
Language Identification for Short Medical Texts
Erick Velazquez Godinez, Zolt
´
an Szl
´
avik, Selene Baez Santamar
´
ıa and Robert-Jan Sips
Artificial Intelligence Department, myTomorrows, Anthony Fokkerweg 61 1059CP, Amsterdam, The Netherlands
Keywords:
Language Detection, Sentence Embedding, Graphotactics, Linguistic Knowledge.
Abstract:
Language identification remains a challenge for short texts originating from social media. Moreover, domain-
specific terminology, which is frequent in the medical domain, may not change cross-linguistically, making
language identification even more difficult. We conducted language identification on four datasets, two of
them with general language, and two of them containing medical language. We evaluated the impact of two
embedding representations and a set of linguistic features based on graphotactics. The proposed linguistic
features reflect the graphotactics of the languages included in the test dataset. For classification, we imple-
mented two algorithms: random forest and SVM. Our findings show that, when classifying general language,
linguistic-based features perform close to the embedding representations of fastText and BERT. However,
when classifying text with technical terms, the linguistic features outperform embedding representations. The
combination of embeddings with linguistic features had a positive impact on the classification task under both
settings. Therefore, our results suggest that these linguistic features could be applied for big and small datasets
keeping the good performances in both general and medical languages. As future work, we want to test the
linguistic features for a more significant set of languages.
1 INTRODUCTION
Language identification (LI) is the task of determining
the language that a piece of text is written (Wehrmann
et al., 2018). Often, Natural Language Processing
(NLP) tools and techniques assume that the language
of texts is already known since their performance is
language-dependent (Jauhiainen et al., 2019). There-
fore, Automatic Language Identification
1
is the first
step in any NLP pipeline to ensure that the appropri-
ate language model is used.
Language identification is considered a solved
problem as stated in (Jauhiainen et al., 2019), how-
ever, the authors also highlight existing challenges.
One of them is related to short texts, as these may
not contain enough information to determine the lan-
guage. Moreover, LI is complicated when dealing
with closely related languages (Molina et al., 2016),
e.g. Portuguese-Spanish-Italian, Danish-Norwegian-
Swedish. Finally, LI can be complicated in the case of
domain-specific language, e.g. words in medicine are
1
The terms Language Identification (LI), Automatic
Language identification (ALI), and Language Detection are
used to describe the same task. In this paper, we use the
term LI, implying that it is a task is conducted by an artifi-
cially intelligent agent.
composed of roots from Latin and Greek, which have
the advantage to be precise and unchanging. These
characteristics make them hold the same meaning in
different languages (Holt et al., 1998).
Nowadays, social media platforms are widely
used to share, discuss, and seek health information
(Pershad et al., 2018). For instance, data from Twitter
may be used to detect adverse drug reactions, which
a medical practitioner may want to know, or be obli-
gated to report, with the purpose of preventing unnec-
essary risks (Manousogiannis et al., 2019). Further-
more, multilingualism, code-switching, dialects, dif-
ferences between patient and health care professional
(HCP) language, and privacy considerations accentu-
ate the need for effective methods that detect the lan-
guage in short medical texts.
In this paper, we address LI of tweets for nine Eu-
ropean languages, using four datasets covering both
the general and medical domains. We compare exist-
ing established LI methods with novel ones based on
multilingual embeddings, as well as on linguistic fea-
tures that we based on graphotactics. We apply two
different classifiers on top of these features.
Godinez, E., Szlà ˛avik, Z., Santamarà a, S. and Sips, R.
Language Identification for Short Medical Texts.
DOI: 10.5220/0008950903990406
In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2020) - Volume 5: HEALTHINF, pages 399-406
ISBN: 978-989-758-398-8; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
399

2 RELATED WORK
Since 2012, langid. py (Lui and Baldwin, 2012) has
been established as a standard baseline for language
identification. It is a multinomial Naive Bayes clas-
sifier based on an n-gram character model. More
recently, language identification has been conducted
with Recurrent Neural Networks (Wehrmann et al.,
2018). Language identification is now one of the
modules available in fastText, a commonly used tool
for text classification with word embeddings, with a
specific LI module (Joulin et al., 2016).
Recently, word embeddings have been success-
fully applied in different NLP tasks (text classifica-
tion (Joulin et al., 2016), next sentence prediction
(Devlin et al., 2018), sentiment analysis (Wehrmann
et al., 2018; Yin and Jin, 2015). Several multi-
lingual word embeddings have been proposed via
cross-lingual transfer, see (Ruder et al., 2019) for a
complete survey. Two of widely used multilingual
word embedding are fastText (Joulin et al., 2016) and
BERT (Devlin et al., 2018). Adaptations for BERT
in the medical domain have been proposed recently
in (Lee et al., 2019) and (Alsentzer et al., 2019), but
none of these are multilingual.
Previous to the word embedding models, tradi-
tional n-gram character models have been used for
language identification at the word level (Barman
et al., 2014), and n-gram character models are also
at the core of word embedding creation (Joulin et al.,
2016; Devlin et al., 2018). Concerning n-grams, it
has been stated in (Bender, 2009) that models based
on n-grams are, in fact, language-dependent, because
they perform well for languages that share similar ty-
pological properties. N-gram strategies process text
as a sequence of symbols, and it is possible to reveal
structures, because of the word distributions in var-
ious contexts. In English-like languages, the effec-
tiveness of n-gram driven models is based mostly on
two properties: relatively low inflectional morphol-
ogy and relatively fixed word order. Inversely, lan-
guages with more complex morphology present more
sparsity (higher inflected forms for lemmata), which
limits the ability of n-gram models to capture depen-
dencies between open word classes and closed word
classes. Thus, n-gram models would not work for
language identification when we confront similar lan-
guages and dialects.
In (Jauhiainen et al., 2019) a survey of language
identification, the authors reported SVM and Con-
ditional Random Fields as the most used algorithms
for LI between 2016 and 2017. In (Mandal et al.,
2018) the authors compared the performance of an
SVM model and an LSTM model for language iden-
tification in tweets that present code-switching. The
authors presented two neural models based on char-
acter and phonetic representations that combined in
stacking and thresholding techniques. However, in
order to create the phonetic representation of a text, it
is paramount to know previously the language since,
a character can be used to encode different sounds
cross-linguistically (De Saussure, 1989). For exam-
ple, the letter v in standard Spanish represents the
sound [b], in English [v], and [ f ] in and German. The
phonetic characteristics of each language seems to be
a good way to tackle language identification, because
every language has its unique set of sounds and rules
to combine them. The way that a language combines
sounds is called phonotactics. However, a sequence
of sounds may be licit for a language, while for other
languages, the same sequence may be illicit. The
phonotactics restrictions vary from language to lan-
guage (Zsiga, 2012). Since it is not possible to access
the phonetic representation of a text without knowing
the language previously, we could use the concept of
graphotactics. Like its counterpart, graphotactics im-
plies that every language has its way to restrict the
combination of the characters (Coulmas, 2003).
In this paper, we propose a set of patterns that
aim to capture the graphotactic restrictions of lan-
guages. The principal advantage of the graphotac-
tic patterns is that they are an aggregated version
of the n-gram models. Thus one single graphotac-
tic pattern can generate string patterns while n-gram
model needs to change the value of n to generate the
same patterns. Some related works attempted to use
grapheme-phoneme information for language detec-
tion at the word level. For example, in (Giwa and
Davel, 2014) the author replaced each character of
a word by a language identifier, then this informa-
tion was used to predict the language of origin of a
new word. For example, the word “#queen#” would
take this representation: E E E E E, where # denotes
the start and the end of a word and E is the language
identifier for English. Also, in (Nguyen and Cornips,
2016) the author used subword information (morphs
or morphemes-like) to detect code-switching within
words of Limburguish, a variant of Dutch. Mor-
phemes are units that convey meaning; they can be
a word, a root, a prefix, suffix, etc. (Zsiga, 2012).
To the best of our knowledge, LI research has not
yet focused specifically on medical-domain texts, and
thus we need to rely on other domains for inspiration
to find out what works for medical LI.
HEALTHINF 2020 - 13th International Conference on Health Informatics
400
3 METHODOLOGY
Our methodology consists of the following compo-
nents: i) obtaining or creating datasets for both the
general and the medical domains, ii) extracting fea-
tures for modelling, iii) training classifiers using these
features, iv) comparison of models built using combi-
nations of dataset-, feature-, and classifier choices.
3.1 Datasets
For our analysis, we used four datasets: the first two
datasets contain tweets in general language; the third
one contains tweets with medical terms, and it was au-
tomatically annotated. We created the fourth dataset
from the tweets where the automatic annotation was
uncertain. This last dataset was manually annotated
by two experts. We focused on the identification of
nine European languages: Danish, Swedish, English,
Dutch, German, Portuguese, Spanish, French, and
Italian. Table 3.1 summarizes the distribution of lan-
guages in the four datasets. As shown in the table 3.1,
the prevalence of the languages varies in each dataset.
The General Topic Dataset (GTD): The first
dataset is on general topics, and it has been built by
the Twitter team to test language identification algo-
rithms. Twitter provides three datasets for language
identification evaluation
2
: the recall dataset (RD), the
precision dataset (PD), and the uniformly sampled
dataset (UD). The datasets consist of tweet IDs with
tags to encode their corresponding language. When
collecting the complete tweets, we found that a con-
siderable number of tweets were not available any-
more. We merged the RD and PD datasets. We fil-
tered the tweets to get only those in the languages that
we targeted. We were able to retrieve 10,420 tweets
for the nine languages.
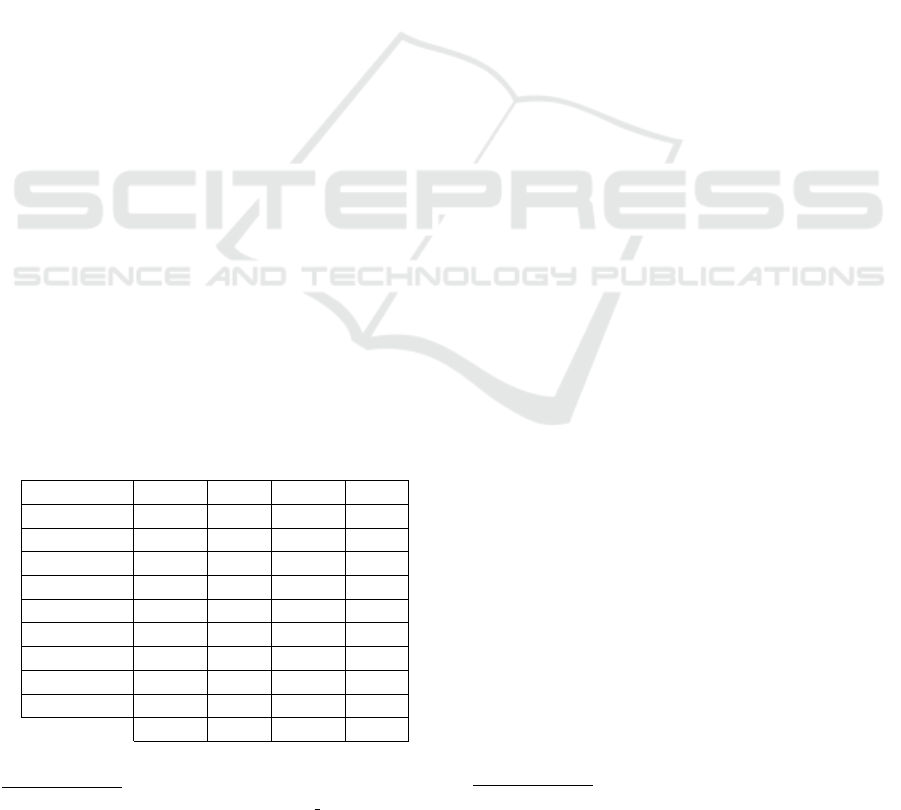
Table 1: Language distribution for the four datasets.
Language GTD TPD MTD SCD
Danish 391 0 106 0
Swedish 420 0 595 0
English 2560 1505 5058 153
Dutch 939 1430 680 0
German 1141 1479 870 0
Portuguese 1101 0 1556 13
Spanish 1727 1562 2298 32
French 1038 1551 1638 22
Italian 1103 1539 557 6
Total 10420 9066 13358 226
2
https://blog.twitter.com/engineering/en us/a/2015/
evaluating-language-identification-performance.html
The (Tromp and Pechenizkiy, 2011) Dataset (TPD):
This balanced dataset is composed of six languages:
German, Dutch, English, Spanish, French, and Ital-
ian. While creating the dataset, (Tromp and Pech-
enizkiy, 2011) removed the messages containing mul-
tiple languages or bilingual terminology. User names
and hashtags were also removed. The authors explain,
in the file of distribution, that several tweets belong
to the same user, but the tweets were anonymized.
While the decision of removing multilingual tweets
may have helped to improve language detection in
short monolingual text, it does not address problem
of language detection in multilingual tweets. This is
important, since around 20% of the tweets are multi-
lingual. Our interest in using this corpus is to test our
proposition in ideal scenario and see how they per-
form in noisier datasets. We expect to have the best
performance on this dataset compared to the rest.
The Medical Topic Dataset (MTD): To collect more
specialized language on Twitter, we implemented the
following strategy. One of our in-house medical ex-
perts gave us a list of the ten most common diseases
in colloquial and medical language. The list was pro-
vided originally in English. For this list, we extracted
their UMLS
3
Concept Unique Identifiers (CUI) and
used them to find corresponding terms in the other
nine languages using Wikidata
4
. We were able to
expand the original list of English terms to include
terms in all languages however, the distribution of
these terms is unbalanced. Thus when using this list
to retrieve tweets, the resulting dataset was unbalance.
The columns MTD and SCD in table show the distri-
bution of tweets in each language. We collected the
tweets in two periods. The first one includes tweets
from the last two weeks of August 2019, and the last
group includes tweets from the first two weeks of
September 2019. We excluded retweets to avoid the
double classification of the same tweet. Originally,
we collected 18483 tweets. After eliminating dupli-
cates and retweets, the final dataset includes 137984
tweets.
Special Cases Dataset (SCD): To label the language
of each tweet, we first used the language tag pro-
vided by Twitter as a candidate language for every
tweet. We then ran the Amazon Comprehend Lan-
guage Identifier to detect the language. If the two tags
are the same for a given tweet, we keep this as the final
tag. When there is disagreement, we selected these
tweets for manual annotation for the language. The
kappa coefficient of the two automatic annotators was
0.9648. From the original number of tweets, we got
440 tweets to be manually annotated. The annotation
3
The Unified Medical Language System
4
https://www.wikidata.org
Language Identification for Short Medical Texts
401

task consisted 1) to identify the language tweets, and
2) to report if a tweet is written with more than one
language. The final dataset contains 231 tweets, con-
taining the tweets where the annotators agreed. We
discuss more details about the annotation process in
section 4.
3.2 Feature Creation
We used two different kinds of features: fist, sen-
tence embeddings, and second, linguistic-based fea-
tures from the text. We used the embeddings and the
linguistic features as inputs for two different classi-
fiers in different configurations.
Embeddings: Embeddings have obtained good re-
sults for language identification and code-switching
(Wehrmann et al., 2018; Xia, 2016). We used two
pre-trained models: fastText (Joulin et al., 2016), and
BERT (Devlin et al., 2018). We based our selection
of tools on the ability to generate embedding for mul-
tilingual texts.
FastText generates embeddings at the word and
sentence level, on a 16 dimensional space. We de-
cided to take only the sentence embeddings, since
we are interested in detecting the language at tweet
level. Then, we used two models of BERT: BERT-
Base multilingual cased and BERT-Base multilingual
uncased. At this moment, there is only a base version
of the multilingual model and no large version, as it
occurs with monolingual versions of BERT. In order
to create the sentence embedding, we used the con-
catenation of the last four layers in BERT because its
performance has been reported to be better with this
configuration (Devlin et al., 2018).
Linguistic Features: As stated in (Bender, 2009),
n-gram models seem to be language-dependent. We
wanted to create a language-independent strategy, but
being able to capture the graphotactics of each lan-
guage. We created regular expressions that would
capture the combination of characters based on some
restrictions. These restrictions aimed to capture
groups of consonants, groups of vowels, groups of
consonants followed by a vowel, groups of vowels
followed by a consonant, etc. These restrictions can
easily reflect the graphotactics of language. While
the patterns are language-independent, they can be
applied to any text an retrieve the combination of
graphemes for each language. The complete list of
these features and their description is in table 2. We
use a TF-IDF strategy to count the incidence of the
patterns. We kept all diacritics and upper cases of the
tweets. These patterns can occur at any place of a to-
ken.
Since the regular expressions generated more
strings patterns, the number of features was high. We
used four different methods for feature selection:
• Variance-based: We fixed a threshold to remove
the features that have a low variance. The thresh-
old was fixed at 0.001.
• Univariate-based: The selection is based on uni-
variate statistical tests. Since we are dealing with
a very sparse matrix, we selected the X
2
test for
this selection, as suggested in the user manual of
the sci-kit learn library.
• Linear model-based (L1): The selection is based
on a regression analysis method: Least Absolute
Shrinkage and Selection Operator (lasso).
• Tree estimators: These methods are commonly
used to compute the importance of features.
3.3 Classifiers for Language
Identification
As a baseline, we selected langID. py (Lui and Bald-
win, 2012), and the language classifier module of
fastText (Joulin et al., 2016). Both tools have been
recently used in the context of LI for short texts
(Wehrmann et al., 2018).
We also used two different classifiers to test the
impact of the features for language identification. As
they are widely used in LI (Jauhiainen et al., 2019),
we used the following two classifiers:
• A random forest with 500 trees, and the maximum
depth was set up at 16,
• A SVM multi-class classifier with a linear kernel.
4 RESULTS
In this section, we provide an analysis of the per-
formance of the three sets of features i.e. fastText
embeddings, BERT embeddings, and linguistic fea-
tures, that we used for language identification on four
datasets i.e the GTD, the TPD, the MTD and the SCD.
In each case, we compare the performance of two
classifiers, random forest, and SVM. We decided to
use precision, recall, and F-measure instead of using
accuracy, as accuracy cannot express where the clas-
sification fails. Table 3 shows the results for the two
baselines. We use this table as a reference to compare
the performance of the each dataset with the features
that we tested.
Next, we performed language identification on the
general Twitter dataset (GTD), the Tromp and Pech-
enizkiy dataset (TPD), see (Tromp and Pechenizkiy,
HEALTHINF 2020 - 13th International Conference on Health Informatics
402
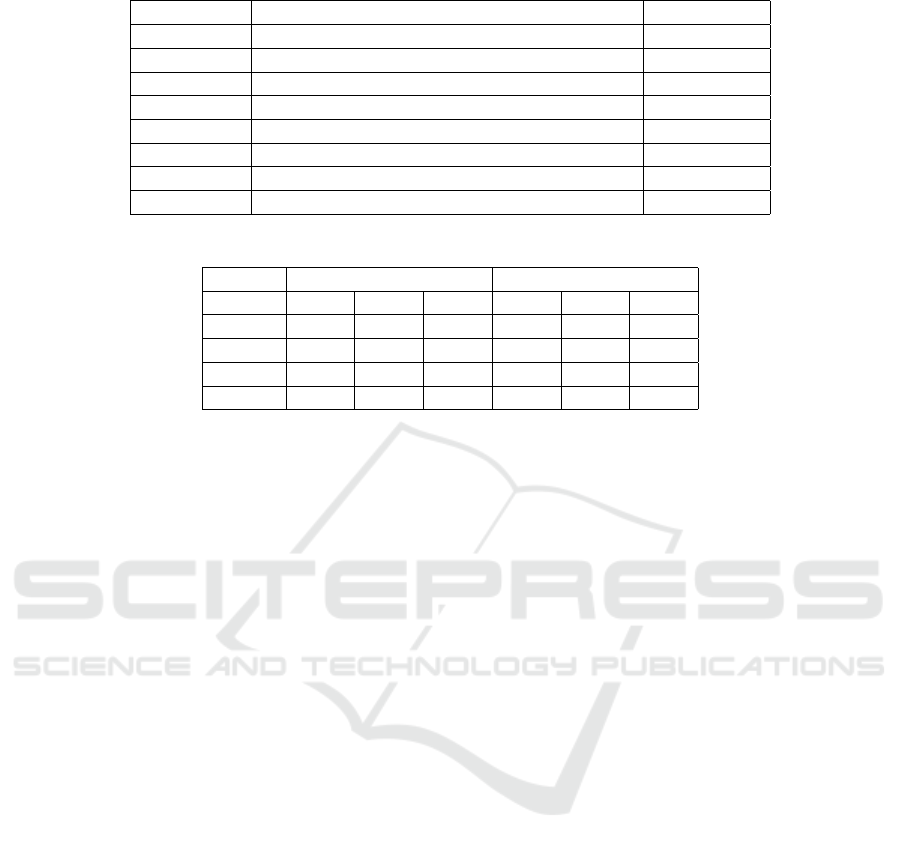
Table 2: Patterns created to capture the graphotactics of languages.
Pattern Description Example
C{2,} Consonant cluster stru- ggl -es
C{2,}V Consonant cluster followed by a vowel dive- rso -s
VC{2,} Consonant cluster preceded by a vowel g- olp -e
V{2,} Vowel cluster eeu -wen
V{2,}C Vowel cluster followed by a consonant famil- ial -es
CV{2,} Vowel cluster preceded by a consonant re-voi-r
V{2,}C{2,} Vowel cluster followed by a consonant cluster or- ient -e
C{2,}V{2,} Consonant cluster followed by a vowel cluster re- spei -to
Table 3: Performance of the baseline tools on the four datasets.
Dataset Langid.py fastText classifier
P R F1 P R F1
GTD 82.24 71.67 76.67 88.06 85.55 86.75
TPD 99.0 97.57 98.26 99.35 99.19 99.27
MTD 97.43 93.34 95.32 96.38 80.54 87.71
SCD 83.34 55.45 66.46 80.89 72.73 76.26
2011), the medical topic dataset (MTD), the special
case dataset (SCD). We evaluated the task using a
weighted average of precision, recall, and F-measure.
The weighting strategy considers the number of true
instances for each class. We also decided to only show
the results with the tree strategy selection for the lin-
guistic features since the performance with the classi-
fier was the best in all cases. We organised the results
in two tables, table 4 for the random forest model and
table 5 for the SVM model.
4.1 The General Twitter Dataset: GTD
In table 4, column GTD, we observe that the ran-
dom forest classifier achieved the highest perfor-
mance with the fastText embeddings (fT) with 84.4%
of F1. For the two other independent sets of features,
BERT and tree linguistic features (TLF), the perfor-
mance exceeded only 53% of F1. We observed that
the combination of the BERT embeddings with the
other two independent sets of features (fastText and
TLF) had a positive impact on the performance of the
classification. However, the combination of fastText
and the TLF is slightly lower. We also noticed that
none of the independent sets of features outperformed
the results of the baseline. These results indicate that
the embedding representation of fastText encodes bet-
ter than the BERT embeddings.
In table 5, column GTD, we oberserve the results
for the SVM classifier. As it occurred with the ran-
dom forest classifier as well, the TLF outperformed
the BERT embeddings; on this occasion, the incre-
ment was 7.42% in precision. We also conducted a
statistical significance test based on approximate ran-
domization (Noreen, 1989). We tested the hypothesis
to know if the performance of the classification with
the TLF was, and we found that the classification with
TLF is significantly better than with BERT embed-
dings (p=0.004). The combination of fastText em-
beddings (fT) and the TLF outperformed both base-
lines. The combination of the embeddings with the
TLF has been found to be a more significant incre-
ment of the performance that the combination of both
embeddings (fT and BERT). We also tested this hy-
pothesis and the resulting p-value is 0.001. Our re-
sults are consistent with previous works where the
SVM classifier outperformed other classifiers for lan-
guage identification (Eldesouki et al., 2016).
If we compare the performance of both algorithms
on the GTD and the TPD datasets (see columns GTD
and TPD in tables 4, and 5), we will see that the F1
measure is lower. Our explanation for this is that
the GTD contains multilingual tweets, while this is
not the case for the TPD. While identifying the lan-
guage for the baselines, both langID. py and the fast-
Text module computed the confidence of determining
the language for each tweet. This value goes from 0.0
to express no confidence and 1.0 to express full con-
fidence. We filtered the tweets in each dataset to see
where the baselines tools had a confidence lower than
0.5. For the GTD, 20.6% of the tweets have a con-
fidence below that threshold. For the TPD, the base-
lines found only 0.96% of the tweets with low confi-
dence. This reflects the fact that the TPD is composed
only by monolingual tweets. Thus, we can infer that
the GTD has tweets with code-switching making the
performance of the classifiers to go down.
Language Identification for Short Medical Texts
403
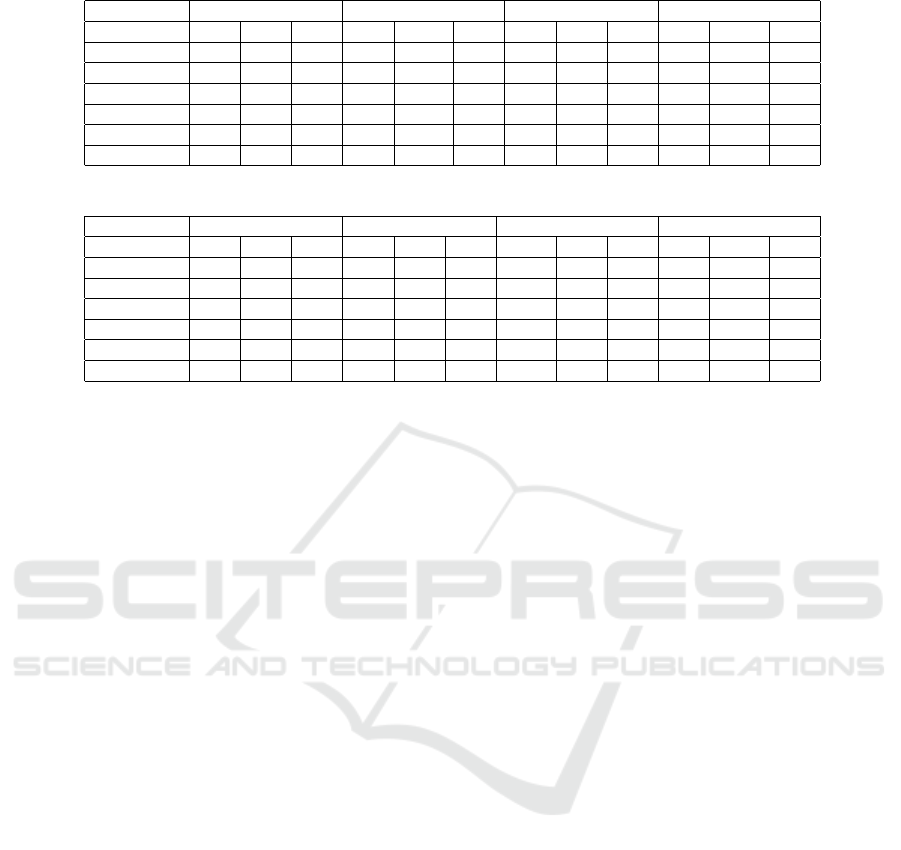
Table 4: Performance with the random forest algorithm.
GTD TPD MTD SCD
Features P R F1 P R F1 P R F1 P R F1
fT 86.2 84.3 84.4 99.6 99.66 99.6 97.9 97.8 98.7 81.9 78.9 74.7
TLF 75.3 55.4 53.3 91.6 91.0 91.1 81.9 76.6 73.7 65.5 76.9 69.4
fT+TLF 86.2 81.9 82.1 99.7 99.7 99.7 95.5 96.1 95.7 76.1 82.4 78.2
BERT 73.3 55.5 53.2 96.9 96.8 96.8 74.3 75.0 69.7 48.7 69.76 57.3
fT+BERT 86.4 81.2 81.6 97.2 99.7 99.7 95.3 95.9 95.5 61.5 74.7 65.9
BERT+TLF 85.1 76.0 76.3 99.7 99.7 99.7 93.9 94.1 93.4 60.1 74.2 64.9
Table 5: Performance with the SVM algorithm.
GTD TPD MTD SCD
Features P R F1 P R F1 P R F1 P R F1
fT 87.6 86.3 86.4 99.7 99.7 99.7 98.7 98.7 98.7 74.7 80.9 76.3
TLF 83.0 81.1 80.7 98.2 98.1 98.1 97.1 97.0 97.0 76.2 81.5 77.0
fT+TLF 90.2 89.4 89.4 99.8 99.8 99.8 99.02 99.0 99.0 78.6 83.81 79.9
BERT 75.6 75.5 75.3 98.7 98.7 98.7 97.4 97.4 97.4 69.3 75.5 71.5
fT+BERT 79.4 79.2 79.1 99.0 99.0 99.0 97.9 97.8 97.8 72.2 76.8 73.3
BERT+TLF 81.7 81.5 81.4 99.1 99.1 99.1 97.9 97.9 97.9 72.7 77.3 73.8
4.2 The Tromp and Pechenizkiy
Dataset: TPD
The good balance between the languages is reflected
in the performance of the algorithms. The two algo-
rithms can learn and classify tweets with a better per-
formance than in any of the other datasets. Our re-
sults show that no matter the features being used, the
performance of both classifiers reaches 91% in terms
of F1. Lower performances were still achieved with
the random forest classifier, table 4, while the SVM
classifier, table 5, reaches higher performances in all
cases. From our results, we can infer that having a
balanced distribution of the classes in a dataset will
help with the learning process of the algorithm.
Regarding the performance with the SVM clas-
sifier (see table 5, column TPD) the TLF features,
the fastText and BERT embeddings reached values
above 98% in terms of F1. The TLF reached a per-
formance very close to the fastText and BERT em-
beddings. However, when comparing with the base-
lines, the TFL did not pass the statistical test to show
that the classification was better with them. For the
fastText and BERT embeddings, the performance is
still statistically significant (p=0.001). We conclude
that the good performance is due to the actual com-
position of the Tromp corpus: it does not contain
tweets with multilingual or bilingual expressions, and
all links and emoticons have been removed. Texts are
clean, without noise that could have played a role in
classification. However, the corpus provides us with
an opportunity to check the robustness of sets of fea-
tures, particularly the linguistic ones.
4.3 The Medical Topic Dataset: MTD
Regarding the random forest classifier, see table 4,
column MTD, fastText embeddings (fT) achieved the
best performance. In this case, the combination of
the TLF and fastText embeddings had no positive
impact. However, the combination of the TLF and
BERT embeddings resulted in an increment of 23.7%
of the F1 value of the BERT embeddings. The perfor-
mance of TLF was still better than the BERT embed-
dings. The statistical test for the previous assumption
was confirmed by p = 0.01. In the case of the SVM
classifier, see table 5, column MTD the results were
above the baselines performance. As with the pre-
vious dataset, the performance of the TLF was very
close to those that we obtained with the word em-
beddings of fastText and BERT. We still observed
that the combination of the TLF with the embed-
dings increased the performance of the classifiers in
terms of F1 values. This observation supports our
hypothesis that the combination of linguistics-based
features with embeddings improve language identifi-
cation performance on short medical texts.
4.4 Special Cases Dataset: SCD
In this case, both classifiers outperformed the base-
lines with all features. Regarding the random for-
est classifier, see table 4, colum SCD, the best per-
formance was reached by fastText embeddings with
74.7% of F1. Concerning the combination of the
sets of features, fastText embeddings(fT) and the TLF
reached 78.2%. For this specific combination, we ob-
served a slight reduction of the F1 measure, 0.73%.
We found that the precision dropped 2.68%, and the
HEALTHINF 2020 - 13th International Conference on Health Informatics
404

recall increase by 0.51%. The performance of the
BERT embeddings is the lowest in the set of features
that we tested for both classifiers. For the SVM clas-
sifier, see table 5, column SCD, we observed that the
TLF are the ones that achieved the best performance
with 77% of the F1 measure. When combining fea-
tures, we observed again that the classification pro-
duced the best performance with fastText embeddings
(fT) and the TLF. Their performance is 79.9% of F1
measure. As we can see in table 5, column SCD the
performance of the classifiers are similar to the per-
formance on the GTD dataset, see its correspondent
column. In fact, when identifying the language with
the baseline tools, the F1-values are low for the GTD
and SCD. The GTD has 20.96% of tweets with low
confidence for language identification. In the SCD,
29.99% of tweets also have low confidence. Another
factor that can explain the low performance of the al-
gorithms and even the low confidence of the baseline
tools is that 41.55% of the tweets contain at least two
languages. Since both datasets share similar charac-
teristics in terms of the confidence of language identi-
fication, we can infer that the GTD includes a similar
number of multilingual tweets.
4.5 The Manual Annotation of the SCD
Regarding our manual annotation, for language iden-
tification, we observed a kappa coefficient of 0.6727.
In (Pustejovsky and Stubbs, 2012), the authors point
out that this value is interpreted as substantial and
highlight the fact that having more than two classes
to annotate could decrease the inter-annotator agree-
ment. For our annotation process, we asked the an-
notators to identify nine different languages. For the
composition of the final dataset, we first kept the
tweets where the two annotators agreed. We also in-
cluded the tweets where one of the annotator’s label
coincided with one of the two automatic labels. This
strategy made drop the number of tweets from 440 to
231.
The annotation was challenging because tweets
can have code-switching, i.e., a tweet is written in
more than two languages. In fact, 44.55% of the final
tweets are written in more than two languages. Thus
when it comes to identifying the principal language
by the annotator, their linguistic knowledge may have
played a role. Our annotators had a different linguistic
background. For annotator one, the linguistic prefer-
ence was Spanish, French, English, and German. For
annotator two, Dutch, English, German, French, and
Spanish. Thus, for a bilingual tweet, let us say half-
Spanish, half-English, the annotator who had Spanish
as the first language would assign Spanish as the prin-
cipal language as opposed to English, as is the case
for a different annotator. The decision to identify the
principal language of a code-switched tweet may have
been subjective.
Regarding the annotation of layman language,
medical topic, and medical language, we observed a
kappa coefficient of 0.4759. 71.86% of the tweets
were tagged as layman language, 9.52% of the tweets
were tagged as a medical topic, and 18.18% of the
tweets were tagged as medical language. 11.68% of
the tweets contain only one word, which can have an
impact on language identification.
Our results on general and medical language
datasets show that they are indeed inherently differ-
ent. Our results indicate that the same approach that
works best for the general language is not the opti-
mal one when dealing with short medical texts. To
process medical tweets, we recommend that embed-
dings, i.e., “big data” based, learned features should
be combined with expert knowledge-based features,
in our case based on linguistics. This is in line with
recent examples in which various sub-domains within
AI are being brought together to result in better, more
complete solutions for the medical domain (Van den
Bercken et al., 2019; Manousogiannis et al., 2019).
Finally, our general recommendation is to bet on hy-
brid AI methods that combine the versatility of sta-
tistical approaches with the strength of symbolic ap-
proaches.
5 CONCLUSIONS AND FUTURE
WORK
In this paper, we analyzed the impact of two types of
features and their combination for language identifi-
cation, with two classifiers and four different datasets.
We found that the combination of embeddings and
linguistic features offered a substantial gain in terms
of F1-score, compared to only one of these types. Yet,
language identification is still a challenge for short
medical texts. Since medical language relies on Latin
and Greek roots, it has different graphotactics from
layman language. This difference needs to be con-
sidered while designing NLP systems for the medical
domain.
As for future work, we plan to continue building
a dataset of tweets with medical terminology, relying
on expert knowledge for labelling. We plan to release
the first version of our dataset with the publication of
this paper. Regarding the language identification task,
we plan to move from sentence level to word-level
language identification to address the code-switching
that is present in social media data.
Language Identification for Short Medical Texts
405

REFERENCES
Alsentzer, E., Murphy, J. R., Boag, W., Weng, W.-H.,
Jin, D., Naumann, T., and McDermott, M. (2019).
Publicly available clinical bert embeddings. arXiv
preprint arXiv:1904.03323.
Barman, U., Das, A., Wagner, J., and Foster, J. (2014).
Code mixing: A challenge for language identification
in the language of social media. In Proceedings of the
first workshop on computational approaches to code
switching, pages 13–23.
Bender, E. M. (2009). Linguistically na
¨
ıve != language in-
dependent: Why NLP needs linguistic typology. In
Proceedings of the EACL 2009 Workshop on the In-
teraction between Linguistics and Computational Lin-
guistics: Virtuous, Vicious or Vacuous?, pages 26–32,
Athens, Greece. ACL.
Coulmas, F. (2003). Writing systems: An introduction to
their linguistic analysis. Cambridge University Press.
De Saussure, F. (1989). Cours de linguistique g
´
en
´
erale:
´
Edition critique, volume 1. Otto Harrassowitz Verlag.
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K.
(2018). Bert: Pre-training of deep bidirectional trans-
formers for language understanding. arXiv preprint
arXiv:1810.04805.
Eldesouki, M., Dalvi, F., Sajjad, H., and Darwish, K.
(2016). QCRI @ DSL 2016: Spoken Arabic dialect
identification using textual features. In Proceedings
of the Third Workshop on NLP for Similar Languages,
Varieties and Dialects (VarDial3), pages 221–226,
Osaka, Japan. The COLING 2016 Organizing Com-
mittee.
Giwa, O. and Davel, M. H. (2014). Language identification
of individual words with joint sequence models. In
Interspeech 2014.
Holt, R. J., Stanaszek, M. J., and Stanaszek, W. F. (1998).
Understanding Medical Terms: A Guide for Phar-
macy Practice. CRC Press.
Jauhiainen, T. S., Lui, M., Zampieri, M., Baldwin, T., and
Lind
´
en, K. (2019). Automatic language identification
in texts: A survey. Journal of Artificial Intelligence
Research, 65:675–782.
Joulin, A., Grave, E., Bojanowski, P., Douze, M., J
´
egou,
H., and Mikolov, T. (2016). Fasttext. zip: Com-
pressing text classification models. arXiv preprint
arXiv:1612.03651.
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H.,
and Kang, J. (2019). Biobert: pre-trained biomedi-
cal language representation model for biomedical text
mining. arXiv preprint arXiv:1901.08746.
Lui, M. and Baldwin, T. (2012). langid. py: An off-the-
shelf language identification tool. In Proceedings of
the ACL 2012 system demonstrations, pages 25–30.
ACL.
Mandal, S., Das, S. D., and Das, D. (2018). Language
identification of bengali-english code-mixed data us-
ing character & phonetic based lstm models. arXiv
preprint arXiv:1803.03859.
Manousogiannis, E., Mesbah, S., Bozzon, A., Baez, S., and
Sips, R. J. (2019). Give it a shot: Few-shot learning
to normalize adr mentions in social media posts. In
Proceedings of the Fourth Social Media Mining for
Health Applications (# SMM4H), pages 114–116.
Molina, G., AlGhamdi, F., Ghoneim, M., Hawwari, A.,
Rey-Villamizar, N., Diab, M., and Solorio, T. (2016).
Overview for the second shared task on language iden-
tification in code-switched data. In Proceedings of
the Second Workshop on Computational Approaches
to Code Switching, pages 40–49. ACL.
Nguyen, D. and Cornips, L. (2016). Automatic detec-
tion of intra-word code-switching. In Proceedings of
the 14th SIGMORPHON Workshop on Computational
Research in Phonetics, Phonology, and Morphology,
pages 82–86.
Noreen, E. W. (1989). Computer-intensive methods for test-
ing hypotheses. Wiley New York.
Pershad, Y., Hangge, P. T., Albadawi, H., and Oklu, R.
(2018). Social medicine: Twitter in healthcare. Jour-
nal of clinical medicine, 7(6):121.
Pustejovsky, J. and Stubbs, A. (2012). Natural Language
Annotation for Machine Learning: A guide to corpus-
building for applications. ” O
´
Reilly Media, Inc.”.
Ruder, S., Vuli
´
c, I., and Søgaard, A. (2019). A survey of
cross-lingual word embedding models. Journal of Ar-
tificial Intelligence Research, 65:569–631.
Tromp, E. and Pechenizkiy, M. (2011). Graph-based n-
gram language identification on short texts. In Proc.
20th Machine Learning conference of Belgium and
The Netherlands, pages 27–34.
Van den Bercken, L., Sips, R.-J., and Lofi, C. (2019). Eval-
uating neural text simplification in the medical do-
main. In The World Wide Web Conference, pages
3286–3292. ACM.
Wehrmann, J., Becker, W. E., and Barros, R. C. (2018).
A multi-task neural network for multilingual senti-
ment classification and language detection on twitter.
In Proceedings of the 33rd Annual ACM Symposium
on Applied Computing - SAC 18, pages 1805–1812.
ACM Press.
Xia, M. X. (2016). Codeswitching language identification
using subword information enriched word vectors. In
Proceedings of The Second Workshop on Computa-
tional Approaches to Code Switching, pages 132–136.
Yin, Y. and Jin, Z. (2015). Document sentiment classifi-
cation based on the word embedding. In 2015 4th
International Conference on Mechatronics, Materials,
Chemistry and Computer Engineering. Atlantis Press.
Zsiga, E. C. (2012). The sounds of language: An introduc-
tion to phonetics and phonology. John Wiley & Sons.
HEALTHINF 2020 - 13th International Conference on Health Informatics
406
