
Multimodal Fusion Strategies for Outcome Prediction in Stroke
Esra Zihni
1,3 a
, Vince Madai
1
, Ahmed Khalil
2
, Ivana Galinovic
2
, Jochen Fiebach
2 b
,
John D. Kelleher
3 c
, Dietmar Frey
1
and Michelle Livne
1
1
Predictive Modelling in Medicine Research Group, Department of Neurosurgery,
Charit
´
e - Universit
¨
atsmedizin Berlin, Berlin, Germany
2
Centre for Stroke Research Berlin, Charit
´
e - Universit
¨
atsmedizin Berlin, Berlin, Germany
3
ADAPT Research Center, Technological University Dublin, Dublin, Ireland
Keywords:
Machine Learning, Multimodal Fusion, Neural Networks, Predictive Modeling, Acute-ischemic Stroke.
Abstract:
Data driven methods are increasingly being adopted in the medical domain for clinical predictive modeling.
Prediction of stroke outcome using machine learning could provide a decision support system for physicians
to assist them in patient-oriented diagnosis and treatment. While patient-specific clinical parameters play an
important role in outcome prediction, a multimodal fusion approach that integrates neuroimaging with clinical
data has the potential to improve accuracy. This paper addresses two research questions: (a) does multimodal
fusion aid in the prediction of stroke outcome, and (b) what fusion strategy is more suitable for the task at hand.
The baselines for our experimental work are two unimodal neural architectures: a 3D Convolutional Neural
Network for processing neuroimaging data, and a Multilayer Perceptron for processing clinical data. Using
these unimodal architectures as building blocks we propose two feature-level multimodal fusion strategies: 1)
extracted features, where the unimodal architectures are trained separately and then fused, and 2) end-to-end,
where the unimodal architectures are trained together. We show that integration of neuroimaging information
with clinical metadata can potentially improve stroke outcome prediction. Additionally, experimental results
indicate that the end-to-end fusion approach proves to be more robust.
1 INTRODUCTION
Stroke
1
is a major cause of death and long-term dis-
abilities worldwide. In the clinical setting, physi-
cians decide which patients will benefit from treat-
ment on the basis of likely long-term outcomes if
treated. Currently, a time-window approach based
on the time from stroke onset to treatment is being
used as the main treatment decision criteria, together
with subjective assessment of acute stroke imaging
acquired in routine examination. Outcome predic-
tion in stroke aims to develop a machine learning
based decision support system that provides reliable
information to physicians to assist them for better di-
agnosis and treatment for ischemic stroke patients.
It is known that patient-specific clinical parameters
play an important role in creating a baseline for out-
come prediction, while combining imaging informa-
tion has the potential to improve the predictive accu-
a
https://orcid.org/0000-0003-2288-2406
b
https://orcid.org/0000-0002-7936-6958
c
https://orcid.org/0000-0001-6462-3248
1
The code for this project can be found here: https://
github.com/prediction2020/multimodal-classification
racy (Asadi et al., 2014; Whiteley et al., 2012; Vora
et al., 2011). We hypothesize that using state-of-the-
art machine learning algorithms to train a model on
both data modalities (i.e. clinical metadata and neu-
roimaging) would increase outcome prediction accu-
racy. We aim to develop an automated method to
predict a binary 3 months post-stroke outcome, us-
ing time-of-flight (TOF) magnetic resonance angiog-
raphy (MRA) images and clinical metadata.
Deep learning methods have achieved state-of-
the-art performance compared to classical machine
learning methods in predictive modeling, which has
led to their increased adoption in medical applications
(Kelleher, 2019). A number of studies have presented
models using Multilayer Perceptrons (MLPs) for out-
come prediction based on clinical parameters (Asadi
et al., 2014; Heo et al., 2019). Additionally, using
Convolutional Neural Networks (CNNs) on imaging
data has been proven to give promising results in tis-
sue outcome prediction (Nielsen et al., 2018; Pinto
et al., 2018), as well as predicting final stroke out-
come (Hilbert et al., 2019). Hence, we propose two
unimodal architectures based on deep learning meth-
ods: a 3D CNN to process neuroimaging data and an
MLP for processing clinical metadata; both tailored
Zihni, E., Madai, V., Khalil, A., Galinovic, I., Fiebach, J., Kelleher, J., Frey, D. and Livne, M.
Multimodal Fusion Strategies for Outcome Prediction in Stroke.
DOI: 10.5220/0008957304210428
In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2020) - Volume 5: HEALTHINF, pages 421-428
ISBN: 978-989-758-398-8; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
421

to the requirements of the data and the final outcome
prediction task.
Our motivation for this work is that the fusion of
multiple data modalities that observe the same phe-
nomenon may allow for more robust predictions by
capturing complementary information. Recently, us-
ing brain imaging data together with clinical informa-
tion has become a popular target to replace the cur-
rent time-window approach. There are several mod-
els that combine information from clinical and neu-
roimaging data to create a joint feature space (Cui
et al., 2018; Dharmasaroja and Dharmasaroja, 2012;
Johnston et al., 2002). However, these methods are
limited to the manual extraction of predetermined fea-
tures from brain images and therefore do not have the
capacity to account for data-driven features. To the
best of our knowledge, the pilot study conducted by
Bacchi et al. is the only data-driven multimodal ar-
chitecture developed so far that combines clinical and
imaging information to predict stroke outcome.
There are mainly two types of multimodal fusion
in multimodal machine learning: feature-level and
decision-level. Feature-level fusion integrates fea-
tures extracted from various modalities, whereas in
decision level fusion the integration is performed on
the final decisions of each modality. Feature-level
fusion is widely used by researchers due to 1) the
possibility to exploit the correlation and interactions
between low level features of each modality and 2)
the increasing popularity of deep learning methods
for feature extraction (Poria et al., 2017; Baltrusaitis
et al., 2019). In this paper we consider two strategies
of feature-level fusion: extracted features and end-to-
end. Most existing research on feature-level fusion
adopts the extracted features strategy, where separate
learning of modality features is followed by learn-
ing from the combined feature space (Wang et al.,
2017; Oramas et al., 2017; Slizovskaia et al., 2017;
Mouzannar et al., 2018; Kim and McCoy, 2018).
However, Goh et al. point out that these existing
models operate primarily on different streams of syn-
chronous raw data (e.g. a video stream and its corre-
sponding audio stream, or an image and its respective
text caption), whereas for clinical and imaging data
this synchronization does not exist. Recent work in
the medical domain showed that the end-to-end strat-
egy involving simultaneous learning of imaging and
clinical modalities yielded promising results both for
the diagnosis of Alzheimer’s disease (Esmaeilzadeh
et al., 2018) and the prediction of stroke outcome
(Bacchi et al., 2019). These end-to-end studies, how-
ever, do not compare their method to the widely used
extracted features approach in literature. Here, we
conduct a comparative study to explore the advan-
tages and disadvantages of these two feature-level
fusion approaches, i.e extracted features and end-to-
end, with regards to stroke outcome prediction.
In this comparative study we use the CNN and
MLP unimodal architectures (mentioned above) as
fundamental building blocks, we propose two multi-
modal feature-level fusion strategies: 1) an extracted
features strategy where the unimodal architectures are
trained separately and then frozen and fused at the
extracted feature-level, and 2) an end-to-end strat-
egy where the unimodal architectures are fused at the
feature-level and then trained simultaneously.
In this paper we address two research questions:
(a) does the fusion of clinical and neuroimaging
modalities aid in the prediction of stroke outcome,
and (b) which fusion strategy is better suited for the
task and data at hand.
2 DATA
The data used in this project was of patients from the
1000Plus study (Hotter et al., 2009). Examinations
on patients at admission included National Institute
of Health Stroke Scale (NIHSS) scoring and stroke
MRI including time-of-flight (TOF) magnetic reso-
nance angiography (MRA). TOF-MRA imaging en-
ables the analysis of the anatomy of blood vessels,
which may provide an important measure for bet-
ter understanding of the vessel status and blood flow
throughout the vasculature. Modified Rankin Scale
(mRS), that quantifies the degree of disability or de-
pendence in daily activities, was rated 90 days after
symptoms onset. The available database consisted of
514 patients and additionally included information on
patients’ demographics and medical history. Of these
106 were excluded due to missing mRS score and 92
were excluded due to missing or distorted acute TOF-
MRA imaging.
2.1 Clinical Data
The following seven clinical predictors were selected
as clinical input: age, sex, initial NIHSS, cardiac his-
tory, diabetes, hypercholesterolemia, and thromboly-
sis treatment. Inclusion criteria were for categorical
predictors to have at least 1 to 4 ratio of absence /
existence and for all predictors to have no more than
5% missing values. Table 1 gives a summary of the
clinical predictors and their distribution.
Missing values were imputed using mean impu-
tation. The continuous variables (age, NIHSS) were
centered over patients using zero-mean unit-variance.
HEALTHINF 2020 - 13th International Conference on Health Informatics
422
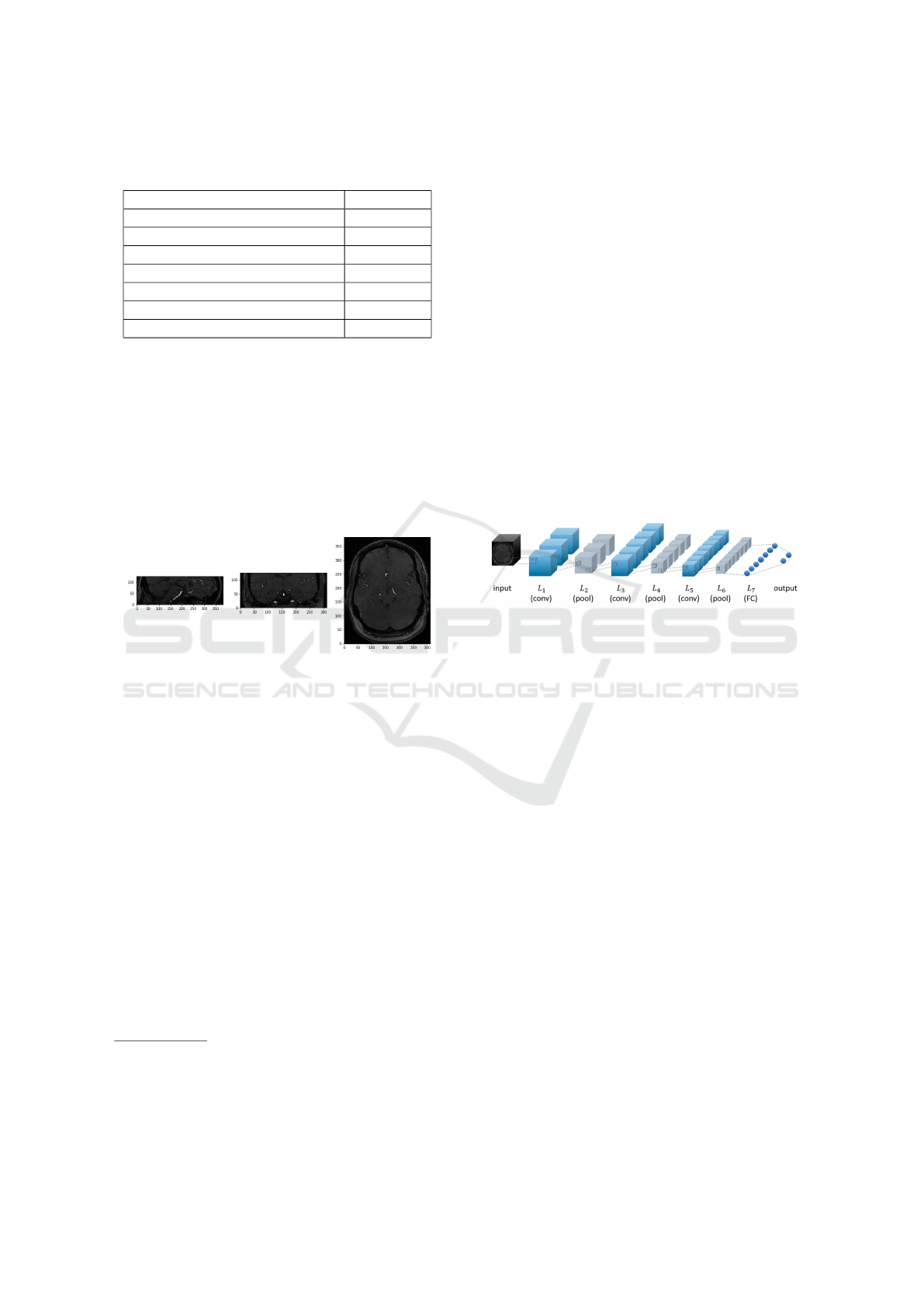
Table 1: Statistics of clinical predictors. IQR: interquartile
range; NIHSS: National Institutes of Health Stroke Scale.
Clinical information Value
Median age (IQR) 72.0 (16.0)
Median initial NIHSS (IQR) 3.0 (4.0)
Sex (females/males) 116 / 197
Thrombolysis treatment (yes/no) 58 / 255
Cardiac history (yes/no) 87 / 226
Diabetes (yes/no) 84 / 229
Hypercholesterolemia (yes/no) 187 / 126
2.2 Imaging Data
3D volumes of acute TOF-MRA images in NIfTI for-
mat were used as imaging input
2
. The scans were
gray-scale with voxel intensity values [0,255]. Figure
1 shows an example image. Images were resized from
312x384x127 to 156x192x64 voxels due to memory
constraints. After resizing, the voxel intensity values
were centered using zero mean and unit variance.
Figure 1: The middle slices of a time-of-flight magnetic res-
onance angiography (TOF MRA) image taken from the (A)
sagittal (B) coronal and (C) horizontal planes.
3 ARCHITECTURE
All frameworks were trained on a binary classifica-
tion task using binary cross-entropy loss. A softmax
output layer consisting of two fully connected (FC)
neurons was used as output.
3.1 Unimodal Frameworks
In the scope of this project, the unimodal frameworks
were designed to 1) provide baseline performance in
order to assess the value in multiple modality inte-
gration and 2) extract separately trained clinical and
imaging features (Figure 3A-3B).
2
All imaging is performed with a 3T MRI scanner (Tim
Trio; Siemens AG, Erlangen, Germany) dedicated to clini-
cal research. TOF vessel imaging had the following param-
eters: repetition time (TR) = 22 ms; echo time (TE) = 3.86
ms; time of acquisition = 3:50 minutes.
3.1.1 Multilayer Perceptron
The clinical data was modeled using a multilayer per-
ceptron (MLP) with a single fully connected (FC)
hidden layer. The number of neurons in the hidden
layer was fine tuned during model selection (see sec-
tion 4.2). The hidden layer neurons were rectified
linear units (ReLUs). In order to prevent over-fitting
1) `
2
norm regularization was introduced to penalize
weights in the hidden and output layer neurons and 2)
dropout was used on the hidden layer neurons.
3.1.2 Convolutional Neural Network
The 3D imaging data was modeled using a 3D con-
volutional neural network (CNN) consisting of three
convolutional blocks followed by a single FC layer. A
convolutional block refers to a set of consecutive con-
volutional and max pooling layers. The architecture
of the CNN framework is given in Figure 2.
Figure 2: Illustration of the convolutional neural network
used in this paper. The architecture consists of three convo-
lutional (L
1
,L
3
,L
5
) and three max pooling (L
2
,L
4
,L
6
) lay-
ers followed by a fully connected (L
7
) layer.
Filter size, filter stride and pooling size as well
as number of filters in the convolutional layers and
number of neurons in the FC layer were fine tuned
during model selection (see section 4.2). All convo-
lutional layer neurons as well as all FC layer neurons
were ReLUs. `
2
norm regularization was introduced
to penalize weights in the convolutional and FC lay-
ers. Dropout was used only on the FC layer neurons.
3.2 Multimodal Frameworks
We developed two multimodal frameworks that have
the same architectural design: In each unimodal
pipeline, the output layer was dropped and the penul-
timate layer output was fed into an FC layer. This
embedded the high dimensional imaging data into a
lower dimension and vice versa for the clinical data.
The outputs from these embeddings were then con-
catenated and fed to a final FC layer followed by the
output layer (Figure 3C-3D).
The embedding layers allow for weighting of fea-
ture vectors from the two data modalities. This is
done by adjusting the number of neurons in each em-
bedding layer. For both multimodal frameworks, two
Multimodal Fusion Strategies for Outcome Prediction in Stroke
423
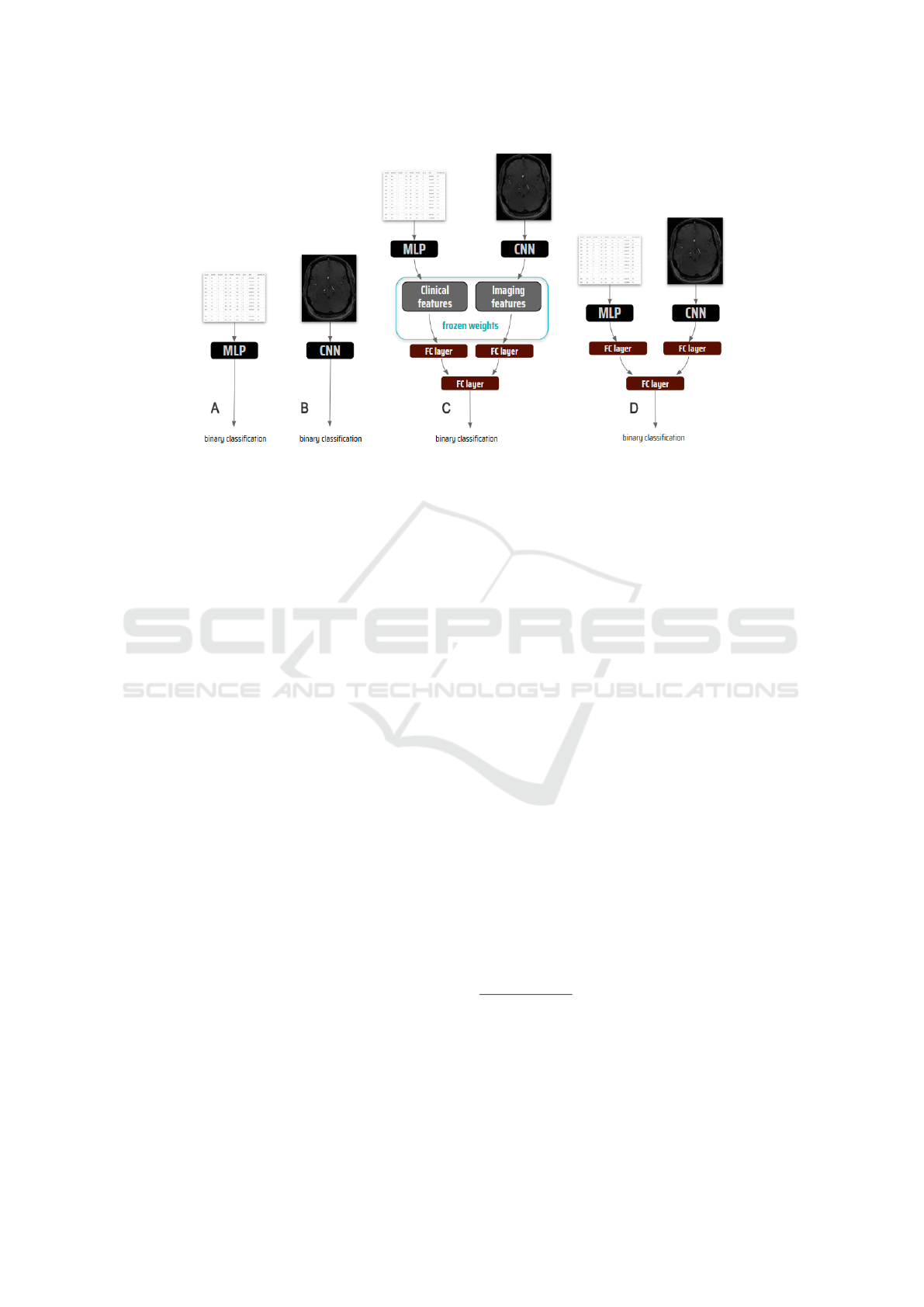
Figure 3: The frameworks: (A) Multilayer Perceptron (MLP) for modeling clinical data, (B) Convolutional Neural Network
(CNN) for modeling imaging data, (C) Extracted features and (D) End-to-end strategies for modeling multimodal data.
schemes for assigning the number of neurons for fea-
ture embedding were tested: 1) assigning equal num-
ber of neurons to each modality and 2) assigning dou-
ble the number of neurons for clinical feature embed-
ding. The second scheme was chosen based on the
unimodal results indicating better performance of the
clinical data-based model, i.e MLP. While the archi-
tectural designs were the same for both fusion strate-
gies, they differed in the training process.
3.2.1 Extracted Features Strategy
In this framework, the clinical and imaging features
are first learned separately. The whole framework is
then trained using the learned features as input, i.e the
weights in the penultimate layers of the trained CNN
and MLP are frozen and only the following FC lay-
ers are trained. All FC layer neurons were ReLUs.
`
2
norm regularization and dropout was introduced in
the FC layers for regularization. The number of neu-
rons in the embedding layers and the final FC layer
were fine tuned during model selection (section 4.2).
3.2.2 End-to-End Strategy
In this strategy, the whole framework was trained end-
to-end on both data modalities simultaneously (Figure
3.D), i.e the two modalities were trained together on
the prediction task. All convolutional layer and all FC
layer neurons were ReLUs. `
2
norm regularization
was used in the convolutional and FC layers, whereas
dropout was only used on the FC layer neurons. The
process for setting the filter size, filter stride, pooling
size and number of filters of the CNN part is described
in section 4.2.2. We chose a filter size of (3x3x3),
filter stride of (1x1x1), pooling size of (3x3x3) and
number of filters of L
1
: 16, L
3
: 32, L
5
: 64 followed
by a FC layer of L
7
: 128 neurons (see Figure 2 for ar-
chitecture). The number of neurons in the MLP hid-
den layer, the embedding layers and the final FC layer
were fine tuned during model selection (section 4.2).
4 EXPERIMENTAL SETUP
Supervised machine learning methods were used to
predict 90 days post-stroke mRS scores. The mRS
range of [0,6] was dichotomized, consistently with the
standard applied models in the field (Heo et al., 2019;
Wouters et al., 2018). A score between [0-2] indicates
good outcome (87 patients) and [3-6] indicates bad
outcome (226 patients). All frameworks were trained
for the same binary classification task
3
.
4.1 Model Training
Binary cross-entropy loss, which quantifies how dif-
ferent two probability distributions are, was selected
as the loss function, as it is a common choice for bi-
nary classification tasks. Loss was minimized using
the Adaptive Moment Estimation (Adam) optimizer.
3
All frameworks were developed using Python (v3.6.5)
and all models were trained using Keras (v2.2.4) running
on a Tensorflow (v1.12.0) backend. Nibabel (v2.3.0) li-
brary was used for reading imaging data and Scikit-learn
(v0.20.3) library was used for pre-processing both clinical
and imaging data. All training and evaluation was done
on a workstation with Intel
R
Core
TM
i7-6950X CPU @
3.00GHz x 20 and TITAN RTX GPU x 2.
HEALTHINF 2020 - 13th International Conference on Health Informatics
424

Adam was recently recommended to be used as the
default optimization algorithm in deep learning be-
cause of its fast convergence (Ruder, 2016). Initial
weights were sampled from a Glorot uniform distri-
bution. A Softmax function was used as the output
layer activation to calculate the final class probabili-
ties of the good and bad outcome classes.
Early stopping was introduced during training in
order to prevent over-fitting: training stopped once the
improvement in validation loss was below a specified
value. The value for each framework was set depend-
ing on the appropriate range of the validation loss.
4.2 Model Selection
In addition to the architectural hyper-parameters se-
lected in each framework for fine tuning (sections
3.1 and 3.2) the following hyper-parameters were fine
tuned: batch size, ratio of the `
2
norm regularization,
dropout rate and the learning rate of the optimizer.
4.2.1 Training-validation-test Splits
The data was randomly split into three subsets: train-
ing, validation and test with 200, 50 and 63 patients
in each respectively. Patients in each set corresponded
for both input modalities, i.e. the clinical and imag-
ing information in each set was from the same pa-
tients. Additionally, the sets were consistently used
for training, validation and testing of each of the four
frameworks, in order to achieve comparable results.
To account for the variance between subsets,
which is likely to be higher in small datasets, the
random selection of training-, validation- and test
sets was repeated five times resulting in five different
splits. Model selection using grid search was there-
fore repeated for each split. This aims to reduce bias
and variance of the models. Since grid search resulted
in different hyper-parameters in different splits, final
performance was assessed for each split individually.
4.2.2 Grid Search
Model selection, i.e the best choice of hyper-
parameters, was done using an exhaustive search
method called grid search. Using grid search, mod-
els were trained and evaluated on the training and
validation sets respectively for each hyper-parameter
combination. The hyper-parameter combination that
yielded best model performance on the validation set
were chosen for final training. Model performance on
the validation set was evaluated using area under the
receiver operator characteristics curve (AUC) score.
Typically the cross-validation method is used to over-
come variance in model performance; however, due to
computational limitations (e.g. long training times) it
was not adopted in this project. Finally, using the se-
lected hyper-parameters a final model was trained on
the combined training and validation data. The same
model selection process was carried out for all five
training-validation-test splits, resulting in five models
for each framework.
The only exception to this model selection pro-
cess was the CNN in the end-to-end fusion frame-
work. In order to save computational power and time,
rather than fitting the architecture hyper-parameters
(filter size, filter stride, pooling size and number of
filters) as part of a grid search, they were pre-set to
the most frequently occurring combination of these
hyper-parameters found across the five splits of fitting
the unimodal CNN architecture.
Table 2: (a) Test and (b) training performances by median and interquartile range (IQR) calculated over 100 training and
test runs. Columns represent the different splits, last column shows the average over the other five. Rows represent the 1)
convolutional neural network (CNN), 2) multilayer perceptron (MLP), 3) feature extraction and 4) end-to-end frameworks.
(a) Test performance
Framework
median AUC score (iqr)
Split 1 Split 2 Split 3 Split 4 Split 5
Splits
average
CNN 0.61 (0.05) 0.76 (0.03) 0.68 (0.03) 0.68 (0.04) 0.67 (0.2) 0.68
MLP 0.70 (0.05) 0.76 (0.02) 0.80 (0.01) 0.71 (0.06) 0.78 (0.05) 0.75
Extracted features 0.60 (0.02) 0.78 (0.01) 0.78 (0.02) 0.76 (0.01) 0.83 (0.01) 0.75
End-to-end 0.71 (0.04) 0.78 (0.02) 0.79 (0.03) 0.73 (0.05) 0.81 (0.04) 0.76
(b) Training performance
Framework
median AUC score (iqr)
Split 1 Split 2 Split 3 Split 4 Split 5
Splits
average
CNN 0.97 (0.04) 0.99 (0.004) 1.00 (0) 0.94 (0.08) 0.89 (0.3) 0.96
MLP 0.81 (0.05) 0.85 (0.01) 0.87 (0.004) 0.82 (0.03) 0.83 (0.03) 0.84
Extracted features 0.97 (0.004) 0.96 (0.006) 0.99 (0.001) 0.88 (0.007) 0.99 (0.001) 0.96
End-to-end 0.90 (0.06) 0.90 (0.07) 0.91 (0.06) 0.92 (0.08) 0.87 (0.08) 0.90
Multimodal Fusion Strategies for Outcome Prediction in Stroke
425

4.3 Model Evaluation
Model performances were measured using AUC. To
evaluate overfitting, the AUC score was calculated on
both the respective training (merged training and val-
idation sets) and test sets of the trained models.
Variations in model performance due to random
processes such as dropout and parallel computing was
investigated by repeated training and evaluation. The
repetition was 100 times for each training-validation-
test split. The median and interquartile range (IQR)
over the 100 training and test AUC scores was calcu-
lated and used as the final performance measure. Ad-
ditionally, variation in model performance due to data
variability was investigated by calculating the mean
AUC score over the median of five splits.
Finally, a non-parametric paired t-test, i.e.
Wilcoxon signed rank test, was performed in order to
determine if the multimodal frameworks (i.e. end-to-
end and feature extraction) significantly outperformed
the clinical data driven MLP network. The test was
based on the distribution of test performances given
by the 100 runs within each split. Here, multimodal
frameworks were compared to the clinical data based
MLP framework in order to highlight the benefits of
integrating neuroimaging information.
5 RESULTS
Table 2 summarizes performances for the unimodal
and multimodal frameworks. For each split, me-
dian AUC scores calculated over 100 training and
evaluation runs are presented together with the IQR.
Average training and test performance over splits
(calculated as the mean) is given in the last column.
Imaging. CNN showed low performance on the test
sets with a mean AUC score of 0.68 over the splits,
but performed very high on the training sets with
a mean AUC score of 0.96 This result indicates a
strong overfitting in the CNN models.
Clinical. MLP performed well on both the training
and test sets with an average AUC score of 0.84 and
0.75 respectively. The MLP was therefore less prone
to overfitting compared to the CNN.
Multimodal. The feature extraction framework per-
formed on average the same as the MLP framework
and better than the CNN frameworks with an AUC
of 0.75 on test sets. On the other hand, performance
on the training sets reached an average of 0.96, the
same value as the CNN models average. In general
the feature extraction framework was the most stable
in both training and test performances with an IQR of
no more than 0.02 on any of the splits.
The end-to-end framework performed better than
the MLP on average in both training and test sets
with AUC scores of 0.90 and 0.76 respectively.
Additionally end-to-end performed better than the
CNN only in average test performance, thus not
exhibiting the overfitting characteristic of the CNN.
Significance. Table 3 shows the results of the
Wilcoxon signed rank test that was based on the distri-
bution of test performances within each split. The sig-
nificance test showed that the end-to-end framework
performed significantly better compared to the clini-
cal based MLP framework in all splits with the excep-
tion of split 3, which showed the opposite result. The
feature extraction framework yielded inconsistent re-
sults, with significantly improved performance for 3
of the splits (i.e. 2,4,5) and significantly worse per-
formance for the other two splits.
6 DISCUSSION
Our results show that there is potentially clinical value
in TOF-MRA images for stroke outcome prediction.
Both multimodal architectures displayed better test
performance in the majority of the five splits com-
pared to the MLP and CNN models trained only on
clinical and neuroimaging data respectively.
Of the two multimodal feature-level fusion strate-
gies, the end-to-end strategy achieved more consis-
tent improvement over the five splits. Although the
improvements were not substantial, they were shown
to be statistically significant by the Wilcoxon signed
rank test. On the other hand, while showing a lower
averaged performance over splits, the extracted fea-
tures strategy demonstrated higher stability within a
split, i.e a lower variance in performance over the 100
runs within a split, indicated by IQR values. This is
expected, since the extracted features framework only
learns the final FC layer weights, which makes this
strategy less prone to variations caused by random
processes during learning.
The test and training performance patterns demon-
strate that the extracted features strategy enforces a
strong prediction bias towards one of the modali-
ties. This bias is well exemplified in the first split,
where the effect of the low performing CNN model
is reflected in the test performance of the extracted
features model. Here, since the imaging features
were extracted from a low performing model and
were not introduced as trainable parameters in the ex-
HEALTHINF 2020 - 13th International Conference on Health Informatics
426

Table 3: The Wilcoxon signed rank test p values on test performances over 100 runs. The test compares a) end-to-end against
the MLP and b) extracted features against the MLP. The splits where the multimodal frameworks outperformed the unimodal
MLP (in terms of median AUC score) are highlighted in bold.
Frameworks
p values
Split 1 Split 2 Split 3 Split 4 Split 5
Extracted features vs. MLP 7e-18 3e-13 6e-16 4e-16 1e-17
End-to-end vs. MLP 5e-04 3e-11 5e-03 6e-04 4e-08
tracted features framework, the inherent shortcomings
of the learned imaging features could not be mitigated
through additional representation learning in the MLP
network. Whereas in the end-to-end framework, since
the features from both modalities are extracted and
learned simultaneously, the network seems to adapt
itself to the better performing modality (e.g clinical
metadata), hence alleviating the poor feature repre-
sentation of the CNN pipeline. The same effect is ex-
pressed in the training performances of all splits. The
extracted features strategy displays the near-to-perfect
training performance of the CNN framework more
profoundly than the end-to-end strategy. Following
these findings we can suggest that the extracted fea-
tures approach may be a stronger strategy when both
modalities perform well for the task at hand sepa-
rately, but not when one modality suffers from low
performance. We show that for the case at hand, the
end-to-end strategy works better.
In the scope of this project imaging data was hy-
pothesized as a means to improve the performance of
clinical-based outcome prediction models, rather than
providing reliable outcome prediction by itself. Nev-
ertheless, the CNN framework trained only on imag-
ing data for the outcome prediction task showed com-
parable results to the data-efficient method of Hilbert
et al., 2019. At the same time, performance was rel-
atively high on the training sets compared with the
test sets, indicating that the model was suffering from
overfitting. Overfitting could not be overcome by the
introduced regularization methods such as `
1
,`
2
norm
regularizations, dropout, batch normalization or de-
creasing number of model parameters by using less
convolutional blocks or less number of filters. This
shows that even when the architecture is tailored to
the needs of the data and task at hand, the features dis-
covered during training are not representative of the
actual classification task but rather tailored to the cor-
relation between the input and output of the training
set. This may be resulting from the properties of the
imaging data, the complexity of the model class and
the coarse definition of the classification problem.
Our study has several limitations. First, the small
sample size of the given cohort limits the generaliz-
ability of our models. Although many clinical predic-
tors were recorded in the 1000Plus study, only seven
predictors could be included in our project due to the
high percentage of missing data. In this case, the
ratio of features to sample size showed to be suffi-
cient enough to prevent overfitting, but having more
features might have been beneficial in utilizing the
full capacity of a complex model, such as an MLP.
Similarly for imaging, several patients had to be ex-
cluded due to incomplete scans. In this case, since
every voxel in the input image is considered as a fea-
ture, the feature space was very large in compari-
son to the sample size. This can explain the strong
overfitting behaviour of the CNN models. Addition-
ally, a small sample size resulted in high data vari-
ability between training-validation-test sets. This was
demonstrated by the performance inconsistency be-
tween splits. Furthermore, limitations in computa-
tional power restricted the training of the CNN and
end-to-end to small mini-batches. Additionally since
model training is longer with imaging data, cross val-
idation was not used during model selection and the
number of training-validation-test sets were limited to
five. Performing cross validation for selecting the best
hyper-parameters may provide more stability in over-
all model performances, i.e. variance between and
within splits will be reduced. The same argument is
valid if more training-validation-test sets can be used
for model selection and evaluation. Improved stabil-
ity by using cross-validation and increased number of
splits may allow for a more reliable comparison be-
tween the two multimodal fusion approaches.
7 CONCLUSION
We developed and evaluated two multimodal feature-
level fusion frameworks to predict final outcome in
acute ischemic stroke patients using clinical data and
neuroimaging. We showed that a multimodal ap-
proach achieves better results and neuroimaging may
hold beneficial information for outcome prediction
when used with clinical metadata. We demonstrated
how a multimodal approach using simultaneous end-
to-end learning of modalities, outperforms learning
from the combination of separately learned features.
Multimodal Fusion Strategies for Outcome Prediction in Stroke
427

ACKNOWLEDGEMENTS
This research was supported by the PRECISE4Q
project, funded through the European Union’s Hori-
zon 2020 research and innovation program under
grant agreement No. 777107, and the ADAPT Re-
search Centre, funded by Science Foundation Ireland
(Grant 13/RC/2106) and is co-funded by the Euro-
pean Regional Development fund.
REFERENCES
Asadi, H., Dowling, R., Yan, B., and Mitchell, P. (2014).
Machine Learning for Outcome Prediction of Acute
Ischemic Stroke Post Intra-Arterial Therapy. PLoS
ONE, 9(2):e88225.
Bacchi, S., Zerner, T., Oakden-Rayner, L., Kleinig, T., Pa-
tel, S., and Jannes, J. (2019). Deep Learning in the
Prediction of Ischaemic Stroke Thrombolysis Func-
tional Outcomes: A Pilot Study. Academic Radiology,
pages 1–5.
Baltrusaitis, T., Ahuja, C., and Morency, L. P. (2019). Mul-
timodal Machine Learning: A Survey and Taxonomy.
IEEE Transactions on Pattern Analysis and Machine
Intelligence, 41(2):423–443.
Cui, H., Wang, X., Bian, Y., Song, S., and Feng, D. D.
(2018). Ischemic stroke clinical outcome prediction
based on image signature selection from multimodal-
ity data. In 2018 40th Annual International Confer-
ence of the IEEE Engineering in Medicine and Biol-
ogy Society (EMBC), pages 722–725. IEEE.
Dharmasaroja, P. and Dharmasaroja, P. A. (2012). Pre-
diction of intracerebral hemorrhage following throm-
bolytic therapy for acute ischemic stroke using multi-
ple artificial neural networks. Neurological Research,
34(2):120–128.
Esmaeilzadeh, S., Belivanis, D. I., Pohl, K. M., and Adeli,
E. (2018). End-To-End Alzheimer’s Disease Diagno-
sis and Biomarker Identification. pages 337–345.
Heo, J., Yoon, J. G., Park, H., Kim, Y. D., Nam, H. S., and
Heo, J. H. (2019). Machine Learning–Based Model
for Prediction of Outcomes in Acute Stroke. Stroke,
50(5):1263–1265.
Hilbert, A., Ramos, L. A., van Os, H. J., Olabarriaga, S. D.,
Tolhuisen, M. L., Wermer, M. J., Barros, R. S., van der
Schaaf, I., Dippel, D., Roos, Y. B., van Zwam, W. H.,
Yoo, A. J., Emmer, B. J., Lycklama
`
a Nijeholt, G. J.,
Zwinderman, A. H., Strijkers, G. J., Majoie, C. B., and
Marquering, H. A. (2019). Data-efficient deep learn-
ing of radiological image data for outcome prediction
after endovascular treatment of patients with acute is-
chemic stroke. Computers in Biology and Medicine,
115(October):103516.
Hotter, B., Pittl, S., Ebinger, M., Oepen, G., Jegzentis, K.,
Kudo, K., Rozanski, M., Schmidt, W. U., Brunecker,
P., Xu, C., Martus, P., Endres, M., Jungeh
¨
ulsing, G. J.,
Villringer, A., and Fiebach, J. B. (2009). Prospective
study on the mismatch concept in acute stroke patients
within the first 24 h after symptom onset - 1000Plus
study. BMC Neurology, 9(1):60.
Johnston, K. C., Wagner, D. P., Haley, E. C., and Connors,
A. F. (2002). Combined Clinical and Imaging Infor-
mation as an Early Stroke Outcome Measure. Stroke,
33(2):466–472.
Kelleher, J. D. (2019). Deep Learning. MIT Press.
Kim, E. and McCoy, K. F. (2018). Multi modal deep
learning using images and text for information graphic
classification. ASSETS 2018 - Proceedings of the 20th
International ACM SIGACCESS Conference on Com-
puters and Accessibility, pages 143–148.
Mouzannar, H., Rizk, Y., and Awad, M. (2018). Dam-
age Identification in Social Media Posts Using Mul-
timodal Deep Learning. Proceedings of the Inter-
national ISCRAM Conference, 2018-May(May):529–
543.
Nielsen, A., Hansen, M. B., Tietze, A., and Mouridsen, K.
(2018). Prediction of Tissue Outcome and Assessment
of Treatment Effect in Acute Ischemic Stroke Using
Deep Learning. Stroke, 49(6):1394–1401.
Oramas, S., Nieto, O., Sordo, M., and Serra, X. (2017). A
deep multimodal approach for cold-start music recom-
mendation. ACM International Conference Proceed-
ing Series, Part F1301:32–37.
Pinto, A., Mckinley, R., Alves, V., Wiest, R., Silva, C. A.,
and Reyes, M. (2018). Stroke Lesion Outcome Predic-
tion Based on MRI Imaging Combined With Clinical
Information. Frontiers in Neurology, 9.
Poria, S., Cambria, E., Bajpai, R., and Hussain, A. (2017).
A review of affective computing: From unimodal
analysis to multimodal fusion. Information Fusion,
37:98–125.
Ruder, S. (2016). An overview of gradient descent opti-
mization algorithms.
Slizovskaia, O., Gomez, E., and Haro, G. (2017). Mu-
sical instrument recognition in user-generated videos
using a multimodal convolutional neural network ar-
chitecture. ICMR 2017 - Proceedings of the 2017
ACM International Conference on Multimedia Re-
trieval, pages 226–232.
Vora, N. A., Shook, S. J., Schumacher, H. C., Tievsky,
A. L., Albers, G. W., Wechsler, L. R., and Gupta, R.
(2011). A 5-Item Scale to Predict Stroke Outcome Af-
ter Cortical Middle Cerebral Artery Territory Infarc-
tion. Stroke, 42(3):645–649.
Wang, D., Mao, K., and Ng, G.-W. (2017). Convolutional
neural networks and multimodal fusion for text aided
image classification. In 2017 20th International Con-
ference on Information Fusion (Fusion), pages 1–7.
IEEE.
Whiteley, W. N., Slot, K. B., Fernandes, P., Sandercock,
P., and Wardlaw, J. (2012). Risk Factors for Intracra-
nial Hemorrhage in Acute Ischemic Stroke Patients
Treated With Recombinant Tissue Plasminogen Ac-
tivator. Stroke, 43(11):2904–2909.
Wouters, A., Nysten, C., Thijs, V., and Lemmens, R. (2018).
Prediction of Outcome in Patients With Acute Is-
chemic Stroke Based on Initial Severity and Improve-
ment in the First 24 h. Frontiers in Neurology, 9.
HEALTHINF 2020 - 13th International Conference on Health Informatics
428
