
Evaluating Cross-lingual Semantic Annotation for Medical Forms
Ying-Chi Lin
1 a
, Victor Christen
1 b
, Anika Groß
2 c
, Toralf Kirsten
3,4 d
,
Silvio Domingos Cardoso
5,6 e
, C
´
edric Pruski
5 f
, Marcos Da Silveira
5 g
and Erhard Rahm
1 h
1
Department of Computer Science, Leipzig University, Germany
2
Department Computer Science and Languages, Anhalt University of Applied Sciences in Koethen/Anhalt, Germany
3
Faculty Applied Computer Sciences and Biosciences, Mittweida University of Applied Sciences, Germany
4
LIFE Research Centre for Civilization Diseases, Leipzig University, Germany
5
LIST, Luxembourg Institute of Science and Technology, Luxembourg
6
LRI, University of Paris-Sud XI, France
Keywords:
Cross-lingual Semantic Annotation, Medical Forms, UMLS, Machine Translation.
Abstract:
Annotating documents or datasets using concepts of biomedical ontologies has become increasingly important.
Such ontology-based semantic annotations can improve the interoperability and the quality of data integration
in health care practice and biomedical research. However, due to the restrictive coverage of non-English
ontologies and the lack of comparably good annotators as for English language, annotating non-English doc-
uments is even more challenging. In this paper we aim to annotate medical forms in German language. We
present a parallel corpus where all medical forms are in both German and English languages. We use three
annotators to automatically generate annotations and these annotations are manually verified to construct an
English Silver Standard Corpus (SSC). Based on the parallel corpus of German and English documents and
the SSC, we evaluate the quality of different annotation approaches, mainly 1) direct annotation using German
corpus and German ontologies and 2) integrating machine translators to translate German corpus and annotate
the translated corpus with English ontologies. The results show that using German ontologies only produces
very restricted results, whereas translation achieves better annotation quality and is able to retain almost 70%
of the annotations.
1 INTRODUCTION
Ontologies are an established means to formally
represent domain knowledge and they are used in-
tensely in the life sciences, in particular to seman-
tically describe or annotate biomedical documents.
Such semantic annotations are helpful to improve
interoperability and the quality of data integration,
e.g., in health care practice and biomedical research
(Hoehndorf et al., 2015). Biomedical annotations can
a
https://orcid.org/0000-0003-4921-5064
b
https://orcid.org/0000-0001-7175-7359
c
https://orcid.org/0000-0002-2684-8427
d
https://orcid.org/0000-0001-7117-4268
e
https://orcid.org/0000-0003-2643-9890
f
https://orcid.org/0000-0002-2103-0431
g
https://orcid.org/0000-0002-2604-3645
h
https://orcid.org/0000-0002-2665-1114
also enhance retrieval quality for semantic document
search (Jovanovi
´
c and Bagheri, 2017). For instance,
PubMed
1
, the search engine for MEDLINE database,
uses MeSH (Medical Subject Headings) terms to re-
trieve more relevant results. The Text Information Ex-
traction System (TIES)
2
of University of Pittsburgh
utilizes concepts from the NCI Metathesaurus and
their synonyms to obtain better recall of documents.
Further, using the hierarchy information within the
ontologies can further expand the potential matches.
The TIES has been applied to retrieve clinical data
of the Cancer Genome Atlas (TCGA) dataset and is
able to find cases based on the semantic features of
the pathology report.
Annotating data across multiple disconnected
databases using concepts of the same ontologies sup-
1
PubMed https://www.ncbi.nlm.nih.gov/pubmed
2
TIES http://ties.dbmi.pitt.edu
Lin, Y., Christen, V., Groß, A., Kirsten, T., Cardoso, S., Pruski, C., Da Silveira, M. and Rahm, E.
Evaluating Cross-lingual Semantic Annotation for Medical Forms.
DOI: 10.5220/0008979901450155
In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2020) - Volume 5: HEALTHINF, pages 145-155
ISBN: 978-989-758-398-8; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
145

ports improved data integration. One example is the
German Biobank Alliance (GBA)
3
. The alliance con-
sists of biobanks from 15 German university hospitals
and two IT expert centres. The aim is to establish uni-
form quality standards and to make their biomaterials
available for biomedical research throughout Europe.
The information about the specimens, such as the di-
agnosis of the patients, are annotated with LOINC
4
(McDonald et al., 2003) and ICD-10
5
(World Health
Organization, 2004) codes. A search interface that in-
tegrates these codes allows researchers to extract in-
formation across all biobanks.
We use the term cross-lingual semantic annotation
task to denote the process of assigning concepts of
English ontologies to text segments of non-English
documents. This process is a necessity mainly be-
cause many biomedical ontologies are most well de-
veloped in English while ontologies in other lan-
guages are by far less comprehensive and do not cover
as much knowledge (Schulz et al., 2013; N
´
ev
´
eol et al.,
2014b; Starlinger et al., 2017; N
´
ev
´
eol et al., 2018 and
as we will also show later in this paper). For instance,
in the release version 2019AA of the Unified Medi-
cal Language System (UMLS) Metathesaurus, among
the total of 14.6 million terms, 71% of them are in
English, followed by 10% in Spanish. The other lan-
guages, such as French, Japanese or Portuguese, each
covers less than 3% of the total term amounts. In addi-
tion, the tools for annotating non-English biomedical
documents are not as well developed as tools for En-
glish documents (Jovanovi
´
c and Bagheri, 2017), such
as MetaMap (Aronson and Lang, 2010) or cTAKES
(Savova et al., 2010). For instance, there have been ef-
forts to adapt cTAKES for German corpora but further
improvement is still needed (Becker and B
¨
ockmann,
2016). Overall, before UMLS has been adequately
extended to cover its concepts in non-English and
good quality annotators have been developed, cross-
lingual annotation is the current way to overcome
such deficiencies.
LIFE, stands for Leipzig Research Center for Civ-
ilization Diseases and conducts several epidemiolog-
ical studies including LIFE Adult Study (Loeffler
et al., 2015) and LIFE Heart Study (Beutner et al.,
2011). The goal of LIFE Adult Study is to investigate
the influence of genetic, environmental, social and
lifestyle factors on the healthy state of the local popu-
lation in the city of Leipzig, Germany. Since 2005, the
project has recruited more than 10,000 participants.
All participants took part at an extensive core assess-
3
GBA https://www.bbmri.de
4
Logical Observation Identifier Names and Codes
5
International Statistical Classification of Diseases:
tenth revision
ment program including structured interviews, ques-
tionnaires, physical examinations, and biospecimen
collection. In this paper, we aim to annotate some
of the medical forms used in the LIFE Adult Study.
Since these LIFE forms are in German and we try
to produce annotations as comprehensively and well-
covered as possible, we decided to tackle the task as a
cross-lingual semantic annotation problem.
In this study, we report different strategies to an-
notate German forms and also investigate the quality
change of using machine translation in such annota-
tion workflow. Our work has the following main con-
tributions:
1) We manually build a parallel corpus in both En-
glish and German of medical forms used in a large
epidemiological study.
2) We manually build a Silver Standard Corpus (SSC)
that provides good quality annotations to evaluate
cross-lingual semantic annotation tasks.
3) Based on 1) and 2), we are able to compare the an-
notation quality for using German ontologies on Ger-
man forms versus the use of English ontologies.
4) We also investigate the annotation quality when us-
ing machine translation.
5) While questions in medical forms are typically an-
notated with several concepts we also consider the
special case where questions correspond to a single
concept. We determine a Gold Standard Corpus for
such question-as-concept annotations and provide an
initial evaluation for them.
In the following Section 2 we present related work
on building our SSC and previous studies on cross-
lingual semantic annotations. Section 3 describes
firstly the parallel corpus and the ontologies we used
for annotation. We then explain the methods used
to build the Silver Standard and the various annota-
tion tasks we applied to annotate non-English medical
forms. In Section 4 we present the SSC. We further
show the results of the various annotation tasks and
compare them to the SSC. Finally, in Section 5 we
summarize our findings and discuss directions for fu-
ture research.
2 RELATED WORK
Silver Standard Corpus (SSC)
Building a manually annotated Gold Standard Corpus
(GSC) is laborious, costly and needs human exper-
tise. Hence, GSC can only cover a small number of
semantic groups and is limited in the number of doc-
uments (Rebholz-Schuhmann et al., 2011). To over-
come these shortcomings, the construction of Silver
Standard Corpora (SSC) has been proposed. The term
HEALTHINF 2020 - 13th International Conference on Health Informatics
146

”Silver Standard” has been referred to corpora that
are generated using several annotation tools and their
outputs are then harmonized automatically (Rebholz-
Schuhmann et al., 2010; Lewin et al., 2013; Oellrich
et al., 2015).
The ”Collaborative Annotation of a Large Scale
Biomedical Corpus” (CALBC) is one of the first
attempts to generate a large-scale automated anno-
tated biomedical SSC (Rebholz-Schuhmann et al.,
2010). The researchers applied four annotation tools
on 150,000 MEDLINE abstracts and the resulting
annotations are included in the final SSC based on
their cosine similarity and at least two annotators
agree on the same annotation (2-vote-agreement).
In 2013, an English SSC was developed for the
multilingual CLEF-ER named entity recognition
challenge (Lewin et al., 2013). In total six annotation
tools were used to generate the annotations. The
annotations were chosen with a voting threshold of
3 tools and by applying a centroid algorithm (Lewin
et al., 2012, 2013). Simpler methods were applied in
(Oellrich et al., 2015) where 2-vote-agreement was
used as inclusion criterion by using four annotators
and the annotations have to be exact or partially
matched. To be able to determine the annotation
quality of the cross-lingual annotation tasks, we
also built an English SSC. As in previous works, we
use automatic annotation tools to generate a set of
annotations first. But differently, we then manually
verified the generated annotations. In this way we can
avoid the problem that using voting agreement for
inclusion might only reveal the closeness of the tools
but not necessarily the correctness of the annotations
(Oellrich et al., 2015).
Biomedical Cross-lingual Semantic Annotation
The CLEF (Conference and Labs of the Evaluation
Forum) has hosted several challenges on cross-lingual
annotation of biomedical named entities. The CLEF-
ER 2013 evaluation lab (Rebholz-Schuhmann et al.,
2013) was organized as part of the EU project ”Multi-
lingual Annotation of Named Entities and Terminol-
ogy Resources Acquisition” (MANTRA). The orga-
nizers provided parallel corpora including MEDLINE
titles, EMEA (European Medicines Agency) drug la-
bels and patent claims in English, German, French,
Spanish and Dutch. These documents were annotated
using terms from ten UMLS Semantic Groups, in both
English and non-English concepts. An English SSC
was also prepared by the organizers (Lewin et al.,
2013). The seven systems submitted to the challenge
showed high heterogeneity (Hellrich et al., 2014).
The CLEF-ER 2013 resulted in a small multilin-
gual gold standard corpus (the Mantra GSC) that con-
tains 5530 annotations in the above mentioned five
languages (Kors et al., 2015) and the QUAERO cor-
pus, a larger GSC with 26,281 annotations in French
(N
´
ev
´
eol et al., 2014a). The QUAERO corpus was
used in CLEF eHealth 2015 Task 1b (N
´
ev
´
eol et al.,
2015) and 2016 Task 2 (N
´
ev
´
eol et al., 2016), the fur-
ther cross-lingual annotation challenges. The best so-
lution in 2015 (Afzal et al., 2015) used the intersec-
tion of two translators, i.e. Google Translator and
Bing Translate, to expand the UMLS terminology into
French. The annotations were generated by a rule
based dictionary lookup system, Peregrine (Schuemie
et al., 2007). The generated annotations went through
several post-processing steps to reduce false positives.
The challenge in 2016 used the same training set as in
2015 but took the test set provided in 2015 as the de-
velopment set. The test set in 2016 was a new one,
hence, the results between the two years cannot be
compared directly. The winning team in 2016 (Cabot
et al., 2016) used ECMT (Extracting Concepts with
Multiple Terminologies) that relies on bag-of-words
and pattern-matching to extract concept. The system
integrates up to 13 terminologies that were translated
into French.
In 2018, Roller et al. (2018) proposed a sequential
concept normalization system that outperformed the
winning teams in the CLEF challenges in 2015 and
2016 in most of the corpora. They use Solr to lookup
concepts sequentially, i.e. the French term to be an-
notated is firstly searched in the French UMLS, then
the English version of the UMLS and finally the term
is translated into English using a self-developed neu-
ral translation model for further searching in the En-
glish UMLS. Similar post-processing procedures as
in (Afzal et al., 2015) were also applied.
Most recently, Perez et al. (2019) evaluated two
approaches to annotate Spanish biomedical docu-
ments with concepts in the Spanish UMLS. The
first approach integrates a NLP pipeline in the pre-
processing, Apache Lucene for concept indexing and
a word-sense disambiguation component. The sec-
ond approach uses machine translation to obtain En-
glish documents and these documents are annotated
using MetaMap with the same Spanish UMLS sub-
set but the English version. Finally, the generated
English annotations are transferred back to Spanish.
The combination of the results using union of these
two approaches performed best (F-measure of 67.1%)
on the MEDLINE sub-corpus in the Spanish Mantra
GSC. On the other hand, Roller et al. (2018) achieved
69.1% in F-measure on the same sub-corpus.
As mentioned above, machine translators have
been mostly applied to translate the ontologies into
the corresponding language in the cross-lingual anno-
Evaluating Cross-lingual Semantic Annotation for Medical Forms
147

example_1
Page 1
Question Associated UMLS concepts
English Feeling nervous, anxious, or on edge
1 C0027769 nervous
2 C0003467 anxious
3 C3812214 Feeling nervous, anxious or on edge
German Nervosität, Ängstlichkeit oder Anspannung
1 C0027769 Nervosität
2 C0087092 Ängstlichkeit
DeepL nervousness, anxiety or tension
1 C0027769 nervous
2 C0003467 anxious
Figure 1: An example question from the form ”Generalized Anxiety Disorder” in original English, German and after DeepL
translation. The associated UMLS concepts to each version of the question are also presented.
tation tasks (e.g. Hellrich and Hahn, 2013; van Mul-
ligen et al., 2013; Afzal et al., 2015). In contrast to
previous work, we choose to translate the corpus but
not the ontologies. This is mainly because with our
parallel corpus and the advantage of having manually
verified SSC, we are able to examine the impact on
annotation quality of utilizing machine translators.
3 METHODS
3.1 Corpus and Ontology
The corpus used in this study includes 37 forms and
728 questions in German. These forms were used in
the LIFE Adult Study (see Section 1). We selected
the assessment forms that are available in multiple
languages including English and German or are orig-
inally in English and have been translated into Ger-
man. Hence, we can build a parallel corpus consisting
of forms in both English and German. Some exam-
ples of these forms are the standardized instruments
used in clinical studies such as the Patient Health
Questionnaire (PHQ, Kroenke et al., 2002) and the
form ”Generalized Anxiety Disorder” (GAD-7, L
¨
owe
et al., 2008). Figure 1 shows an example question
from GAD-7 in three different versions: English, Ger-
man and the German-translated version using the ma-
chine translator DeepL (see Section 3.3 for further de-
tail). It also presents the associated UMLS concepts
for each version of the question.
The goal of the annotation task is to find as
many annotations as possible per question of a form
so that we can maximize semantic interoperabil-
ity for our corpus. We choose UMLS Metathe-
saurus as ontology source and use three annota-
tion tools, namely MetaMap (Aronson and Lang,
2010), cTAKES (Becker and B
¨
ockmann, 2016) and
AnnoMap (Christen et al., 2015, 2016), to annotate
the forms. UMLS integrates many biomedical ontolo-
gies. We use its version 2017AA which contains 201
source vocabularies with approximately 3.47 million
concepts. For different annotation tasks we utilize dif-
ferent subsets of UMLS as explained in Section 3.3
below.
3.2 The Silver Standard Corpus
The SSC is constructed using the English corpus and
the English version of UMLS. Since UMLS is very
large and not all contained ontologies are relevant, we
select a subset of it to optimize efficiency. In a pre-
vious study, Lin et al. (2017) compared the three an-
notators that we also use in this work. The results
showed that cTAKES generates the best recall. Con-
sequently, we use cTAKES to determine a subset of
the UMLS that covers approximately 99% of the an-
notations that were found by using the entire UMLS.
The subset includes 1.03 million concepts from six
ontologies: 1) UMLS Metathesaurus
6
, 2) MeSH, 3)
NCI Thesaurus, 4) LOINC, 5) SNOMED CT US, and
6) Consumer Health Vocabulary. We call this subset
selected UMLS subset.
We use three tools: MetaMap, cTAKES and
AnnoMap to annotate the English forms with the
selected UMLS subset. A detailed description and
tests of the parameters of these three tools can be
found in our previous publication (Lin et al., 2017).
Since we will manually verify the pre-annotations
produced by the automatic annotators, we set up the
tools in a way that they can produce good recall but
not extremely bad precision. For each annotation can-
didate, MetaMap computes a score considering lin-
guistic metrics and the scores range between 0 and
1000. We use a filtering score of 700 for MetaMap,
i.e. only candidates having scores of more than 700
will be included in the result set. Based on our initial
study, the annotation results using a filtering score 700
also gained better F-measure than those using filtering
scores 800 and 900 (which are subsets of 700). Be-
cause cTAKES tends to return many false positives,
we use longestMatch and without overlap to get the
best precision. With longestMatch setting, we allow
cTAKES to return only the concept with the longest
6
This is a subset of the UMLS Metathesaurus with the
Root Source Abbreviation (RSAB) as ”MTH”
HEALTHINF 2020 - 13th International Conference on Health Informatics
148
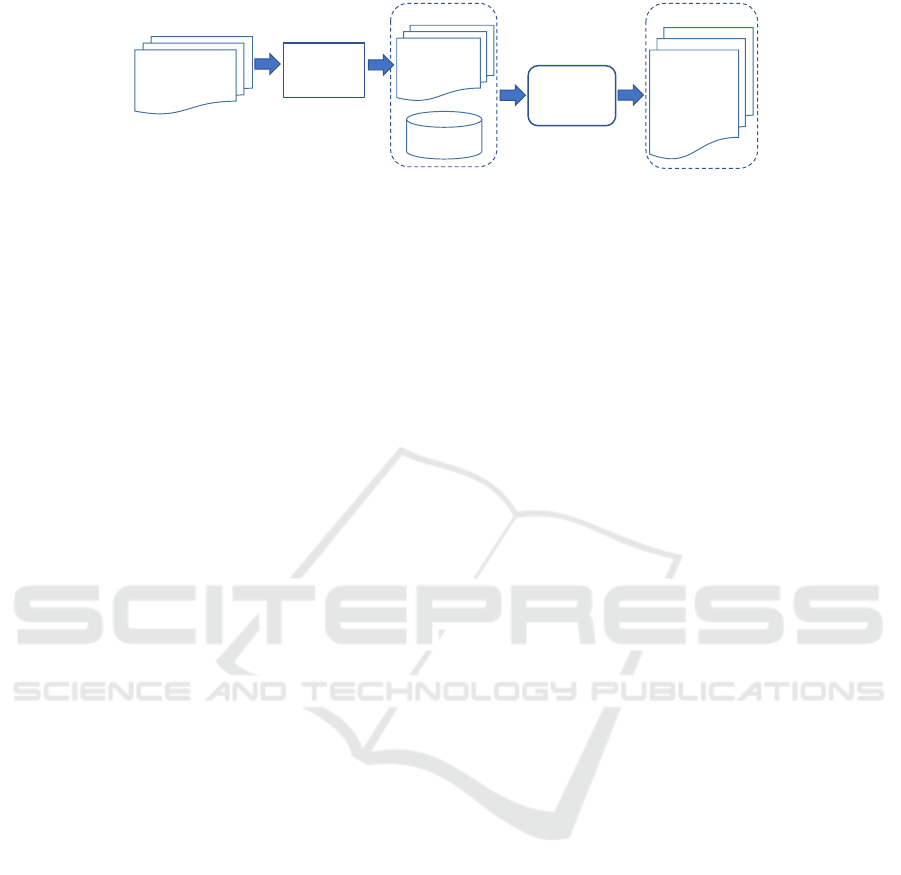
– MetaMap
– cTAKES
– AnnoMap
– DeepL
– Microsoft
Translator
Output
Annotation
mappings
!ℳ
#
$
,&'()
…
!ℳ
#
*
,&'()
Set of
German forms
f
1
…
f
m
UMLS
Translated
English forms
f
1
…
f
m
Machine
translator
Annotator
Figure 2: Workflow for cross-lingual semantic annotation using machine translators.
matched span and without overlap avoids the matches
on discontiguous spans.
For AnnoMap, we investigate two configu-
rations: 1) AnnoMapQuestion that uses whole
questions to find matching concepts for annota-
tion and 2) AnnoMapWindow that matches frag-
ments of questions within a defined window size.
AnnoMapQuestion constructs a similarity score from
three string similarity functions: TF/IDF, Trigram and
LCS (longest common substring) similarity. It re-
tains all annotation candidates with a score above
a given threshold δ. For this work we set three
filtering thresholds (δ) of 0.5, 0.6, and 0.7 for the
AnnoMapQuestion. AnnoMapWindow uses Soft
TF/IDF to match words/phrases within a defined win-
dow size. We consider three window sizes of 2, 3,
and 5 words. An annotation is kept if its Soft TF/IDF
similarity is larger than 0.7. A further filtering mecha-
nism in AnnoMap called group-based selection is also
applied to improve precision (Christen et al., 2015).
The annotated results are verified manually to
build the SSC. The manual verification is done by
two human annotators with assistance of a GUI
application for annotation selection (Geistert, 2018).
An annotation is included in the SSC if both human
annotators consented. For the manual selection of the
annotations, we use the following principles:
1. Use the context to select the most appropriate
concept: if there are several concepts assigned to the
same segment of text, choose the one with the most
precise description based on definition, synonym and
semantic types.
2. If more than one concept fits perfectly to the same
mention, keep all of those concepts: this is the case
when concepts are of different but suitable semantic
types. For instance, for the question ”Shortness
of breath” in the form PHQ, the UMLS concept
with CUI (Concept Unique Identifier) C0013404 of
semantic type ”Sign or Symptom” and C3274920 of
semantic type ”Intellectual Product” are both correct
and are both retained as annotations.
3. If the whole question matches to a UMLS concept,
we also include concepts that match and thus anno-
tate fragments of the question. For example, UMLS
concept C3812214 is named ”Feeling nervous,
anxious or on edge” and corresponds to a whole
question in form GAD-7 (see Figure 1). We call
such concepts questionAsConcept-type annotations
(QaC-annotations) in this paper. In the manual
verification process, we not only keep such concepts,
but also the concepts which covers fragments of the
question, e.g. the concepts C0027769 for ”nervous”
and C0003467 for ”anxious”.
We calculate the Inter-Annotator Agreement be-
tween the two human annotators using Cohen’s
Kappa (Cohen, 1960).
3.3 Annotating German Medical Forms
We investigate two approaches to annotate German
forms. Firstly, we explore the feasibility of annotating
the corpus of German forms with the German UMLS.
Secondly, we investigate the annotation of machine-
translated corpora with English UMLS versions.
Using the SSC as the reference mapping, we are able
to comparatively evaluate the annotation quality of
both approaches. We note that the annotation quality
is also influenced by the translation from the original
English corpus to the German version. However, it
is difficult to quantify this quality change due to the
lack of corresponding subset of German UMLS.
Annotating German Corpus: To annotate the
German corpus, we use all available German on-
tologies in UMLS (German UMLS). They include
1) DMDICD10 (ICD-10), 2) ICPCGER (ICPC),
3) LNC-DE (LOINC), 4) MDRGER (MedDRA),
5) MSHGER (MeSH), 6) DMDUMD (UMDNS)
and 7) WHOGER (WHOART). Since cTAKES and
MetaMap are designed to annotate English corpora,
we use AnnoMapQuestion to annotate the German
corpus. The annotations found by AnnoMapQuestion
are verified manually to obtain only correct annota-
tions. Further, we use the same method and the same
seven ontologies but in English (German UMLS in
English) to annotate the parallel English corpus to
produce comparable results as to the German corpus.
Evaluating Cross-lingual Semantic Annotation for Medical Forms
149

These annotations are also manually verified.
Annotation using Machine Translated Corpora:
Figure 2 shows our workflow for cross-lingual an-
notation using machine translators. We translate the
German forms using machine translators and annotate
the translated forms using the English UMLS. To be
comparable, we use the same method to annotate the
translated corpora as for the SSC, i.e. use selected
UMLS subset as ontologies and the three annotation
tools with the same parameter settings.
For the selection of suitable machine translators,
we first randomly selected 50 questions from our Ger-
man corpus and translated them into English using
five machine translators: DeepL
7
, Microsoft Trans-
lator
8
, Google Translate
9
, Yandex
10
and Moses
11
.
We manually checked the translated results. The
translators that produced the best translations of a
question (most similar to the original English text)
was graded with one point. Finally, we selected
the best two translators with the highest number of
points for further experiments. They are DeepL and
Microsoft Translator.
Lin et al. (2017) showed that combining anno-
tations generated by three annotators using 2-vote-
agreement (at least found by two of the three anno-
tators) can obtain better F-measure than using single
tools. Hence, we combine the results from MetaMap,
cTAKES, AnnoMapQuestion and AnnoMapWindow
using this method. Further, combining annotation re-
sults using union can improve recall. Hence, we fur-
ther combine the results from the annotators using
union to see how many correct annotations are re-
tained after translation. In addition to combining the
results from the tools, the results from using different
translators can also be combined. For example, Afzal
et al. (2015) took the union of concepts translated by
both Google Translate and Microsoft Bing to achieve
good recall for annotating the QUAERO corpus. Sim-
ilarly, we will consider the combination of DeepL and
Microsoft Translator results using union.
3.4 QuestionAsConcept-type
Annotations
QaC-annotations can be very useful for data integra-
tion applications. For example, to compare the results
of the same question/questionnaire from different re-
search studies, the annotations can be applied in the
7
https://www.deepl.com/translator
8
https://www.microsoft.com/en-us/translator/
9
https://translate.google.com
10
https://translate.yandex.com
11
http://www.statmt.org/moses/
372
446
167
330
873
468
467
AnnoMap cTAKES
MetaMap
Figure 3: Number of annotations generated by each an-
notator in the Silver Standard Corpus. The result of
AnnoMap is the union of the results of AnnoMapQuestion
and AnnoMapWindow.
item matching process. Furthermore, cross-country
comparisons can also be possible if questions of dif-
ferent languages can be mapped to the same UMLS
concept. Therefore, we manually built a Gold Stan-
dard Corpus consisting of questionAsConcept-type of
annotations. We first identified the forms that have
such concepts in UMLS based on the AnnoMap re-
sults in the SSC. We then used keywords of the ques-
tions to search the correct QaC-annotations in the
UMLS UTS Metathesaurus Browser
12
. The questions
with QaC-annotations were included in the GSC. We
then evaluate how the annotators perform on finding
such QaC-annotations. We again use the same way
we pre-annotate the SSC to annotate these questions
and only the QaC-annotations are counted as true pos-
itive. Further, we also investigate the annotation qual-
ity with machine translators using the same methods
mentioned in Section 3.3.
4 EVALUATION
4.1 The Silver Standard Corpus
In total 12,551 unique annotations are found by three
annotators and eight different settings, i.e. same an-
notation found by several annotators or with differ-
ent settings are counted as a unique annotation. From
those, we identified 3,123 manually verified anno-
tations as the Silver Standard. The Inter-Annotator
Agreement (IAA) between the two human annotators
is 93.9 Kappa. Since the annotation principles are
concise and the human annotators only have to decide
if an annotation is correct or not (dichotomous), the
IAA is relatively high. Figure 3 presents the contri-
bution of each annotator in the SSC where the result
of AnnoMap is the union of AnnoMapQuestion and
12
UMLS Terminology Services https://uts.nlm.nih.gov/
metathesaurus.html
HEALTHINF 2020 - 13th International Conference on Health Informatics
150
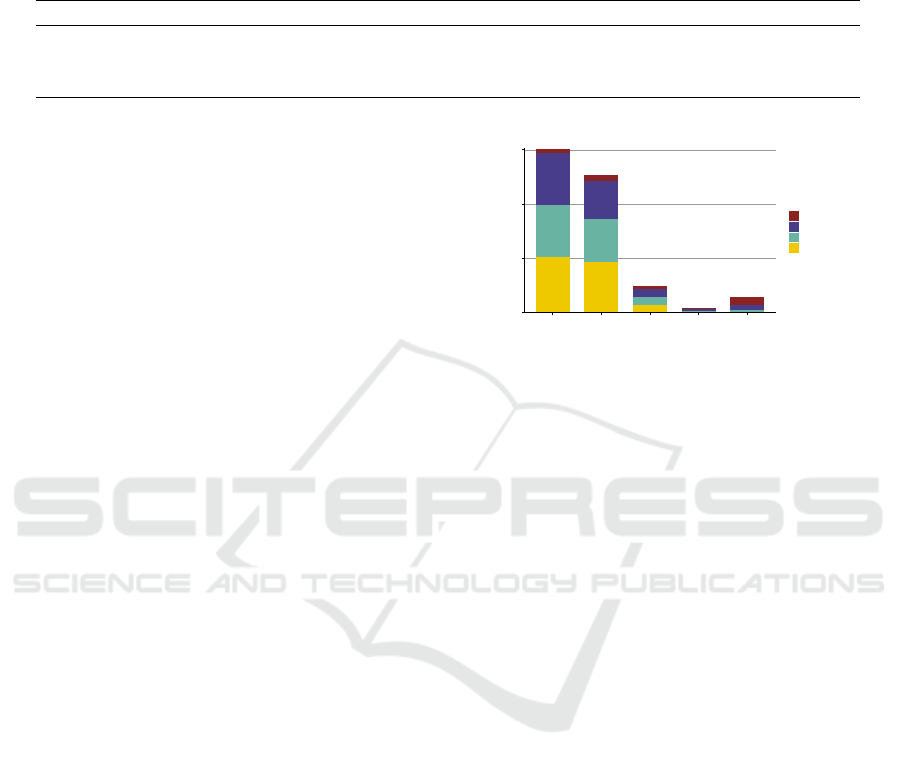
Table 1: Annotating German corpus and the original English forms using different UMLS subsets. The annotations are gener-
ated by AnnoMapQuestion with δ = {0.5, 0.6, 0.7} and then manually verified. German UMLS contains all available German
ontologies in 2017AA UMLS. German UMLS in English are the same ontologies as in German UMLS but the English ver-
sion. The selected UMLS subset is the subset of UMLS that used to build SSC. Hence, the results in the last row equates
to the results of SSC.
Corpus UMLS subset No. of concepts in ontologies No. of annotations
German German UMLS 111,079 81
Original English German UMLS in English 604,452 249
Original English selected UMLS subset 1,031,097 395
AnnoMapWindow. In total, MetaMap contributes the
most (2,138 annotations) while cTAKES the fewest
(1,954 annotations) to the SSC. More than one-fourth
of the annotations in SSC are found by all three
tools (873 annotations, 28.0%). Importantly, approx-
imately one-third of the annotations are only found
by single tools (1,006 annotations, 32.2%). These
annotations would have been missed out if a 2-vote-
agreement was applied for the inclusion in the SSC.
Notably, MetaMap, which is designed for annotation
tasks using UMLS, is able to find 467 annotations that
are not found by the other two tools.
Figure 4 shows the number of annotations found
by each annotator in the SSC with respect to the num-
ber of words of the found annotations. The number
of words of an annotation is calculated by averaging
the number of words of its concept name and all its
synonyms. The annotations found by MetaMap are
mostly consisting of less than 2 or 3 words and it can
not find any annotations that are longer than 5 words.
The reason of not finding longer annotations is be-
cause MetaMap splits the input text into phrases first
and these phrases are the basic units for the annota-
tion generation. cTAKES is able to find annotations
longer than 5 words but relatively few. The results
of AnnoMapWindow on the distribution of number
of annotations against annotation length is similar to
those of cTAKES but AnnoMapWindow is able to
find more annotations longer than 5 words. In con-
trast, AnnoMapQuestion generates few short annota-
tions but contributes the most for the annotations with
5 or more words due to its focus to annotate entire
questions.
4.2 Annotating German Corpus
In this section we investigate the annotation results of
the German corpus using German UMLS and com-
pare them to the results from the parallel English cor-
pus. Table 1 shows the number of concepts in the
different UMLS subsets and the number of manually
verified annotations. In total, 81 correct annotations
are obtained when the 37 German forms are anno-
0
1000
2000
3000
1 2 3 4 5
No. of words
No. of annotations
AnnoMapQuestion
AnnoMapWindow
cTAKES
MetaMap
Figure 4: Number of annotations generated by each annota-
tor for different number of words of the annotations in the
SSC. Note that no. of words = 1 in x-axis represents the
average word number less than 2 and no. of words = 2 rep-
resents the average word number larger than 2 but less than
3 and so on. No. of words = 5 includes all annotations with
average word number larger than 5.
tated using all UMLS ontologies that are available in
German (German UMLS). Using the same UMLS on-
tologies but in English (German UMLS in English)
to annotate the English corpus produces 249 correct
annotations, i.e., about three times as many as for the
German corpus. This is because the German UMLS
version contains much fewer concepts (111,079) than
those in English version (604,452) despite the restric-
tion to the same set of seven ontologies. Similarly,
German version has only 1/7 number of entries as
in the English version (217,171 vs. 1,463,453). For
instance, the German version of MeSH is translated
from the English version. The main keyword of each
concept is 1:1 translated but the number of synonyms
differ in two languages. Further, many of the de-
scriptions, references and definitions are not trans-
lated from the original version (DIMDI, 2019). A
further observation is that the German annotations are
mostly short concepts with one or two words and do
not include any QaC-annotations. It indicates that
such concepts do not exist in the German UMLS, as
opposed to the results we see from the respective En-
glish UMLS.
We also annotated the English corpus using the
selected UMLS subset that was used to build the
SSC. Using this subset, we obtained 395 correct an-
notations. This number reveals that the respective
Evaluating Cross-lingual Semantic Annotation for Medical Forms
151

Table 2: Annotation quality by using the translated corpora, DeepL and Microsoft Translator (MT). The results of MetaMap
are obtained using filtering score of 700. The AnnoMapWindow (AnnoMapW) results are from three window sizes 2, 3 and
5 and the AnnoMapQuestion (AnnoMapQ) results are from three similarity thresholds, i.e. δ = {0.5, 0.6, 0.7}. The results
of SSC (in gray) are also presented for comparison. In addition, the annotation quality of the combination of tools using 2-
vote-agreement (2VoteAgree) and union are shown as rows and that of the combination of both translators (union) are shown
in column.
Annotator
Precision Recall F-Measure
SSC DeepL MT union SSC DeepL MT union SSC DeepL MT union
MetaMap 57.2 26.3 24.9 22.7 68.5 40.5 38.9 45.1 62.3 31.9 30.4 30.2
cTAKES 41.5 18.3 16.9 15.9 62.6 36.9 35.7 42.7 49.9 24.5 23.0 23.1
AnnoMapW 23.9 9.5 9.3 8.0 62.6 35.2 32.6 41.5 34.6 15.0 14.4 13.4
AnnoMapQ 36.8 14.3 12.0 11.8 12.6 6.1 4.8 7.5 18.8 8.5 6.9 9.2
2VoteAgree 59.3 25.6 23.7 22.7 71.1 40.0 38.2 46.6 64.7 31.3 29.3 30.6
union 24.9 10.6 10.1 9.3 100.0 58.0 54.1 68.3 39.8 17.9 17.0 16.4
German UMLS subset in English (German UMLS in
English) lacks many of the most relevant ontologies,
such as NCI Thesaurus or SNOMED CT US. There-
fore, it is crucial to include all ontologies in UMLS
that are relevant for a given annotation task.
4.3 Using Machine Translators
In this section we report the annotation results of
using the two selected machine translators, DeepL
and Microsoft Translator. DeepL, which produced
the best translation result in the machine trans-
lator selection process, also performs better than
Microsoft Translator (Table 2). Annotating using
DeepL translated corpus results in better precision
and recall and consequently also the F-measure is bet-
ter. This indicates that using only a small amount of
translated samples (in our case, 50 questions) can al-
ready determine the suitability of a translator. (Meth-
ods described in Section 3.3).
By translating the German corpus into English
and annotate them using the English UMLS, we are
able to find more correct annotations than by anno-
tating the German forms directly using the German
UMLS. In contrast to the 81 annotations found us-
ing German corpus by AnnoMapQuestion (reported
in Section 4.2), translating the German forms using
DeepL and Microsoft Translator, AnnoMapQuestion
obtained 190 and 150 correct annotations, respec-
tively (data not shown). Combining the results from
different tools using 2-vote-agreement on the results
from original English corpus (SSC in Table 2) im-
proves precision, recall and F-measure of single tools.
The best single tool result is generated by MetaMap
with F-measure of 62.3% while 2-vote-agreement is
able to achieve 64.7%. Interestingly, this is differ-
ent for the translated corpora where MetaMap alone
produces slightly better results than the combina-
tion 2-vote-agreement. The best F-measure for trans-
lated corpora is obtained by MetaMap using DeepL
(31.9%) and that of the 2-vote-agreement is 31.3%.
The recall results of the row union in Table 2
shows how many annotations are retained from all
tools after translation. In total, 58.0% of the anno-
tations in SSC are retained using DeepL and 54.1%
by using Microsoft Translator and even 68.3% if we
take the union for the two translators. This is a very
promising result and substantially better than using
only one of the annotators or only one of the transla-
tor tools (e.g. MetaMap using DeepL is restricted to
a recall of 40.5%).
The precision results (and as a consequence the F-
measure results) in Table 2 are relatively low when
using the translated forms. However, this is of less
importance if we apply a manual selection and veri-
fication of the automatically found annotation candi-
dates as we did in building the SSC. With such a man-
ual verification, we can ideally achieve a precision for
the translated corpora of 100%. In combination with
the recall of 68.3% this would result in a F-measure
of as good as 81.2%.
4.4 QuestionAsConcept-type
Annotations
Finding questionAsConcept-type of annotations is of
highest priority in terms of meta-anlysis or data in-
tegration across multiple databases. We built a Gold
Standard Corpus that contains 205 questions and 214
QaC-annotations. Since QaC-annotations are defined
to map a whole question, there is deviation in the
number of questions and the number of annotations
in the GSC. The main reason is that there are multi-
ple QaC-annotations that are of different but suitable
semantic types mapped to the same questions.
We then examine the three annotators for their
HEALTHINF 2020 - 13th International Conference on Health Informatics
152

MetaMap
cTAKES
AmW
AmQ(0.5)
AmQ(0.6)
AmQ(0.7)
0
25
50
75
100
%
Precision
Recall
F−measure
Figure 5: Annotation quality of three annotators for
QaC-annotations using original English corpus. MetaMap
is with filtering score of 700. AmQ denotes the annotator
AnnoMapQuestion and the numbers in the parentheses in-
dicate the threshold δ used in post-processing. AmW refers
to AnnoMapWindow.
ability to find such annotations. Figure 5 shows the
results. AnnoMapQuestion is the most suitable tool
to find annotations that have longer text. With the in-
crease of the threshold δ from 0.5 to 0.7, the preci-
sion improves significantly (from 50.8% to 82.7%).
On the other hand, the recall results do not vary
much, with 85.6% using δ = 0.5 and 82.3% using
δ = 0.7. As a consequence, the best F-measure,
82.5%, is achieved using δ = 0.7. AnnoMapWindow
is able to find 159 QaC-annotations (74.0% recall)
while cTAKES only found 86 (40.0% recall). Since
MetaMap can only annotate short phrases, it only
found 23 QaC-annotations (10.7% recall).
We also investigate the annotation quality on
QaC-annotations if machine translators are involved.
Since AnnoMapQuestion is the most suitable tool,
we report only the best results from it, i.e. with
δ = 0.7 (Figure 6). Again, the results using DeepL
are better than those of Microsoft Translator. Trans-
lation has more impact on recall than on preci-
sion. This might be explained by the fact that even
with AnnoMapQuestion that already considers simi-
larity measures such as trigram, TF/IDF and a LCS
(Longest Common Substring) (Christen et al., 2015),
the change of the wording and the sequence of the
words after translation hinder the annotator to find
the correct match. Many true positives have become
false negatives. Using DeepL alone, we are able to
retain only 26.5% recall. Even by combining the
Microsoft Translator results, the recall of the union
can only reach 30.7%.
4.5 Result Summary
The presented evaluation showed that the three con-
sidered annotator tools are able to find both concepts
for short phrases (i.e. using MetaMap, cTAKES and
AnnoMapWindow) and also concepts that annotate
0
25
50
75
100
Precision Recall F−measure
%
GSC
DeepL
Microsoft
Union
Figure 6: Annotation quality of QaC-annotations using
translated corpora from DeepL, Microsoft Translator and
the union of them. The results of using original English
corpus (GSC) is also shown for reference. Used annotator
is AnnoMapQuestion with δ = 0.7.
whole questions (using AnnoMapQuestion). Based
on a parallel corpus and the determined SSC, we
could evaluate the annotation quality of different an-
notation approaches. We investigated two possible
ways to annotate the German forms, i.e. 1) direct
annotation using German UMLS ontologies and 2)
integrating machine translators into annotation work-
flow. Due to the current restriction on the coverage of
UMLS in other languages than English, direct anno-
tations using the original non-English corpus can only
produce very restricted results. The cross-lingual an-
notation approach provides two advantages. Firstly, it
gives the possibility to use the English version of the
UMLS, which is much more comprehensive than in
other languages. Secondly, one is able to apply many
annotators as most of them are designed to annotate
English corpora. This is a better option than using
just the non-English UMLS, as we have shown in this
study. With translation, we are able to obtain a recall
of 68.3%. This indicates an F-measure of 81.2% is
possible using machine translators.
5 CONCLUSIONS
In this study we present a parallel corpus of medical
forms used in epidemiological study or clinical inves-
tigations. We use three annotators to automatically
generate annotations first and then manually verified
them to build an English SSC. The obtained annota-
tions in the SSC have been integrated into the LIFE
Datenportal
13
and will be used to enhance the search
function in the future. For future work, we seek to de-
velop further methods to improve the annotation qual-
ity on such cross-lingual annotation tasks. As we have
shown in this study, due to translation the paraphrase
of the questions significantly decreases the chance of
annotators to find the correct concepts. We will there-
13
https://ldp.life.uni-leipzig.de
Evaluating Cross-lingual Semantic Annotation for Medical Forms
153

fore include additional semantic matching mechanism
to enhance matching probability and hence to obtain
even better recall and good precision.
ACKNOWLEDGEMENTS
This work is funded by German Federal Min-
istry of Education and Research (BMBF) (grant
031L0026, ”Leipzig Health Atlas”), the German
Research Foundation (DFG) (grant RA 497/22-1,
”ELISA - Evolution of Semantic Annotations”), and
National Research Fund Luxembourg (FNR) (grant
C13/IS/5809134).
REFERENCES
Afzal, Z., Akhondi, S. A., van Haagen, H., van Mulligen,
E. M., and Kors, J. A. (2015). Biomedical concept
recognition in French text using automatic translation
of English terms. In CLEF (Working Notes).
Aronson, A. R. and Lang, F.-M. (2010). An overview of
MetaMap: historical perspective and recent advances.
Journal of the American Medical Informatics Associ-
ation, 17(3):229–236.
Becker, M. and B
¨
ockmann, B. (2016). Extraction of
UMLSr concepts using Apache cTAKES
TM
for Ger-
man language. Studies in Health Technology and In-
formatics, 223:71–76.
Beutner, F., Teupser, D., Gielen, S., Holdt, L. M., Scholz,
M., Boudriot, E., Schuler, G., and Thiery, J. (2011).
Rationale and design of the Leipzig (LIFE) Heart
Study: phenotyping and cardiovascular characteristics
of patients with coronary artery disease. PLoS One,
6(12):e29070.
Cabot, C., Soualmia, L. F., Dahamna, B., and Darmoni, S. J.
(2016). SIBM at CLEF eHealth Evaluation Lab 2016:
Extracting concepts in French medical texts with ecmt
and cimind. In CLEF (Working Notes), pages 47–60.
Christen, V., Groß, A., and Rahm, E. (2016). A reuse-based
annotation approach for medical documents. In Inter-
national Semantic Web Conference, pages 135–150.
Springer.
Christen, V., Groß, A., Varghese, J., Dugas, M., and Rahm,
E. (2015). Annotating medical forms using UMLS. In
International Conference on Data Integration in the
Life Sciences, pages 55–69. Springer.
Cohen, J. (1960). A coefficient of agreement for nominal
scales. Educational and Psychological Measurement,
20(1):37–46.
DIMDI (2019). Medical Subject Headings. In
https://www.dimdi.de/dynamic/de/klassifikationen/weitere-
klassifikationen-und-standards/mesh/.
Geistert, D. (2018). Visualisierung von Annotation - Map-
pings f
¨
ur klinische Formulare. Bachelor’s Thesis,
Universit
¨
at Leipzig, Germany.
Hellrich, J., Clematide, S., Hahn, U., and Rebholz-
Schuhmann, D. (2014). Collaboratively annotating
multilingual parallel corpora in the biomedical do-
main—some MANTRAs. In LREC, pages 4033–
4040.
Hellrich, J. and Hahn, U. (2013). The JULIE LAB
MANTRA System for the CLEF-ER 2013 Challenge.
In CLEF (Working Notes).
Hoehndorf, R., Schofield, P. N., and Gkoutos, G. V. (2015).
The role of ontologies in biological and biomedical
research: a functional perspective. Briefings in Bioin-
formatics, 16(6):1069–1080.
Jovanovi
´
c, J. and Bagheri, E. (2017). Semantic annota-
tion in biomedicine: the current landscape. Journal
of Biomedical Semantics, 8(1):1–18.
Kors, J. A., Clematide, S., Akhondi, S. A., Van Mulligen,
E. M., and Rebholz-Schuhmann, D. (2015). A mul-
tilingual gold-standard corpus for biomedical concept
recognition: the Mantra GSC. Journal of the Ameri-
can Medical Informatics Association, 22(5):948–956.
Kroenke, K., Spitzer, R. L., and Williams, J. B. W. (2002).
The PHQ-15: validity of a new measure for evaluating
the severity of somatic symptoms. Psychosom Med,
64(2):258–66.
Lewin, I., Clematide, S., Forner, P., Navigli, R., and Tu-
fis, D. (2013). Deriving an english biomedical silver
standard corpus for CLEF-ER. University of Zurich.
Lewin, I., Kafkas, S¸., and Rebholz-Schuhmann, D. (2012).
Centroids: Gold standards with distributional vari-
ation. In Proceedings of the Eighth International
Conference on Language Resources and Evaluation
(LREC-2012), pages 3894–3900, Istanbul, Turkey.
European Languages Resources Association (ELRA).
Lin, Y.-C., Christen, V., Groß, A., Cardoso, S. D., Pruski,
C., Da Silveira, M., and Rahm, E. (2017). Evaluat-
ing and improving annotation tools for medical forms.
In Proc. Data Integration in the Life Science (DILS
2017), pages 1–16. Springer.
Loeffler, M., Engel, C., Ahnert, P., Alfermann, D., Are-
lin, K., Baber, R., Beutner, F., Binder, H., Br
¨
ahler,
E., Burkhardt, R., et al. (2015). The LIFE-Adult-
Study: objectives and design of a population-based
cohort study with 10,000 deeply phenotyped adults in
Germany. BMC Public Health, 15(1):691.
L
¨
owe, B., Decker, O., M
¨
uller, S., Br
¨
ahler, E., Schellberg,
D., Herzog, W., and Herzberg, P. Y. (2008). Valida-
tion and standardization of the Generalized Anxiety
Disorder Screener (GAD-7) in the general population.
Medical Care, 46(3):266–274.
McDonald, C. J., Huff, S. M., Suico, J. G., Hill, G.,
Leavelle, D., Aller, R., Forrey, A., Mercer, K., De-
Moor, G., Hook, J., et al. (2003). LOINC, a universal
standard for identifying laboratory observations: a 5-
year update. Clinical Chemistry, 49(4):624–633.
N
´
ev
´
eol, A., Cohen, K. B., Grouin, C., Hamon, T., Lavergne,
T., Kelly, L., Goeuriot, L., Rey, G., Robert, A., Tan-
nier, X., and Zweigenbaum, P. (2016). Clinical infor-
mation extraction at the CLEF eHealth Evaluation lab
2016. CEUR Workshop Proceedings, 1609:28–42.
HEALTHINF 2020 - 13th International Conference on Health Informatics
154

N
´
ev
´
eol, A., Dalianis, H., Velupillai, S., Savova, G., and
Zweigenbaum, P. (2018). Clinical natural language
processing in languages other than English: opportu-
nities and challenges. Journal of Biomedical Seman-
tics, 9(1):12.
N
´
ev
´
eol, A., Grosjean, J., Darmoni, S. J., Zweigenbaum, P.,
et al. (2014a). Language resources for French in the
biomedical domain. In LREC, pages 2146–2151.
N
´
ev
´
eol, A., Grouin, C., Leixa, J., Rosset, S., and Zweigen-
baum, P. (2014b). The QUAERO French medical cor-
pus: A ressource for medical entity recognition and
normalization. In In Proc BioTextM, Reykjavik. Cite-
seer.
N
´
ev
´
eol, A., Grouin, C., Tannier, X., Hamon, T., Kelly, L.,
Goeuriot, L., and Zweigenbaum, P. (2015). CLEF
eHealth Evaluation Lab 2015 Task 1b: Clinical named
entity recognition. In CLEF (Working Notes).
Oellrich, A., Collier, N., Smedley, D., and Groza, T.
(2015). Generation of silver standard concept anno-
tations from biomedical texts with special relevance
to phenotypes. PloS one, 10(1):e0116040.
Perez, N., Accuosto, P., Bravo,
`
A., Cuadros, M., Mart
´
ınez-
Garc
´
ıa, E., Saggion, H., and Rigau, G. (2019). Cross-
lingual semantic annotation of biomedical literature:
experiments in Spanish and English. Bioinformatics.
Rebholz-Schuhmann, D., Clematide, S., Rinaldi, F.,
Kafkas, S., van Mulligen, E. M., Bui, C., Hellrich,
J., Lewin, I., Milward, D., Poprat, M., et al. (2013).
Entity recognition in parallel multi-lingual biomedical
corpora: The CLEF-ER laboratory overview. In Inter-
national Conference of the Cross-Language Evalua-
tion Forum for European Languages, pages 353–367.
Rebholz-Schuhmann, D., Jimeno-Yepes, A. J., van Mul-
ligen, E., Kang, N., Kors, J., Milward, D., Cor-
bett, P., Buyko, E., Tomanek, K., Beisswanger, E.,
et al. (2010). The CALBC silver standard corpus
for biomedical named entities-a study in harmonizing
the contributions from four independent named en-
tity taggers. Proceedings of the Seventh Conference
on International Language Resources and Evaluation
(LREC’10).
Rebholz-Schuhmann, D., Yepes, A. J., Li, C., Kafkas, S.,
Lewin, I., Kang, N., Corbett, P., Milward, D., Buyko,
E., and Beisswanger, E. (2011). Assessment of NER
solutions against the first and second CALBC silver
standard corpus. Journal of Biomedical Semantics,
2(5):1.
Roller, R., Kittner, M., Weissenborn, D., and Leser, U.
(2018). Cross-lingual candidate search for biomedi-
cal concept normalization. CoRR, abs/1805.01646.
Savova, G. K., Masanz, J. J., Ogren, P. V., Zheng, J., Sohn,
S., Kipper-Schuler, K. C., and Chute, C. G. (2010).
Mayo clinical Text Analysis and Knowledge Extrac-
tion System (cTAKES): architecture, component eval-
uation and applications. Journal of the American Med-
ical Informatics Association, 17(5):507–513.
Schuemie, M. J., Jelier, R., and Kors, J. A. (2007). Pere-
grine: Lightweight gene name normalization by dic-
tionary lookup. In Proc of the Second BioCreative
Challenge Evaluation Workshop, pages 131–133.
Schulz, S., Ingenerf, J., Thun, S., and Daumke, P. (2013).
German-language content in biomedical vocabularies.
In CLEF (Working Notes).
Starlinger, J., Kittner, M., Blankenstein, O., and Leser, U.
(2017). How to improve information extraction from
german medical records. it-Information Technology,
59(4):171–179.
van Mulligen, E. M., Bui, Q.-C., and Kors, J. A. (2013).
Machine translation of Bio-Thesauri. In CLEF (Work-
ing Notes).
World Health Organization (2004). ICD-10 : international
statistical classification of diseases and related health
problems : tenth revision.
Evaluating Cross-lingual Semantic Annotation for Medical Forms
155
