journal of
biomedical informatics, 58:247–259.
Tagaris, A., Chondrogiannis, E., Andronikou, V., Tsatsa-
ronis, G., Mourtzoukos, K., Roumier, J., Matska-
nis, N., Schroeder, M., Massonet, P., Koutsouris, D.,
et al. (2012). Semantic interoperability between clin-
ical research and healthcare: the ponte approach. In
Extended Semantic Web Conference, pages 191–203.
Springer.
Vuppalapati, C., Ilapakurti, A., and Kedari, S. (2016). The
role of big data in creating sense ehr, an integrated
approach to create next generation mobile sensor and
wearable data driven electronic health record (ehr).
In 2016 IEEE Second International Conference on
Big Data Computing Service and Applications (Big-
DataService), pages 293–296. IEEE.
Wang, X., Zhang, D., Gu, T., Pung, H. K., et al. (2004).
Ontology based context modeling and reasoning us-
ing owl. In Percom workshops, volume 18, page 22.
Citeseer.
Weber, G. M., Murphy, S. N., McMurry, A. J., MacFad-
den, D., Nigrin, D. J., Churchill, S., and Kohane, I. S.
(2009). The shared health research information net-
work (shrine): a prototype federated query tool for
clinical data repositories. Journal of the American
Medical Informatics Association, 16(5):624–630.
A Flexible Semantic Integration Framework for Fully-integrated EHR based on FHIR Standard
691
