
Data and Sessions Management in a Telepathology Platform
Pedro Nunes
1
, Rui Jesus
1
, Rui Lebre
1,2 a
and Carlos Costa
1
1
Institute of Electronics and Informatics Engineering of Aveiro, University of Aveiro, Portugal
2
University of A Coru
˜
na, Spain
Keywords:
Whole-Slide Imaging, Collaborative Platform, Digital Pathology, PACS, DICOM, RBAC.
Abstract:
Digital pathology refers to the acquisition, storage, and interpretation of pathological data gathered by scanners
and displayed in a digital environment, using a distributed network system. This paper discusses the challenges
and opportunities of a collaborative platform applied to digital pathology considering the advantages that it
may carry on regarding education and training but also new paradigms of telepathology and telemedicine.
Furthermore, it proposes the implementation of a secure collaborative platform that integrates a web pathology
viewer with personal areas and virtual archives. The described approach introduces a modern collaborative
concept into the digital pathology workflow supported by a customized medical imaging infrastructure where
data management is ensured by an innovator DICOM standard multi-repository server. The solution was
designed to serve distinct usage contexts, including telepathology and e-academy.
1 INTRODUCTION
Digital pathology refers to the whole workflow since
the scanning of a microscopic image and the associ-
ated metadata, until its distribution and visualization.
It became popular in the last years due to techno-
logical developments and to the increasing trend of
adoption of digital scanners. The digital pathology
scanners produce what is named Whole-Slide Images
(WSI) (Pantanowitz, 2010).
WSI designates the image digitization by scanning
microscopy glass slides (Pantanowitz, 2010; Pan-
tanowitz et al., 2011; Saco et al., 2016). The process
of digitization comprises multiple magnifications and
focal planes, producing high-resolution digital images
that can aggregate several gigabytes of data (Farahani
et al., 2015). Digital pathology is replacing the con-
ventional light microscopy, potentiating new applica-
tions in education, training and diagnosis (Saco et al.,
2016; Triola and Holloway, 2011).
Opposing a traditional pathology environment, the
specimen storage is cheaper as the samples do not
require specialized protection carried out by trained
staff.
A wide variety of advantages emerge from this
modern branch of medical imaging in production and
clinical environments as, for instance, the process of
a
https://orcid.org/0000-0002-3230-0732
storing and remote viewing, annotation, and report-
ing. Unlike the traditional slides, the whole-slide
images do not deteriorate over time and it is possi-
ble to assure homogeneity of the display quality of
the images (Foster, 2010). Moreover, further pros
come across, for instance, improvements in the diag-
nostic accuracy, integration with hospital information
systems or availability of distributed work processes
as collaborative work and telepathology (Pantanowitz
et al., 2011; Bueno et al., 2016a).
WSI has introduced new methods to teach his-
tology and pathology in the academic field that was
not possible until nowadays (Saco et al., 2016; Pan-
tanowitz et al., 2012). The teaching of pathology is
simplified by the digital trend as the slide can be ac-
cessed simultaneously by as many teachers and stu-
dents as required, and it is only necessary to hold a
computer or a smartphone to operate the image, un-
like the physical specimen which can only be handled
by a handler at a time. Additionally, the availabil-
ity of a slide in the digital format allows the remote
access from anywhere and from any device instantly,
including previous examinations that could be hard to
find in the traditional physical archives. Yet, systems
like these demand strong access security since they
can be operated in open networks. Many of the cases
require confidentiality considering the sample private
data related to the patient and the regulations in force
(Abouelmehdi et al., 2018).
Nunes, P., Jesus, R., Lebre, R. and Costa, C.
Data and Sessions Management in a Telepathology Platform.
DOI: 10.5220/0008969904550462
In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2020) - Volume 5: HEALTHINF, pages 455-462
ISBN: 978-989-758-398-8; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
455

Concerning the clinical environment, WSI and
digital pathology make the diagnosis simpler and
faster as the slide can be dynamically viewed, navi-
gated and magnified on screens in a network (Bueno
et al., 2016b). Professional WSI viewers can be in-
tegrated into the hospital databases and be accessed
within the network or made available for research,
remote diagnosis and consultation (second opinions,
among others) (Bueno et al., 2016b; Pantanowitz
et al., 2015). Some authors (Pantanowitz et al.,
2015; Chordia et al., 2016) report a crescent need
for telepathology and digital pathology in the daily
routine practice. In (Chordia et al., 2016), 98% of
the 247 histopathologists interrogated, felt the need
for telepathology and digital pathology. However,
only 34% declared the use of telepathology in medical
practice. These facts may reveal demand for solutions
of telepathology and collaborative platforms not only
for clinical but also for educational purposes.
However, the development of a capable user inter-
face to aid in the adoption of these new technologies
is one of the improvements that can help the teaching
and collaborative diagnosis nowadays. The integra-
tion of new features impossible to include in the tra-
ditional methods became a reality, as the presence of a
thumbnail to allow a fast and immediate panning, the
possibility of annotate regions of interest directly in
the sample and live discussion platform, for instance.
This paper proposes a solution
1
for the lack of
collaborative platforms suitable for both teaching and
clinical paradigms by deploying a secure architecture
integrated with a digital pathology viewer
2
. The plat-
form integrates a pure web pathology viewer (screen-
shot in figure 1) fully compliant with Digital Imaging
and Communications and Medicine (DICOM) stan-
dard and a security layer to restrict and control the ac-
cess to studies (Lebre et al., 2018b), taking advantage
of the Dicoogle open-source project (Valente et al.,
2016) support for WSI storage.
The collaborative viewer supports new features
like an image handling toolbox, shared pointers, and
synchronized actions. In the persistence layer, the
archive server was redesigned to support an innova-
tor multi-repository concept. Data management se-
curity is ensured through the integration of an ac-
counting mechanism specifically developed to med-
ical imaging environments, allowing the creation of
virtual archives for each user. For instance, it al-
lows the restriction of access to only particular studies
within a group domain. The mechanism allows the
centralization of the storage, creating different per-
missions of access to different institution resources.
1
Demo Video: https://youtu.be/Mmsb25edcOo
2
Demo: http://demo.dicoogle.com/pathobox
2 BACKGROUND
2.1 PACS-DICOM Universe
Implementations of Picture Archiving and Communi-
cation Systems (PACS) have risen with the move to-
wards digital radiology (Beckwith, 2016). A PACS
is a set of distinct hardware and software technolo-
gies comprising medical image and data acquisition
equipment, storage equipment, and display worksta-
tion, all of which are integrated by digital networks.
PACS relies on DICOM. DICOM is one of the most
popular standards in the medical imaging field (Silva
et al., 2019). DICOM provides a guide to support
interoperability between multiple vendor equipment
and systems (Bidgood et al., 1997).
In the early time of the launching of DICOM, the
propose was to create a standard format for storing
and handling of radiology images (Godinho et al.,
2017). Notwithstanding, other modalities were in-
troduced, such as nuclear medicine or Breast to-
mosynthesis (Beckwith, 2016) and, more recently,
microscopy, as a result of the rapid spread and adop-
tion of the standard. Later on, the publication of
the supplement 145, addressing digital pathology,
boosted the development of the automated whole-
slide scanners (Bueno et al., 2016a).
DICOM standard is being continuously updated
by the addition of new extensions with the goal of
keeping it up-to-date regarding the needs. However,
unlike the traditional patient-centric DICOM infor-
mation model, WSI are considered specimen centric
as the specimen is the most relevant object (Daniel
et al., 2009; Daniel et al., 2011a).
NEMA, the organization behind DICOM, intro-
duced adjustments to the standard foreseeing the sup-
port for WSI, taking into account the necessary adap-
tations to hold the high-resolution images. Nowa-
days, the standard integrates the data elements, the
definition of the workflow of the preparation, acquisi-
tion, handling, and storage of WSI (Beckwith, 2016;
Daniel et al., 2011b).
2.2 WSI Visualization
One of the key challenges of the display and visual-
ization of the whole-slide images is the big amount
of pixels produced by the scanners so the information
may have the required quality. With the increase of
the number of pixels, also the file size increases, mak-
ing the remote display via a web browser of the sam-
ples difficult. Once their size is much greater than the
regular medical images and cannot be displayed lo-
cally due to the amount of memory available in most
HEALTHINF 2020 - 13th International Conference on Health Informatics
456
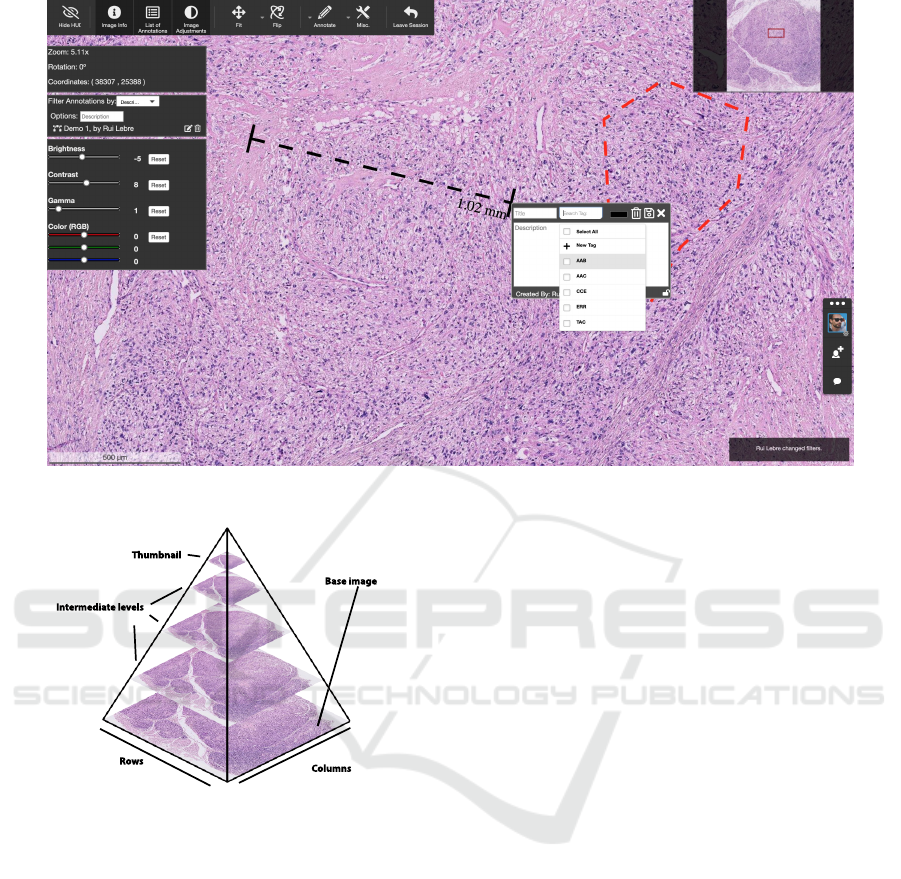
Figure 1: Collaborative web pathology framework screenshot.
Columns
Rows
Intermediate levels
Thumbnail
Base image
Figure 2: DICOM supplement 145 proposes storing Whole-
Slide Images in tiles from a multi resolution hierarchy in
multi-frame object. Adapted from (Daniel et al., 2011a).
computers (Goode et al., 2013). Nowadays, as a result
of the presented challenges, navigation in the digital
slide is performed using a pan and zoom approach.
The solution comprises a pyramid approach (Figure
2). In this pyramid approach, intermediate levels are
generated until the thumbnail is obtained. Figure 3
shows the workflow where pathologists must browse
and select a region of interest in the thumbnail for fur-
ther zoom in to a higher resolution.
Nowadays, solutions like Google Maps or Li-
braries Digital Catalogs (e.g., New York Public Li-
brary, National Library of Australia) adopted this ap-
proach for image and maps handling.
2.3 Related Work
In this scope, solutions that support digital pathology
with a collaborative platform on both commercial ap-
plications and scientific literature were taken into ac-
count. An in-depth analysis of the retrieved studies
was carried out and allowed us to conclude that no so-
lution, commercial or not, is available publicly, sup-
porting a collaborative viewer of DICOM WSI on a
standard PACS.
However, the literature revised reports works
where collaborative systems where multiple users can
access and work on the same image. Daniel et
al. (Daniel et al., 2011a) that Collaborative Digi-
tal Anatomic Pathology can only be achieved using
medical informatics standards. Some authors (Beck-
with, 2016; Daniel et al., 2011a; Tuominen and Isola,
2010) refer to DICOM supplements 122 and 145 as
the standards focusing the whole-slide imaging han-
dling. They also describe how WSI can be integrated
into standards like DICOM and HL7.
In (Lauro et al., 2013), the authors propose a set
of web-based tools to support the workflow of digital
pathology consulting and allow the viewing of WSI
over the web. The authors also state that the digital
pathology segment is dominated by a set of vendors
who have developed their own proprietary format and
viewing solutions. Furthermore, it is reported that
most of WSI vendors provide their own viewing so-
lutions. In the cases where the viewer is available, it
only supports the proprietary formats of each vendor,
Data and Sessions Management in a Telepathology Platform
457
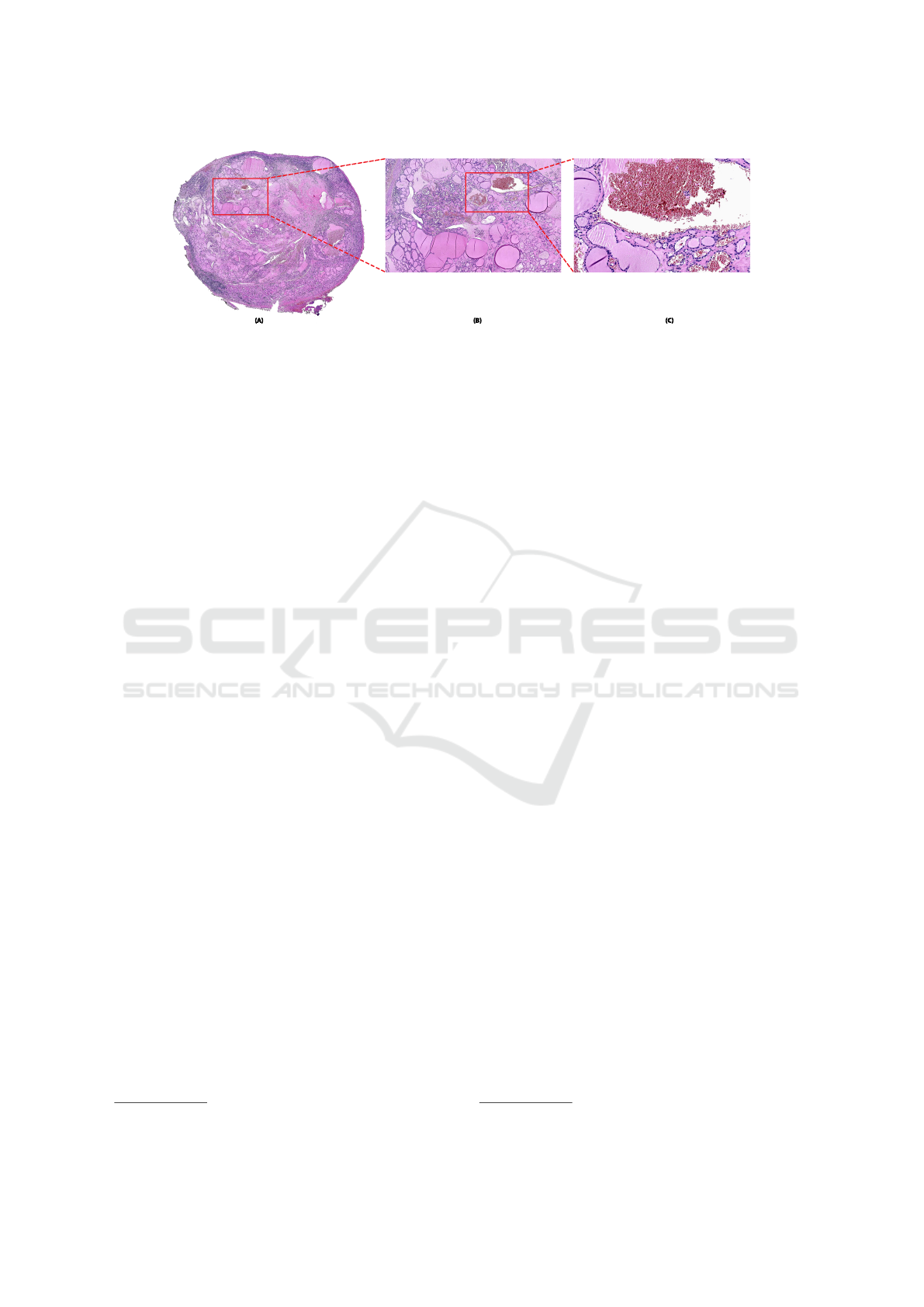
(A) (B) (C)
Figure 3: Example of zooming a Whole-Slide Image in the viewer presented. (A) Thumbnail of the digital slide; (B) Interme-
diate level of zoom; (C) Full zoom level of the image.
in a standalone usage.
Chandrakanth et al. (Bernard et al., 2014) presents
guidelines and problems related to the reason why
telepathology for primary diagnosis has not yet be-
come current practice. In the same paper, the au-
thors reveal iPath (Brauchli and Oberholzer, 2005),
a Web-based telepathology platform that permits the
online presentation and discussion of cases within
user groups.
D
´
ıaz et al. propose in (D
´
ıaz et al., 2018) a
web-based telepathology framework for collaborative
work of pathologists. However, this platform does not
comply with the DICOM standard neither addresses
the academic scenarios.
Moreover, the research came across with similar
platforms in the area of radiology (Zhang et al., 2000).
Bankhead et al. (Bankhead et al., 2017) propose an
extensible open-source software for digital imaging
named QuPath, powering users with scripting tools.
However, it does not address the collaborative envi-
ronment and DICOM support.
Carestream
3
(Snyder et al., 2001; Weiss, 2012) is
a commercial platform built on a consulting basis and
not suitable for collaborative environments. Besides,
Carestream is only applicable to radiology and cardi-
ology modalities.
3 ARCHITECTURE
This section describes the modules that compose the
collaborative platform and the integration with the Di-
coogle PACS repository, intending to provide real-
time collaboration between users. The architecture
was designed to be as flexible as possible, hence why
it is divided into multiple independent modules, mak-
ing it possible to develop and add features to each
module, without affecting the remaining, as well as
completely swap one of the parties while retaining the
3
https://www.carestream.com/en/gb/medical
functionality.
The system is divided into the following three ma-
jor components: a PACS archive where the case stud-
ies are stored; a web viewer that fetches the slides
from the case studies in the archive; the collabora-
tive platform that informs the viewer about the ses-
sion details and their participants and where the user
can manage their sessions. These interactions are
depicted in Figure 4. The PACS archive and the
Viewer had already been developed in a previous
work (Godinho et al., 2017), however, it was neces-
sary to redesign those elements and implement new
mechanisms to support real-time collaboration be-
tween users.
TogetherJS
4
is an open-source technology that
provides real-time collaboration features. It is used
by the web viewer to manage the sessions, handling
the synchronization of events across all the participat-
ing users.
The collaborative platform is agnostic to the web
viewer, due to an abstraction layer defined between
the two components that facilitate the use of different
viewers with minimal adaptation efforts. This is pos-
sible because the actions of multiple users throughout
a session can be made abstract, i.e., independent from
the source (viewer) they came from.
3.1 Management of Sessions
A working session is a collaborative session created
over an image from the PACS repository. The col-
laborative platform is responsible for the creation of
these sessions and the management of its participating
users and their permissions. A session is composed by
its creator, a list of users and their respective permis-
sions, the PACS archive image, the list of events that
happened in that session and additional information
to tell the web viewer how to handle this session.
A session is defined by a unique ID that allows the
sessions to be uniquely identifiable across the system.
4
https://togetherjs.com/
HEALTHINF 2020 - 13th International Conference on Health Informatics
458
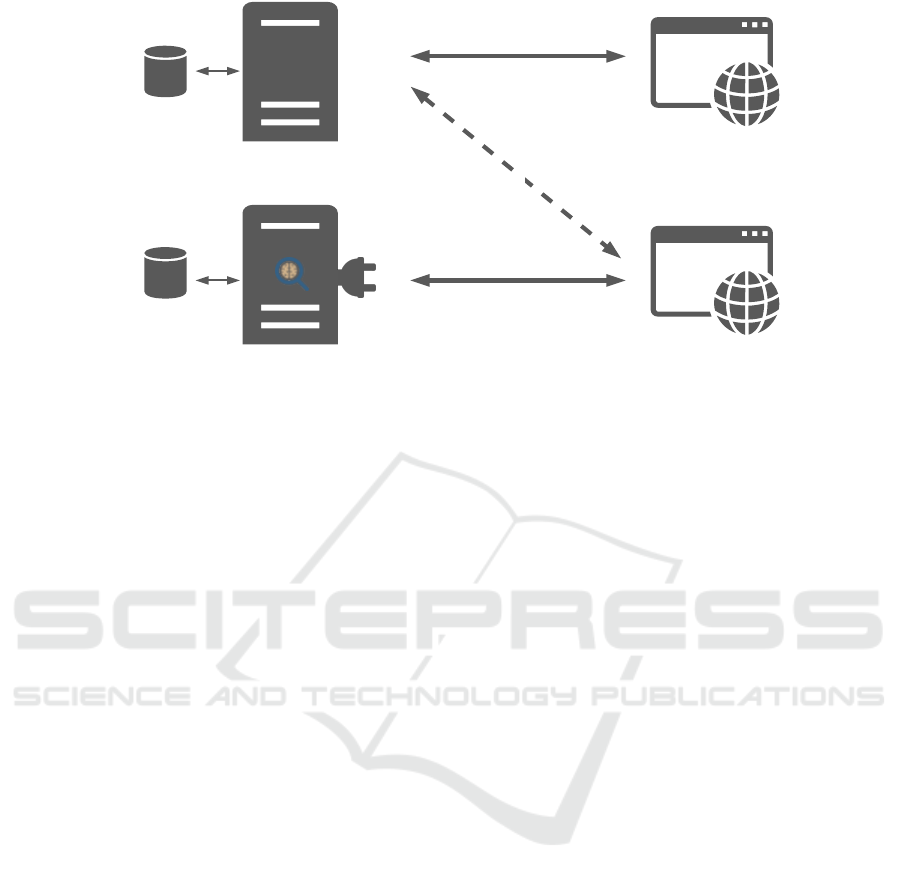
WADO-RS
Collaborative
Platform Server
RBAC
Database
Dicoogle
Users and
Sessions Database
Collaborative Platform
Management Interface
Digital Pathology Viewer
HTTP
Web Socket
Digital
Pathology
Plugin
Figure 4: System general architecture. Dicoogle and its plugins serve the Web Viewer with the WSI. At the same time, the
viewer is connected to the collaborative platform server via WebSockets to retrieve the session details and users’ permissions.
The collaborative platform management interface provides a dashboard to manage all the sessions, users, and groups.
This aspect is crucial since one image can have
multiple sessions associated with it, with each ses-
sion having its own users and events. Furthermore,
one user can create multiple sessions using the same
image.
The creation of sessions was designed to facili-
tate the invitation of other users. Therefore, a unique
web link is generated for every user in a session. The
uniqueness of this weblink is guaranteed by combin-
ing the ID of the session with the ID of the user.
Weblinks allow access to a session without having
to use the platform itself. Only the creator is required
to be logged into the platform since it is still necessary
to define the configurations of the session. For further
automation of the system, emails are used as IDs to
identify the users on the platform. By using emails, it
is possible to share the web links of the session auto-
matically, turning the process of the invitation of new
users transparent and agile.
Additionally, by sending the invitation links
through email, it is naturally secured the access to this
link, since only the owner of the email has access to
it.
By having the user IDs on the web link, it is pos-
sible to register the sessions that each of the potential
participants were invited to, so that if a user did not
sign in into the platform yet, the access to all the ses-
sions he was asked to is still granted, as long as the
email used to sign in is the same as the one sent with
the invitation.
On the viewer side, since the user will be access-
ing it through the web link and the link contains both
the user ID and the session ID, it is possible to query
the collaborative platform for the details of the session
and the permissions of the user. At the same time, it
can also verify if this user already is connected to the
session.
Since sessions are based on web links, the concur-
rent access control is assured by which link is in use
at the current time, due to notifications sent from the
viewer to the platform, preventing the same link from
being used simultaneously in another viewer, in the
case if a user shared the unique and non-transferable
link. It is, therefore, possible to keep control of the
number of users in a session, and it is assumed that
their identity is also correct since the unique access
links were sent via email.
On the other hand, it is possible to use a pub-
lic link, which is missing the user ID. This modality
seeks to give the creator permissions to invite users
without the need for an email. A limit can be defined
on the number of people that can access a given ses-
sion through a public link. The generation of this link
is optional.
This way, the creator can invite a specific group
of people without knowing their emails by creating a
public link, with a possible max limit for its usage,
and manually share it. In this scenario, the invited
users can access the viewer session, but would not be
able to access the managing platform, which requires
their email to be added to the session.
3.2 User Actions and Synchronization
To maintain synchronicity between the users, each
event performed upon the image in the Web Viewer,
such as zooming in or changing the saturation, is
broadcast to the remaining users, using TogetherJS.
Data and Sessions Management in a Telepathology Platform
459

This structure allows the separation between the en-
tity which records the actions of the session and the
entity that replicates the session.
Additionally, the events are stored in a database,
ensuring that users that lose the connection temporar-
ily, or are joining in an ongoing session, retrieve the
missing events, keeping up with the rest of the users.
This also allows users to open a standalone copy of a
session, a replay session, replicating chronologically
the events, in which a user can inspect the actions per-
formed.
Since many users can interact with the image at
the same time, certain events can happen in a differ-
ent order from user to user, leading to inconsistent
states throughout the session. Therefore, an asyn-
chronous system is used in which all the users will
eventually be in the same state. Despite having mul-
tiple sessions states across the different viewers, the
database is keeping only one of those versions, which
ultimately will be the final one.
Furthermore, only certain actions, like zooming or
panning the image, create these different states, and
all users are sent to the same state right after one of
these events happen, without conflict. I.e., when two
different zooms on the image are executed at the same
time, the users might end up on different zoom lev-
els. If a third zoom is effectuated, all of the users will
end up in the same zoom level, keeping all the session
users synchronized. A study was conducted to eval-
uate the impact on the usability of the viewer with
these inconsistencies. Since a user will only be in an
inconsistent state for a very short time and since the
most crucial actions, like color filters or annotations,
do not cause an inconsistent state, it was considered
that no delay should be added to the propagation of
the action. So, it was concluded that the user actions
should be responded immediately after the execution,
rather than wait for the server confirmation.
Due to the nature of the working sessions and due
to the dimensions of the whole-slide images, many
zooming or panning type of events are generated,
even though they have minimal if any, visual impact.
Thus, despite all events being broadcast to keep the
synchronization between the users, when storing in
the database, the events are filtered, guaranteeing that
only the events that make a noticeable difference are
stored. This prevents a heavy workflow of messages
being sent between the components and improves the
loading time of an ongoing session in the web viewer,
as well as, improves the replay experience, due to the
presence of only relevant information.
3.3 Access Control Mechanism
This article proposes a collaborative platform archi-
tecture that supports multiple users with associated
resources, including personal data archives with sen-
sitive information. General Data Protection Regula-
tion (GDPR), applied from May 2018, defines spe-
cific guidelines in healthcare and the use of best prac-
tices to minimize the risk of a security breach (art.
32). It requires that personal data needs to be pro-
tected against illegitimate processing, accidental loss,
destruction or damage. Following this regulatory
obligation, the platform was provided with resources
ownership mechanisms that protect resources from
unauthorized access. Those resources may be image
objects, management or organizational services.
As a result, the collaborative system is obligated
to work with a multi-archive paradigm that must be
supported at the persistence layer, i.e., at the stan-
dard medical imaging repository without interfering
with regular DICOM workflows. In practice, it is re-
quested an accounting mechanism that must be capa-
ble of associate the repository resources permissions,
and delegation of rights, to third entities. The solu-
tion adopted is based on the Lebre et al. (Lebre et al.,
2018a) RBAC (Role-Based Access Control) mecha-
nism for standard DICOM archives.
The solution provides ownership concept and ac-
cess control over medical imaging resources, allow-
ing to deploy different permissions to multiple users
and institutions. It opens doors to the creation of nu-
merous virtual archives with various studies each that
can be shared between users of different realms. For
instance, digital pathology studies can be shared be-
tween distinct institutions and a working session can
be created with a study shared among multiple users
from different institutions.
The proposed access control model is an abstrac-
tion of real-work medical imaging environments. It
allows the management of Organizations, Facilities,
Users, Permissions, Resources, and Permission Shar-
ing. For instance, in the academic context, the organi-
zation shall be the University. Bellow the hierarchic
position of the organization, the Schools are repre-
senting the Facilities. Belonging to the schools, there
are the Students A, B and C, the Professor and the
Resources 1, 2 and 3. At the collaborative layer, the
security mechanism is allowed to create a session for
a limited number of users who have permission to ac-
cess only some of the studies. Assuming that a class is
composed of students from two distinct departments,
where the studies to be analyzed belong to both the
departments, with this accounting mechanism frame-
work, it is possible to share permissions between stu-
HEALTHINF 2020 - 13th International Conference on Health Informatics
460

dents and professors so that everyone can work in a
collaborative workflow. Furthermore, the access can,
hypothetically, be restricted to some hours of the day.
For instance, access can be limited to class time only.
4 USE CASES
This section presents the possible use cases of a col-
laborative pathological viewer. It was already shown
the advantages of using a WSI web viewer to work
on digital pathology samples for diagnosis and aca-
demic purposes. By turning such a viewer into a col-
laborative one, where several users can interact and
discuss one sample at the same time, it extends the
viewer’s utility, improving the concept of telepathol-
ogy. There are two main fields where a collabora-
tive viewer could prove very useful: the educational
field and the clinical environment (Pantanowitz et al.,
2011; Saco et al., 2016; Triola and Holloway, 2011;
Bueno et al., 2016a; Patterson et al., 2011).
In the educational field, the presented system al-
lows, for instance, the following use cases:
• Create groups per class and teach that class on-
line, with the possibility of enforcing a schedule
to attend;
• Create an exam and analyze each student’s re-
sponses through the replay tool;
• Give an open lecture allowing the public to follow
it through the public link;
• Create study sessions amongst students.
On the other hand, the clinical research environ-
ment is, perhaps, the environment that benefited the
most from the implementation of an accounting sys-
tem. The introduction of such a mechanism allows
the restriction, control, and auditing of the access per-
formed to specific patient or study data. Among the
multiple use cases in this field, there are cited below:
• Ask for a consultation on a case by inviting a fel-
low doctor into a session;
• Create a work session to discuss a medical case
with other doctors;
• Review diagnostics performed on cases through
the replay feature (if the diagnostic was done on a
previously created session).
Both the environments benefit from services like
the session-catch replay that consists in the fast re-
play of all the operations performed until the present
moment. Additionally, a session administrator may
revoke access and terminate the session.
5 CONCLUSIONS
Collaborative work is a fundamental improvement in
the modern medical image environments, addressing
educational and diagnosis purposes.Digital pathology
and telepathology are an emerging modality in the
clinical decision-making laboratories and with the
clinical staff supporting the development of new tools.
(Bellis et al., 2013). This paper describes an archi-
tecture for a real-time collaborative digital pathology
viewer with access control mechanisms integrated.
The proposed system is based on the concept of work-
ing sessions that can record the actions performed by
the users over time. The design was created using
merely web technologies to address interoperability
with multiple platforms. The potential benefits and
gains in using a system like the one presented are jus-
tified with the clinical and educational points of view
presented in the use cases section.
ACKNOWLEDGEMENTS
This work has received support from the ERDF Euro-
pean Regional Development Fund through the Oper-
ational Programme for Competitiveness and Interna-
tionalization, COMPETE 2020 Programme, and by
National Funds through the FCT, Fundac¸
˜
ao para a
Ci
ˆ
encia e a Tecnologia within the project PTDC/EEI-
ESS/6815/2014.
REFERENCES
Abouelmehdi, K., Beni-Hessane, A., and Khaloufi, H.
(2018). Big healthcare data: preserving security and
privacy. Journal of Big Data, 5(1):1.
Bankhead, P., Loughrey, M. B., Fern
´
andez, J. A., Dom-
browski, Y., McArt, D. G., Dunne, P. D., McQuaid,
S., Gray, R. T., Murray, L. J., Coleman, H. G., et al.
(2017). Qupath: open source software for digital
pathology image analysis. Scientific Reports.
Beckwith, B. A. (2016). Standards for digital pathology
and whole slide imaging. In Digital Pathology, pages
87–97. Springer.
Bellis, M., Metias, S., Naugler, C., Pollett, A., Jothy, S., and
Yousef, G. M. (2013). Digital pathology: attitudes
and practices in the canadian pathology community.
Journal of Pathology Informatics, 4.
Bernard, C., Chandrakanth, S., Cornell, I. S., Dalton, J.,
Evans, A., Garcia, B. M., Godin, C., Godlewski, M.,
Jansen, G. H., Kabani, A., et al. (2014). Guidelines
from the canadian association of pathologists for es-
tablishing a telepathology service for anatomic pathol-
ogy using whole-slide imaging. Journal of Pathology
Informatics, 5.
Data and Sessions Management in a Telepathology Platform
461

Bidgood, W. D., Horii, S. C., Prior, F. W., and Van Syckle,
D. E. (1997). Understanding and using dicom, the data
interchange standard for biomedical imaging. Jour-
nal of the American Medical Informatics Association,
4(3):199–212.
Brauchli, K. and Oberholzer, M. (2005). The ipath
telemedicine platform. Journal of Telemedicine and
Telecare, 11(2 suppl):3–7.
Bueno, G., Fern
´
andez-Carrobles, M. M., Deniz, O., and
Garc
´
ıa-Rojo, M. (2016a). New trends of emerging
technologies in digital pathology. Pathobiology.
Bueno, G., Fern
´
andez-Carrobles, M. M., Deniz, O., and
Garc
´
ıa-Rojo, M. (2016b). New trends of emerging
technologies in digital pathology. Pathobiology, 83(2-
3):61–69.
Chordia, T., Vikey, A., Choudhary, A., Samdariya, Y., and
Chordia, D. (2016). Current status and future trends in
telepathology and digital pathology. Journal of Oral
and Maxillofacial Pathology, 20(2):178.
Daniel, C., Garc
´
ıa Rojo, M., Bourquard, K., Henin, D.,
Schrader, T., Mea, V. D., Gilbertson, J., and Beck-
with, B. A. (2009). Standards to support information
systems integration in anatomic pathology. Archives
of Pathology & Laboratory Medicine, 133(11):1841–
1849.
Daniel, C., Macary, F., Rojo, M. G., Klossa, J., Lauri-
navi
ˇ
cius, A., Beckwith, B. A., and Della Mea, V.
(2011a). Recent advances in standards for collabora-
tive digital anatomic pathology. In Diagnostic Pathol-
ogy, volume 6, page S17. BioMed Central.
Daniel, C., Rojo, M. G., Klossa, J., Della Mea, V., Booker,
D., Beckwith, B. A., and Schrader, T. (2011b). Stan-
dardizing the use of whole slide images in digi-
tal pathology. Computerized Medical Imaging and
Graphics, 35:496–505.
D
´
ıaz, D., Corredor, G., Romero, E., and Cruz-Roa, A.
(2018). A web-based telepathology framework for
collaborative work of pathologists to support teach-
ing and research in latin america. In Sipaim–Miccai
Biomedical Workshop, pages 105–112. Springer.
Farahani, N., Parwani, A. V., and Pantanowitz, L. (2015).
Whole slide imaging in pathology: advantages, lim-
itations, and emerging perspectives. Pathology and
Laboratory Medicine International, 7:23–33.
Foster, K. (2010). Medical education in the digital age: dig-
ital whole slide imaging as an e-learning tool. Journal
of Pathology Informatics, 1.
Godinho, T. M., Lebre, R., Silva, L. B., and Costa, C.
(2017). An efficient architecture to support digital
pathology in standard medical imaging repositories.
Journal of Biomedical Informatics, 71:190–197.
Goode, A., Gilbert, B., Harkes, J., Jukic, D., and Satya-
narayanan, M. (2013). Openslide: a vendor-neutral
software foundation for digital pathology. Journal of
Pathology Informatics, 4.
Lauro, G. R., Cable, W., Lesniak, A., Tseytlin, E., McHugh,
J., Parwani, A., and Pantanowitz, L. (2013). Digital
pathology consultations—a new era in digital imag-
ing, challenges and practical applications. Journal of
digital imaging, 26(4):668–677.
Lebre, R., Basti
˜
ao, L., and Costa, C. (2018a). Shared med-
ical imaging repositories. In MIE - Medical Informat-
ics Europe, pages 411–415.
Lebre, R., Godinho, T., Silva, L., and Costa, C. (2018b).
A performant and fully dicom compliant web pacs for
digital pathology. In International Journal of Com-
puter Assisted Radiology and Surgery, volume 13,
pages 147–148.
Pantanowitz, L. (2010). Digital images and the future of
digital pathology. Journal of Pathology Informatics,
1.
Pantanowitz, L., Evans, A., Pfeifer, J., Collins, L., Valen-
stein, P., Kaplan, K., Wilbur, D., and Colgan, T.
(2011). Review of the current state of whole slide
imaging in pathology. Journal of Pathology Informat-
ics, 2(1):36.
Pantanowitz, L., Farahani, N., and Parwani, A. (2015).
Whole slide imaging in pathology: advantages, lim-
itations, and emerging perspectives. Pathology and
Laboratory Medicine International, page 23.
Pantanowitz, L., Szymas, J., Yagi, Y., and Wilbur, D.
(2012). Whole slide imaging for educational pur-
poses. Journal of Pathology Informatics, 3.
Patterson, E. S., Rayo, M., Gill, C., and Gurcan, M. N.
(2011). Barriers and facilitators to adoption of soft
copy interpretation from the user perspective: lessons
learned from filmless radiology for slideless pathol-
ogy. Journal of Pathology Informatics.
Saco, A., Bombi, J. A., Garcia, A., Ram
´
ırez, J., and Ordi,
J. (2016). Current status of whole-slide imaging in
education. Pathobiology, 83(2-3):79–88.
Silva, J. M., Godinho, T. M., Silva, D., and Costa, C.
(2019). A community-driven validation service for
standard medical imaging objects. Computer Stan-
dards & Interfaces, 61:121–128.
Snyder, P. D., Hasso, C. A., and Masi, L. P. (2001). Pathol-
ogy dependent viewing of processed dental radio-
graphic film having authentication data. US Patent
6,195,474.
Triola, M. M. and Holloway, W. J. (2011). Enhanced virtual
microscopy for collaborative education. BMC Medi-
cal Education, 11(1):4.
Tuominen, V. J. and Isola, J. (2010). Linking whole-slide
microscope images with dicom by using jpeg2000
interactive protocol. Journal of Digital Imaging,
23(4):454–462.
Valente, F., Silva, L. A. B., Godinho, T. M., and Costa, C.
(2016). Anatomy of an extensible open source pacs.
Journal of Digital Imaging, 29(3):284–296.
Weiss, M. (2012). Apc forum: carestream health’s it trans-
formation. MIS Quarterly Executive, 11(1):8.
Zhang, J., Stahl, J. N., Huang, H. K., Zhou, X., Lou, S.-L.,
and Song, K. S. (2000). Real-time teleconsultation
with high-resolution and large-volume medical im-
ages for collaborative healthcare. IEEE Transactions
on Information Technology in Biomedicine, 4(2):178–
185.
HEALTHINF 2020 - 13th International Conference on Health Informatics
462
