
Data Mining in Clinical Trial Text: Transformers for Classification and
Question Answering Tasks
Lena Schmidt
1,2 a
, Julie Weeds
2 b
and Julian P. T. Higgins
1 c
1
University of Bristol, Bristol Medical School, 39 Whatley Road, BS82PS Bristol, U.K.
2
University of Sussex, Department of Informatics, BN19QJ Brighton, U.K.
Keywords:
BERT, Data mining, Evidence-based Medicine, PICO Element Detection, Natural Language Processing,
Question Answering, Sentence Classification, Systematic Review Automation, Transformer Neural Network.
Abstract:
This research on data extraction methods applies recent advances in natural language processing to evidence
synthesis based on medical texts. Texts of interest include abstracts of clinical trials in English and in multilin-
gual contexts. The main focus is on information characterized via the Population, Intervention, Comparator,
and Outcome (PICO) framework, but data extraction is not limited to these fields. Recent neural network
architectures based on transformers show capacities for transfer learning and increased performance on down-
stream natural language processing tasks such as universal reading comprehension, brought forward by this
architecture’s use of contextualized word embeddings and self-attention mechanisms. This paper contributes
to solving problems related to ambiguity in PICO sentence prediction tasks, as well as highlighting how an-
notations for training named entity recognition systems are used to train a high-performing, but nevertheless
flexible architecture for question answering in systematic review automation. Additionally, it demonstrates
how the problem of insufficient amounts of training annotations for PICO entity extraction is tackled by aug-
mentation. All models in this paper were created with the aim to support systematic review (semi)automation.
They achieve high F1 scores, and demonstrate the feasibility of applying transformer-based classification
methods to support data mining in the biomedical literature.
1 INTRODUCTION
Systematic reviews (SR) of randomized controlled tri-
als (RCTs) are regarded as the gold standard for pro-
viding information about the effects of interventions
to healthcare practitioners, policy makers and mem-
bers of the public. The quality of these reviews is
ensured through a strict methodology that seeks to
include all relevant information on the review topic
(Higgins et al., 2019).
A SR, as produced by the quality standards of
Cochrane, is conducted to appraise and synthesize
all research for a specific research question, there-
fore providing access to the best available medical
evidence where needed (Lasserson et al., 2019). The
research question is specified using the PICO (pop-
ulation; intervention; comparator; outcomes) frame-
work. The researchers conduct very broad literature
a
https://orcid.org/0000-0003-0709-8226
b
https://orcid.org/0000-0002-3831-4019
c
https://orcid.org/0000-0002-8323-2514
searches in order to retrieve every piece of clinical
evidence that meets their review’s inclusion criteria,
commonly all RCTs of a particular healthcare inter-
vention in a specific population. In a search, no piece
of relevant information should be missed. In other
words, the aim is to achieve a recall score of one. This
implies that the searches are broad (Lefebvre et al.,
2019), and authors are often left to screen a large
number of abstracts manually in order to identify a
small fraction of relevant publications for inclusion in
the SR (Borah et al., 2017).
The number of RCTs is increasing, and with it
increases the potential number of reviews and the
amount of workload that is implied for each. Re-
search on the basis of PubMed entries shows that both
the number of publications and the number of SRs in-
creased rapidly in the last ten years (Fontelo and Liu,
2018), which is why acceleration of the systematic
reviewing process is of interest in order to decrease
working hours of highly trained researchers and to
make the process more efficient.
In this work, we focus on the detection and an-
Schmidt, L., Weeds, J. and Higgins, J.
Data Mining in Clinical Trial Text: Transformers for Classification and Question Answering Tasks.
DOI: 10.5220/0008945700830094
In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2020) - Volume 5: HEALTHINF, pages 83-94
ISBN: 978-989-758-398-8; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
83

notation of information about the PICO elements of
RCTs described in English PubMed abstracts. In
practice, the comparators involved in the C of PICO
are just additional interventions, so we often refer
to PIO (populations; interventions; outcomes) rather
than PICO. Focus points for the investigation are the
problems of ambiguity in labelled PIO data, integra-
tion of training data from different tasks and sources
and assessing our model’s capacity for transfer learn-
ing and domain adaptation.
Recent advances in natural language processing
(NLP) offer the potential to be able to automate or
semi-automate the process of identifying information
to be included in a SR. For example, an automated
system might attempt to PICO-annotate large corpora
of abstracts, such as RCTs indexed on PubMed, or
assess the results retrieved in a literature search and
predict which abstract or full text article fits the inclu-
sion criteria of a review. Such systems need to be able
to classify and extract data of interest. We show that
transformer models perform well on complex data-
extraction tasks. Language models are moving away
from the semantic, but static representation of words
as in Word2Vec (Mikolov et al., 2013), hence provid-
ing a richer and more flexible contextualized repre-
sentation of input features within sentences or long
sequences of text.
The rest of this paper is organized as follows. The
remainder of this section introduces related work and
the contributions of our work. Section 2 describes the
process of preparing training data, and introduces ap-
proaches to fine-tuning for sentence classification and
question answering tasks. Results are presented in
section 3, and section 4 includes a critical evaluation
and implications for practice.
1.1 Tools for SR Automation and PICO
Classification
The website systematicreviewtools.com (Marshall,
2019) lists 36 software tools for study selection to
date. Some tools are intended for organisational pur-
poses and do not employ PICO classification, such
as Covidence (Covidence, 2019). The tool Rayyan
uses support vector machines (Ouzzani et al., 2016).
RobotReviewer uses neural networks, word embed-
dings and recently also a transformer for named en-
tity recognition (NER) (Marshall et al., 2017). Ques-
tion answering systems for PICO data extraction ex-
ist based on matching words from knowledge bases,
hand-crafted rules and na
¨
ıve Bayes classification,
both on entity and sentence level (Demner-Fushman
and Lin, 2005), (Niu et al., 2003), but commonly fo-
cus on providing information to practicing clinicians
rather than systematic reviewers (Vong and Then,
2015).
In the following we introduce models related to
our sentence and entity classification tasks and the
data on which our experiments are based. We made
use of previously published training and testing data
in order to ensure comparability between models.
1.2 Sentence Classification Data
In the context of systematic review (semi)automation,
sentence classification can be used in the screen-
ing process, by highlighting relevant pieces of text.
A long short-term memory (LSTM) neural network
trained with sentences of structured abstracts from
PubMed was published in 2018 (Jin and Szolovits,
2018). It uses a pre-trained Word2Vec embedding
in order to represent each input word as a fixed vec-
tor. Due to the costs associated with labelling, its
authors acquired sentence labels via automated anno-
tation. Seven classes were assigned on the basis of
structured headings within the text of each abstract.
Table 1 provides an overview of class abbreviations
and their meaning.
1
In the following we refer to it as
the PubMed data.
The LSTM itself yields impressive results with F1
scores for annotation of up to 0.85 for PIO elements,
it generalizes across domains and assigns one label
per sentence. We were able to confirm these scores
by replicating a local version of this model.
Table 1: Classes for the sentence classification task.
Class Abbreviation
P Population
I Intervention, Comparator
O Outcome
A Aim
M Method
R Result
C Conclusion
1.3 Question Answering Data
1.3.1 SQuAD
The Stanford Question Answering Dataset (SQuAD)
is a reading-comprehension dataset for machine
learning tasks. It contains question contexts, ques-
tions and answers and is available in two versions.
The older version contains only questions that can be
1
Intervention and comparator are commonly combined
in PICO annotation tasks, please see original publication for
more details (Jin and Szolovits, 2018)
HEALTHINF 2020 - 13th International Conference on Health Informatics
84

answered based on the given context. In its newer
version, the dataset also contains questions which can
not be answered on the basis of the given context.
The SQuAD creators provide an evaluation script, as
well as a public leader board to compare model per-
formances (Rajpurkar et al., 2016).
1.3.2 Ebm-nlp
In the PICO domain, the potential of NER was shown
by Nye and colleagues in using transformers, as well
as LSTM and conditional random fields. In the fol-
lowing, we refer to these data as the ebm-nlp corpus.
(Nye et al., 2018). The ebm-nlp corpus provided us
with 5000 tokenized and annotated RCT abstracts for
training, and 190 expert-annotated abstracts for test-
ing. Annotation in this corpus include PIO classes, as
well as more detailed information such as age, gen-
der or medical condition. We adapted the human-
annotated ebm-nlp corpus of abstracts for training our
QA-BERT question answering system.
1.4 Introduction to Transformers
In the following, the bidirectional encoder representa-
tions from transformers (BERT) architecture is intro-
duced (Devlin et al., 2018a). This architecture’s key
strengths are rooted in both feature representation and
training. A good feature representation is essential to
ensure any model’s performance, but often data spar-
sity in the unsupervised training of embedding mech-
anisms leads to losses in overall performance. By em-
ploying a word piece vocabulary, BERT eliminated
the problem of previously unseen words. Any word
that is not present in the initial vocabulary is split into
a sub-word vocabulary. Especially in the biomedical
domain this enables richer semantic representations
of words describing rare chemical compounds or con-
ditions. A relevant example is the phrase ’two drops
of ketorolac tromethamine’, where the initial three
words stay intact, while the last words are tokenized
to ’ket’, ’#oro’, ’#lac’, ’tro’, ’#meth’, ’#amine’, hence
enabling the following model to focus on relevant
parts of the input sequence, such as syllables that indi-
cate chemical compounds. When obtaining a numer-
ical representation for its inputs, transformers apply a
’self-attention’ mechanism, which leads to a contex-
tualized representation of each word with respect to
its surrounding words.
BERT’s weights are pre-trained in an unsuper-
vised manner, based on large corpora of unlabelled
text and two pre-training objectives. To achieve bidi-
rectionality, its first pre-training objective includes
prediction of randomly masked words. Secondly, a
next-sentence prediction task trains the model to cap-
ture long-term dependencies. Pre-training is compu-
tationally expensive but needs to be carried out only
once before sharing the weights together with the vo-
cabulary. Fine-tuning to various downstream tasks
can be carried out on the basis of comparably small
amounts of labelled data, by changing the upper lay-
ers of the neural network to classification layers for
different tasks.
SCIBERT is a model based on the BERT-base ar-
chitecture, with further pre-trained weights based on
texts from the Semantic Scholar search engine (Al-
lenAI, 2019). We used these weights as one of our
three starting points for fine-tuning a sentence clas-
sification architecture (Beltagy et al., 2019). Fur-
thermore, BERT-base (uncased) and Bert multilingual
(cased, base architecture) were included in the com-
parison (Devlin et al., 2018a).
1.5 Weaknesses in the Previous
Sentence Classification Approach
In the following, we discuss weaknesses in the
PubMed data, and LSTM models trained on this type
of labelled data. LSTM architectures commonly em-
ploy a trimmed version of Word2Vec embeddings as
embedding layer. In our case, this leads to 20% of the
input data being represented by generic ‘Unknown’
tokens. These words are missing because they oc-
cur so rarely that no embedding vector was trained
for them. Trimming means that the available embed-
ding vocabulary is then further reduced to the known
words of the training, development and testing data,
in order to save memory and increase speed. The per-
centage of unknown tokens is likely to increase when
predicting on previously unseen and unlabelled data.
We tested our locally trained LSTM on 5000 abstracts
from a study-based register (Shokraneh and Adams,
2019) and found that 36% of all unique input features
did not have a known representation.
In the case of the labelled training and testing data
itself, automatic annotation carries the risk of produc-
ing wrongly labelled data. But it also enables the
training of neural networks in the first place because
manual gold standard annotations for a project on the
scale of a LSTM are expensive and time-consuming
to produce. As we show later, the automated annota-
tion technique causes noise in the evaluation because
as the network learns, it can assign correct tags to
wrongly labelled data. We also show that sentence
labels are often ambiguous, and that the assignment
of a single label limits the quality of the predictions
for their use in real-world reviewing tasks.
We acknowledge that the assignment of classes
Data Mining in Clinical Trial Text: Transformers for Classification and Question Answering Tasks
85

such as ‘Results’ or ‘Conclusions’ to sentences is po-
tentially valuable for many use-cases. However, those
sentences can contain additional information related
to the PICO classes of interest. In the original LSTM-
based model the A, M, R, and C data classes in Table 1
are utilized for sequence optimization, which leads to
increased classification scores. Their potential PICO
content is neglected, although it represents crucial in-
formation in real-world reviewing tasks.
A general weakness of predicting labels for whole
sentences is the practical usability of the predictions.
We will show sentence highlighting as a potential
use-case for focusing reader’s attention to passages
of interest. However, the data obtained through this
method are not fine-grained enough for usage in data
extraction, or for the use in pipelines for automated
evidence synthesis. Therefore, we expand our exper-
iments to include QA-BERT, a question-answering
model that predicts the locations of PICO entities
within sentences.
1.6 Contributions of this Research
In this work we investigate state-of-the-art meth-
ods for language modelling and sentence classifica-
tion. Our contributions are centred around devel-
oping transformer-based fine-tuning approaches tai-
lored to SR tasks. We compare our sentence clas-
sification with the LSTM baseline and evaluate the
biggest set of PICO sentence data available at this
point (Jin and Szolovits, 2018). We demonstrate that
models based on the BERT architecture solve prob-
lems related to ambiguous sentence labels by learning
to predict multiple labels reliably. Further, we show
that the improved feature representation and contex-
tualization of embeddings lead to improved perfor-
mance in biomedical data extraction tasks. These
fine-tuned models show promising results while pro-
viding a level of flexibility to suit reviewing tasks,
such as the screening of studies for inclusion in re-
views. By predicting on multilingual and full text
contexts we showed that the model’s capabilities for
transfer learning can be useful when dealing with di-
verse, real-world data.
In the second fine-tuning approach, we apply a
question answering architecture to the task of data ex-
traction. Previous models for PICO question answer-
ing relied on vast knowledge bases and hand-crafted
rules. Our fine-tuning approach shows that an abstract
as context, together with a combination of annotated
PICO entities and SQuAD data can result in a system
that outperforms contemporary entity recognition sys-
tems, while retaining general reading comprehension
capabilities.
2 METHODOLOGY
2.1 Feature Representation and
Advantages of Contextualization
A language processing model’s performance is lim-
ited by its capability of representing linguistic con-
cepts numerically. In this preliminary experiment, we
used the PubMed corpus for sentence classification to
show the quality of PICO sentence embeddings re-
trieved from BERT. We mapped a random selection
of 3000 population, intervention, and outcome sen-
tences from the PubMed corpus to BERT-base un-
cased and SCIBERT. This resulted in each sentence
being represented by a fixed length vector of 768 di-
mensions in each layer respectively, as defined by
the model architecture’s hidden size. These vectors
can be obtained for each of the network’s layers, and
multiple layers can be represented together by con-
catenation and pooling.
2
We used the t-distributed
Stochastic Neighbour Embedding (t-SNE) algorithm
to reduce each layer-embedding into two-dimensional
space, and plotted the resulting values. Additionally,
we computed adjusted rand scores in order to eval-
uate how well each layer (or concatenation thereof,
always using reduce mean pooling) represents our in-
put sequence. The rand scores quantify the extent to
which a na
¨
ıve K-means (N=3) clustering algorithm in
different layers alone led to correct grouping of the
input sentences.
2.2 Sentence Classification
2.2.1 Preparation of the Data
We used the PubMed corpus to fine-tune a sen-
tence classification architecture. Class names and
abbreviations are displayed in Table 1. The cor-
pus was supplied in pre-processed form, compris-
ing 24,668 abstracts. For more information about
the original dataset we refer to its original publi-
cation (Jin and Szolovits, 2018). Because of the
PICO framework, methods for systematic review
semi(automation) commonly focus on P, I, and O de-
tection. A, M, R, and C classes are an additional fea-
ture of this corpus. They were included in the follow-
ing experiment because they represent important in-
formation in abstracts and they occur in a vast major-
ity of published trial text. Their exclusion can lead to
false classification of sentences in full abstracts. In a
2
A more detailed explanation is given by the BERT au-
thors in their paper on pre-training (Devlin et al., 2018b),
and in the Bert-as-service GitHub repository (Xiao, 2018).
HEALTHINF 2020 - 13th International Conference on Health Informatics
86

preliminary experiment we summarized A, M, R, and
C sentences as a generic class named ’Other’ in order
to shift the model’s focus to PIO classes. This resulted
in high class imbalance, inferior classification scores
and a loss of ability to predict these classes when
supporting systematic reviewers during the screening
process.
In the following, abstracts that did not include a
P, I, and O label were excluded. This left a total of
129,095 sentences for training, and 14,344 for testing
(90:10 split).
2.2.2 Fine-tuning
We carried out fine-tuning for sentence classification
based on BERT-base (uncased), multilingual BERT
(cased), and on SCIBERT. We changed the classifi-
cation layer on top of the original BERT model. It
remains as linear, fully connected layer but now em-
ploys the sigmoid cross-entropy loss with logits func-
tion for optimization. During training, this layer is
optimised for predicting probabilities over all seven
possible sentence labels. Therefore, this architecture
enables multi-class, multi-label predictions. In com-
parison, the original BERT fine-tuning approach for
sentence classification employed a softmax layer in
order to obtain multi-class, single-label predictions of
the most probable class only. During the training pro-
cess the model then predicts class labels from Table 1
for each sentence. After each training step, backprop-
agation then adjusts the model’s internal weights. To
save GPU resources, a maximal sequence length of
64, batch size 32, learning rate of 2 × 10
−5
, a warm-
up proportion of 0.1 and two epochs for training were
used.
2.2.3 Post-training Assignment of Classes
In the scope of the experiments for this paper, the
model returns probabilities for the assignment of each
class for every sentence. These probabilities were
used to show effects of different probability thresh-
olds (or simply assignment to the most probable class)
on recall, precision and F1 scores. The number of
classes was set to 7, thereby making use of the full
PubMed dataset.
2.3 Question Answering
2.3.1 Preparation of the Data
Both the training and testing subsets from the ebm-
nlp data were adapted to fit the SQuAD format. We
merged both datasets in order to train a model which
firstly correctly answers PICO questions on the basis
of being trained with labelled ebm-nlp data, and sec-
ondly retains the flexibility of general-purpose ques-
tion answering on the basis of SQuAD. We created
sets of general, differently phrased P, I, and O ques-
tions for the purpose of training a broad representa-
tion of each PICO element question.
Figure 1: Colour coded example for a population entity an-
notation, converted to SQuAD v.2 format. Combined data
are used to train and evaluate the system.
In this section we describe the process of adapting
the ebm-nlp data to the second version of the SQuAD
format, and then augmenting the training data with
some of the original SQuAD data. Figure 1 shows an
example of the converted data, together with a high-
level software architecture description for our QA-
BERT model. We created a conversion script to au-
tomate this task. To reduce context length, it first
split each ebm-nlp abstract into sentences. For each
P, I, and O class it checked the presence of anno-
tated entity spans in the ebm-nlp source files. Then, a
question was randomly drawn from our set of general
questions for this class, to complete a context and a
span-answer pair in forming a new SQuAD-like ques-
tion element. In cases where a sentence did not con-
tain a span, a question was still chosen, but the answer
was marked as impossible, with the plausible answer
span set to begin at character 0. In the absence of
impossible answers, the model would always return
some part of the context as answer, and hence be of
no use for rarer entities such as P, which only occurs
in only 30% of all context sentences.
For the training data, each context can contain
one possible answer, whereas for testing multiple
question-answer pairs are permitted. An abstract is
represented as a domain, subsuming its sentences
and question answer-text pairs. In this format, our
adapted data are compatible with the original SQuAD
v.2 dataset, so we chose varying numbers of origi-
nal SQuAD items and shuffled them into the training
data. This augmentation of the training data aims to
Data Mining in Clinical Trial Text: Transformers for Classification and Question Answering Tasks
87
reduce the dependency on large labelled corpora for
PICO entity extraction. Testing data can optionally
be enriched in the same way, but for the presentation
of our results we aimed to be comparable with previ-
ously published models and therefore chose to eval-
uate only on the subset of expert-annotated ebm-nlp
testing data.
2.3.2 Fine-tuning
The python Huggingface Transformers library was
used for fine-tuning the question-answering mod-
els. This classification works by adding a span-
classification head on top of a pre-trained transformer
model. The span-classification mechanism learns to
predict the most probable start and end positions of
potential answers within a given context (Wolf et al.,
2019).
The Transformers library offers classes for to-
kenizers, BERT and other transformer models and
provides methods for feature representation and op-
timization. We used BertForQuestionAnswering.
Training was carried out on Google’s Colab, using the
GPU runtime option. We used a batch size of 18 per
GPU and a learning rate of 3
−5
. Training lasted for
2 epochs, context length was limited to 150. To re-
duce the time needed to train, we only used BERT-
base (uncased) weights as starting points, and used a
maximum of 200 out of the 442 SQuAD domains.
To date, the Transformers library includes several
BERT, XLM, XLNet, DistilBERT and ALBERT
question answering models that can be fine-tuned
with the scripts and data that we describe in this paper.
3 RESULTS
3.1 Feature Representation and
Contextualization
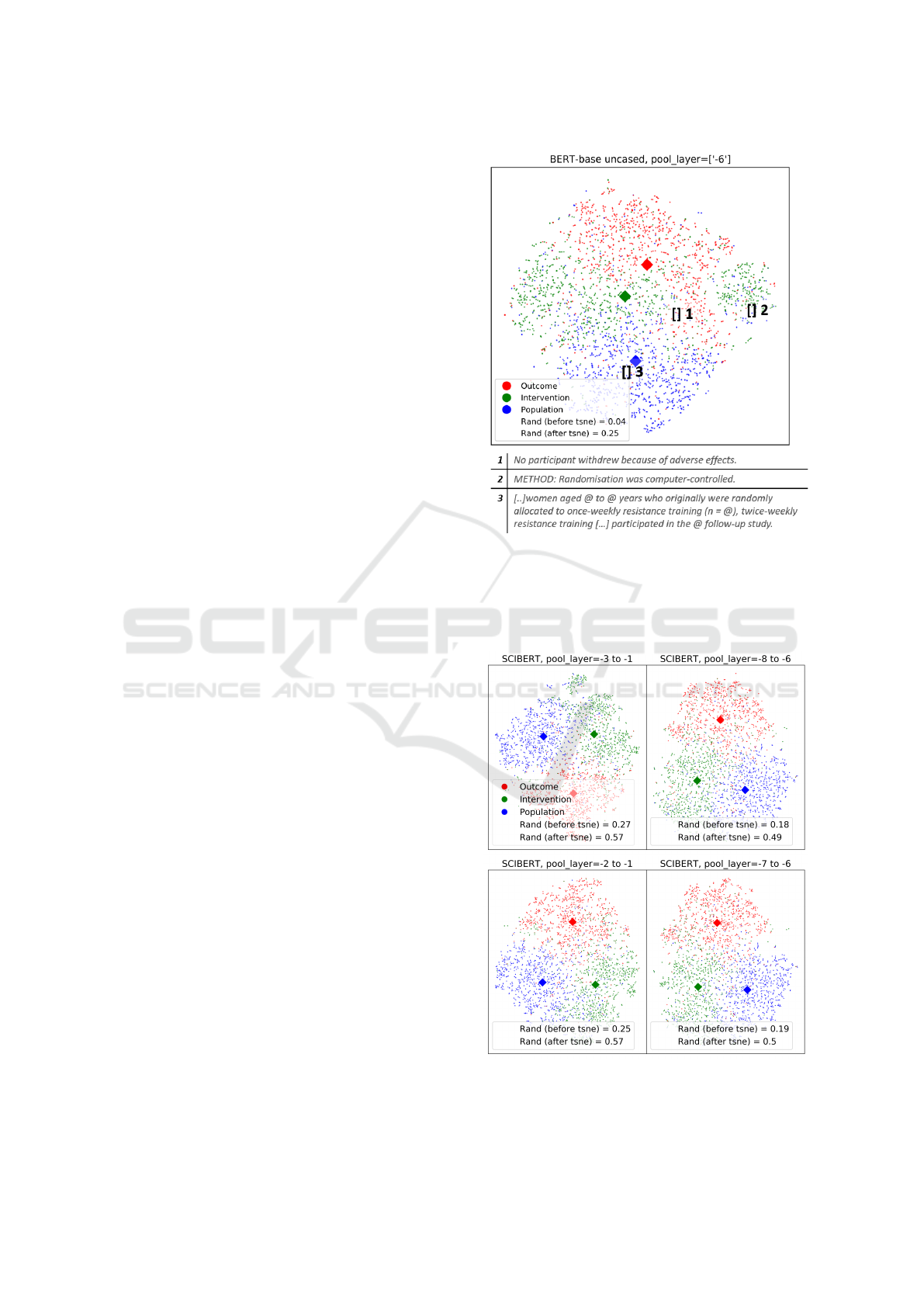
Figure 2 shows the dimensionality-reduced vectors
for 3000 sentences in BERT-base, along with the po-
sitions of three exemplary sentences. All three exam-
ples were labelled as ’P’ in the gold standard. This vi-
sualization highlights overlaps between the sentence
data and ambiguity or noise in the labels.
Sentences 1 and 2 are labelled incorrectly, and
clearly appear far away from the population class cen-
troid. Sentence 3 is an example of an ambiguous case.
It appears very close to the population centroid, but
neither its label nor its position reflect the interven-
tion content. This supports a need for multiple tags
per sentence, and the fine-tuning of weights within
the network.
Figure 2: Visualization of training sentences using BERT-
base. The x and y-axis represent the two most dominant
dimensions in the hidden state output, as selected by the
t-SNE algorithm. This visualization uses the sixth layer
from the top, and shows three examples of labelled P sen-
tences and their embedded positions.
Figure 3: Visualisation of training sentences using SCIB-
ERT. The x and y-axes represent the two most dominant
t-SNE reduced dimensions for each concatenation of lay-
ers.
HEALTHINF 2020 - 13th International Conference on Health Informatics
88

Table 2: Summary of results for the sentence classification task.
Tag Model Precision, Recall, F1 Model Prec., Recall, F1 Model Prec., Recall, F1
Single-label case
P LSTM 0.89, 0.83, 0.86 BERT-base 0.92, 0.87, 0.89 SCIBERT 0.92, 0.88, 0.90
I 0.75, 0.82, 0.78 0.89, 0.88, 0.89 0.90, 0.88, 0.89
O 0.84, 0.83, 0.84 0.88, 0.93, 0.90 0.89, 0.94, 0.92
A 0.98, 0.98, 098 0.93, 0.94, 0.93 0.94, 0.95, 0.95
M 0.87, 0.84, 0.86 0.95, 0.93, 0.94 0.96, 0.93, 0.95
R 0.93, 0.96, 0.95 0.90, 0.94, 0.92 0.91, 0.94, 0.92
C 0.94, 0.91, 0.92 0.90, 0.83, 0.86 0.89, 0.83, 0.86
Multi-label case
P Multilingual 0.87, 0.90, 0.88 SCIBERT 0.81, 0.93, 0.87 SCIBERT 0.97, 0.78, 0.87
I Thresh.: 0.3 0.85, 0.90, 0.88 0.2 0.83, 0.92, 0.87 0.8 0.95, 0.75, 0.84
O 0.87, 0.93, 0.90 0.84, 0.95, 0.89 0.95, 0.83, 0.89
A 0.91, 0.94, 0.93 0.88, 0.96, 0.92 0.98, 0.91, 0.94
M 0.95, 0.94, 0.94 0.91, 0.95, 0.93 0.99, 0.90, 0.94
R 0.88, 0.96, 0.92 0.86, 0.97, 0.91 0.94, 0.85, 0.89
C 0.84, 0.87, 0.86 0.79, 0.90, 0.84 0.95, 0.70, 0.81
Effect of threshold on metrics
Figure 3 shows the same set of sentences, rep-
resented by concatenations of SCIBERT outputs.
SCIBERT was chosen as an additional baseline model
for fine-tuning because it provided the best represen-
tation of embedded PICO sentences. When clustered,
its embeddings yielded an adjusted rand score of 0.57
for a concatenation of the two layers, compared with
0.25 for BERT-base.
3.2 Sentence Classification
Precision, recall, and F1 scores, including a compari-
son with the LSTM, are summarized in Table 2. Un-
derlined scores represent the top score across all mod-
els, and scores in bold are the best results for single-
and multi-label cases respectively. The LSTM assigns
one label only and was outperformed in all classes of
main interest (P, I, and O).
A potential pitfall of turning this task into multi-
label classification is an increase of false-positive pre-
dictions, as more labels are assigned than given in the
single-labelled testing data in the first place. How-
ever, the fine-tuned BERT models achieved high F1
scores, and large improvements in terms of recall and
precision. In its last row, Table 2 shows different
probability thresholds for class assignment when us-
ing the PubMed dataset and our fine-tuned SCIBERT
model for multi-label prediction. After obtaining the
model’s predictions, a simple threshold parameter can
be used to obtain the final class labels. On our la-
belled testing data, we tested 50 evenly spaced thresh-
olds between 0 and 1 in order to obtain these graphs.
Here, recall and precision scores in ranges between
0.92 and 0.97 are possible with F1 scores not drop-
ping below 0.84 for the main classes of interest. In
practice, the detachment between model predictions
and assignment of labels means that a reviewer who
wishes to switch between high recall and high preci-
sion results can do so very quickly, without obtaining
new predictions from the model itself.
More visualizations can be found in this project’s
GitHub repository
3
, including true class labels and a
detailed breakdown of true and false predictions for
each class. The highest proportion of false classi-
fication appears between the results and conclusion
classes.
The fine-tuned multilingual model showed
3
https://github.com/L-ENA/HealthINF2020
Data Mining in Clinical Trial Text: Transformers for Classification and Question Answering Tasks
89

Table 3: Predicting PICOs in Chinese and German. Classes were assigned based on foreign language inputs only. For
reference, translations were provided by native speakers.
Prediction Original sentences with English translations for reference
Chinese
Population ”方法:選擇2004-03/2005-03在惠州市第二人民醫院精神科精神分裂癥住院的患者60例,
簡明精神癥狀量表總分>30分,陰性癥狀量表總分>35分.”(Huang et al., 2005)
Translation: ”Methods: In the Huizhou No. 2 People’s hospital (Mar 2004 - Mar 2005), 60 patients
with psychiatric schizophrenia were selected. Total score of the Brief Psychiatric Rating Scale was
>30, and total score of the Negative Syndrome Scale was >35 in each patient.”
Intervention ”1 隨機分為2組,泰必利組與奎的平組,每組30例,患者家屬知情同意.
Translation: ”1. They were randomly divided into 2 groups, Tiapride group and Quetiapine group, with
30 cases in each group. Patients’ family was informed and their consent was obtained.”
初始劑量25 mg,早晚各1次,以后隔日增加50 mg,每日劑量范圍300~550 mg.”(Huang et al., 2005)
Translation: ”The initial dose was 25 mg, once in the morning and evening. The dose was increased by
50 mg every other day, reaching a daily dose in the range of 300 and 550 mg.”
Outcome ”3 效果評估:使用陰性癥狀量表評定用藥前后療效及陰性癥狀改善情況,使用副反應量表評價藥
物的安全性.”(Huang et al., 2005)
Translation: ”3 Evaluation: the Negative Symptom Scale was used to evaluate the efficacy of the drug
before and after treatment and the improvement of the negative symptoms. The Treatment Emergent
Symptom Scale was used to evaluate the safety of the drug.”
Method ”實驗過程為雙盲.”(Huang et al., 2005)
Translation: ”The experimental process was double-blind.”
German
Aim ”Ziel: Untersuchung der Wirksamkeit ambulanten Heilfastens auf Schmerz, Befindlichkeit und
Gelenkfunktion bei Patienten mit Arthrose.”(Schmidt et al., 2010)
Translation: ”Aim: to investigate outpatient therapeutic fasting and its effects on pain, wellbeing and
joint-function in patients with osteoarthritis.”
Method ”Patienten und Methoden: Prospektive, unkontrollierte Pilotstudie.”(Schmidt et al., 2010)
Translation: ”Patients and methods: prospective, uncontrolled pilot study”
Outcomes ”Anlauf-, Belastungs-, Ruheschmerz (VAS); Druckschmerzschwelle (DSS); [..] (Schmidt et al., 2010)
Translation: ”Pain was measured during warm-up, stress and resting (VAS); Onset of pain under
pressure (DSS);[..]”
Conclusion ”Schlussfolgerung: Heilfasten unter
¨
Arztlicher Aufsicht kann die Symptomatik bei Patienten mit
moderater Arthrose positiv beeinflussen.” (Schmidt et al., 2010)
Translation: ”Conclusions: therapeutic fasting, under supervision of doctors, can have a positive
effect on the symptoms of patients with moderate osteoarthritis.”
marginally inferior classification scores on the exclu-
sively English testing data. However, this model’s
contribution is not limited to the English language be-
cause its interior weights embed a shared vocabulary
of 100 languages, including German and Chinese
4
.
Our evaluation of the multilingual model’s capacity
for language transfer is of a qualitative nature, as
there were no labelled Chinese or German data
available. Table 3 shows examples of two abstracts,
as predicted by the model. Additionally, this table
demonstrates how a sentence prediction model can be
used to highlight text. With the current infrastructure
it is possible to highlight PICOs selectively, to
highlight all classes simultaneously, and to adjust
thresholds for class assignment in order to increase or
decrease the amount of highlighted sentences. When
applied to full texts of RCTs and cohort studies, we
4
For a full list see https://github.com/google-research/
bert/blob/master/multilingual.md
found that the model retained its ability to identify
and highlight key sentences correctly for each class.
We tested various report types, as well as recent
and old publications, but remain cautious that large
scale testing on labelled data is needed to draw solid
conclusions on these model’s abilities for transfer
learning. For further examples in the English lan-
guage, we refer to our GitHub repository.
3.3 Question Answering
We trained and evaluated a model for each P, I, and
O class. Table 4 shows our results, indicated as QA-
BERT, compared with the currently published leader
board for the ebm-nlp data (Nye et al., 2019) and re-
sults reported by the authors of SCIBERT (Beltagy
et al., 2019). For the P and I classes, our models out-
performed the results on this leader board. The index
in our model names indicates the amount of additional
SQuAD domains added to the training data. We never
HEALTHINF 2020 - 13th International Conference on Health Informatics
90

used the full SQuAD data in order to reduce time
for training but observed increased performance when
adding additional data. For classifying I entities, an
increase from 20 to 200 additional SQuAD domains
resulted in an increase of 8% for the F1 score, whereas
the increase for the O domain was less than 1%. Af-
ter training a model with 200 additional SQuAD do-
mains, we also evaluated it on the original SQuAD
development set and obtained a F1 score of 0.72 for
this general reading comprehension task.
In this evaluation, the F1 scores represent the over-
lap of labelled and predicted answer spans on token
level. We also obtained scores for the subgroups of
sentences that did not contain an answer versus the
ones that actually included PICO elements. These re-
sults are shown in Table 5.
For the P class, only 30% of all sentences included
an entity, whereas its sub-classes age, gender, condi-
tion and size averaged 10% each. In the remaining
classes, these percentages were higher. F1 scores for
correctly detecting that a sentence includes no PICO
element exceeded 0.92 in all classes. This indicates
that the addition of impossible answer elements was
successful, and that the model learned a representa-
tion of how to discriminate PICO contexts. The scores
for correctly predicting PICOs in positive scenarios
are lower. These results are presented in Table 5.
Here, two factors could influence this score in a neg-
ative way. First, labelled spans can be noisy. Train-
ing spans were annotated by crowd workers and the
authors of the original dataset noted inter-annotator
disagreement. Often, these spans include full stops,
other punctuation or different levels of detail describ-
ing a PICO. The F1 score decreases if the model pre-
dicts a PICO, but the predicted span includes marginal
differences that were not marked up by the experts
who annotated the testing set. Second, some spans
include multiple PICOs, sometimes across sentence
boundaries. Other spans mark up single PICOS in
succession. In these cases the model might find mul-
tiple PICOs in a row, and annotate them as one or vice
versa.
4 DISCUSSION
In this work, we have shown possibilities for sentence
classification and data extraction of PICO character-
istics from abstracts of RCTs.
For sentence classification, models based on trans-
formers can predict multiple labels per sentence, even
if trained on a corpus that assigns a single label only.
Additionally, these architectures show a great level of
flexibility with respect to adjusting precision and re-
Table 4: Question Answering versus entity recognition re-
sults.
Class Model F1 score
P lstm-crf
0
0.78
lstm-crf-BERT
0
0.68
QA-BERT
20
0.87
I lstm-crf-pattern
1
0.7
lstm-crf-BERT
0
0.57
QA-BERT
20
0.67
QA-BERT
200
0.75
O lstm-crf-BERT
0
0.78
QA-BERT
100
0.67
PIO SCIBERT mean 0.72
QA-BERT mean 0.75
P subclasses when annotated:
Age, Gender, Condition, Size
All lstm-crf
0
0.4
All QA-BERT
20
0.53
Performance in general SQuAD task
All QA-BERT
200
0.72
call scores. Recall is an important metric in SR tasks
and the architectures proposed in this paper enable
a post-classification trade-off setting that can be ad-
justed in the process of supporting reviewers in real-
world reviewing tasks.
However, tagging whole sentences with respect
to populations, interventions and outcomes might not
be an ideal method to advance systematic review au-
tomation. Identifying a sentence’s tag could be help-
ful for highlighting abstracts from literature searches.
This focuses the reader’s attention on sentences, but
is less helpful for automatically determining whether
a specific entity (e.g. the drug aspirin) is mentioned.
Our implementation of the question answering
task has shown that a substantial amount of PICO en-
tities can be identified in abstracts on a token level.
This is an important step towards reliable system-
atic review automation. With our provided code and
data, the QA-BERT model can be switched with more
advanced transformer architectures, including XLM,
XLNet, DistilBERT and ALBERT pre-trained mod-
els. More detailed investigations into multilingual
predictions (Clef, 2018) pre-processing and predict-
ing more than one PICO per sentence are reserved for
future work.
4.1 Limitations
Limitations in the automatically annotated PubMed
training data mostly consist of incomplete detection
or noise P, I, and O entities due to the single la-
Data Mining in Clinical Trial Text: Transformers for Classification and Question Answering Tasks
91
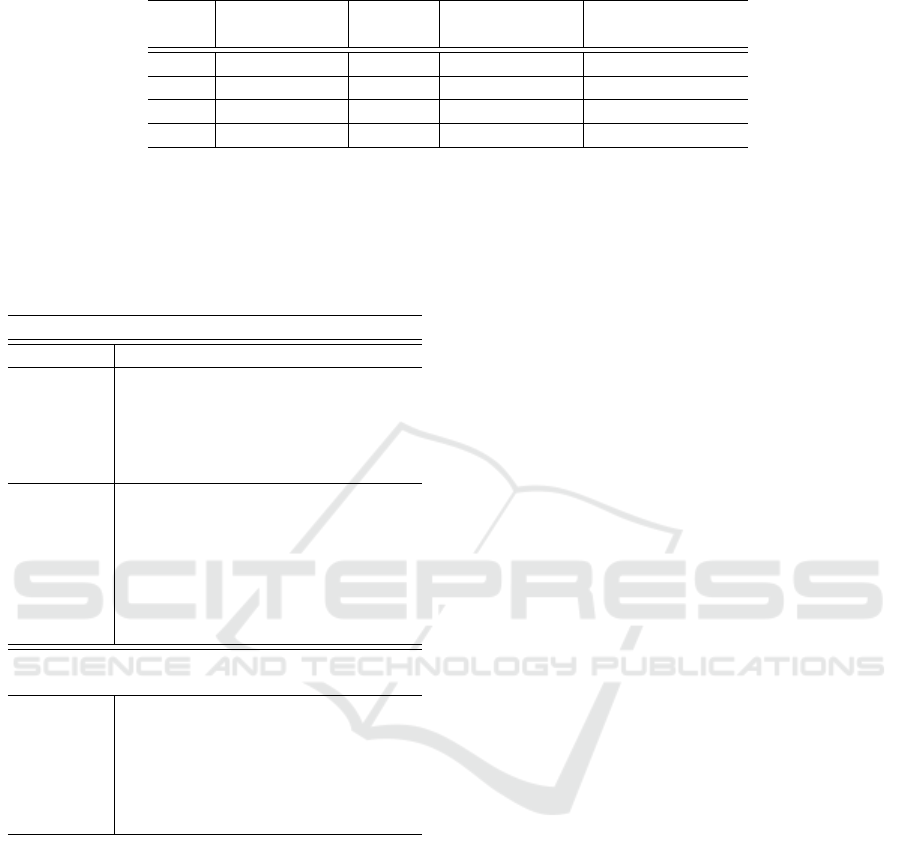
Table 5: Subgroups of possible sentences versus impossible sentences.
Class Model % F1 F1
Possible When Possible When Impossible
P QA-BERT
20
30% 0.74 0.92
I QA-BERT
200
53% 0.60 0.94
0 QA-BERT
100
60% 0.52 0.92
P
all
QA-BERT
20
10% 0.53 0.97
Table 6: This table shows two examples for intervention
span predictions in QA-BERT
200
. On the official SQuAD
development set, the same model achieved a good score, an
exemplary question and prediction for this is given in the
bottom row.
Which intervention did the participants receive?
Prediction Context
auditory
inte-
grative
training
To explore the short-term treatment
effect of the auditory integrative
training on autistic children and
provide them with clinical support
for rehabilitative treatment.
ketorolac
trometha-
mine 10
mg and
30 mg
supposi-
tories
The analgesia activity of ketorolac
tromethamine 10 mg and 30 mg
suppositories were evaluated after
single dose administration by as-
sessing pain intensity and pain re-
lief using a 4 point scale ( VRS ).
What do power station steam turbines use as a
cold sink in the absence of CHP?
surface
con-
densers
Where CHP is not used, steam tur-
bines in power stations use surface
condensers as a cold sink. The con-
densers are cooled by water flow
from oceans, rivers, lakes, and of-
ten by cooling towers [. . . ]
belling. We did not have access to multilingual an-
notated PICO corpora for testing, and therefore tested
the model on German abstracts found on PubMed,
as well as Chinese data provided by the Cochrane
Schizophrenia Group.
For the question answering, we limited the use of
original SQuAD domains to enrich our data. This was
done in order to save computing resources, as an addi-
tion of 100 SQuAD domains resulted in training time
increases of two hours, depending on various other
parameter settings. Adjusted parameters include in-
creased batch size, and decreased maximal context
length in order to reduce training time.
5 CONCLUSION
With this paper we aimed to explore state-of-the-
art NLP methods to advance systematic review
(semi)automation. Both of the presented fine-tuning
approaches for transformers demonstrated flexibility
and high performance. We contributed an approach
to deal with ambiguity in whole-sentence predictions,
and proposed the usage of a completely different ap-
proach to entity recognition in settings where training
data are sparse.
In conclusion we wish to emphasize our argument
that for future applications, interoperability is impor-
tant. Instead of developing yet another stand-alone
organizational interface with a machine learning clas-
sifier that works on limited data only, the focus should
be to develop and train cross-domain and neural mod-
els that can be integrated into the backend of existing
platforms. The performance of these models should
be comparable on standardized datasets, evaluation
scripts and leader boards.
The logical next step, which remains less explored
in the current literature because of its complexity, is
the task of predicting an RCT’s included or excluded
status on the basis of PICOs identified in its text.
For this task, more complex architectures that include
drug or intervention ontologies could be integrated.
Additionally, information from already completed re-
views could be re-used as training data.
ACKNOWLEDGEMENTS
We would like to thank Clive Adams for provid-
ing testing data and feedback for this project. We
thank Vincent Cheng for the Chinese translation. Fur-
thermore, we thank the BERT team at Google Re-
search and Allenai for making their pre-trained model
weights available. Finally, we acknowledge the Hug-
gingface team and thank them for implementing the
SQuAD classes for Transformers.
HEALTHINF 2020 - 13th International Conference on Health Informatics
92

FUNDING
LS was funded by the National Institute for Health
Research (NIHR Systematic Review Fellowship,
RM-SR-2017-09-028). The views expressed in this
publication are those of the author(s) and not neces-
sarily those of the NHS, the NIHR or the Department
of Health and Social Care.
REFERENCES
AllenAI (2019). Semantic scholar. https://www.
semanticscholar.org/. [Last accessed 16. October
2019].
Beltagy, I., Cohan, A., and Lo, K. (2019). Scibert: Pre-
trained contextualized embeddings for scientific text.
ArXiv, abs/1903.10676.
Borah, R., Brown, A. W., Capers, P. L., and Kaiser, K. A.
(2017). Analysis of the time and workers needed to
conduct systematic reviews of medical interventions
using data from the prospero registry. BMJ Open,
7(2).
Clef (2018). Clef ehealth challenge multilingual informa-
tion extraction. http://clef-ehealth.org/.
Covidence (2019). Covidence systematic review software,
Veritas Health Innovation. Melbourne, Australia.
[Available at: www.covidence.org, Last accessed 07.
October 2019].
Demner-Fushman, D. and Lin, J. (2005). Knowledge ex-
traction for clinical question answering : Preliminary
results.
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K.
(2018a). Bert: Pre-training of deep bidirectional trans-
formers for language understanding. arXiv preprint
arXiv:1810.04805.
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K.
(2018b). Bert: Pre-training of deep bidirectional
transformers for language understanding.
Fontelo, P. and Liu, F. (2018). A review of recent publica-
tion trends from top publishing countries. Systematic
Reviews, 7(1).
Higgins, J. P. T., Thomas, J., Chandler, J., Cumpston, M.,
Li, T., Page, M. J., and Welch, V. A., editors (2019).
Cochrane Handbook for Systematic Reviews of Inter-
ventions. John Wiley & Sons, Chichester, UK, 2 edi-
tion.
Huang, Q.-m., Zhang, Z.-q., Luo, Q.-h., and CHEN, H.-h.
(2005). Emotion and social function of patients with
schizophrenia and the interventional effect of tiapride:
A double-blind randomized control study. Chinese
Journal of Clinical Rehabilitation, (24):30–32.
Jin, D. and Szolovits, P. (2018). PICO element detection in
medical text via long short-term memory neural net-
works. In Proceedings of the BioNLP 2018 workshop,
pages 67–75, Melbourne, Australia. Association for
Computational Linguistics.
Lasserson, T. J., Thomas, J., and Higgins, J. P. T. (2019).
Chapter 1: Starting a review. In Higgins, J. P. T.,
Thomas, J., Chandler, J., Cumpston, M., Li, T., Page,
M. J., and Welch, V. A., editors, Cochrane Handbook
for Systematic Reviews of Interventions. John Wiley &
Sons, Chichester, UK, 2 edition.
Lefebvre, C., Glanville, J., Briscoe, S., Littlewood, A., Mar-
shall, C., Metzendorf, M. I., Noel-Storr, A., Rader, T.,
Shokraneh, F., Thomas, J., and Wieland, L. S. (2019).
Chapter 4: Searching for and selecting studies. In
Higgins, J. P. T., Thomas, J., Chandler, J., Cump-
ston, M., Li, T., Page, M. J., and Welch, V. A., ed-
itors, Cochrane Handbook for Systematic Reviews of
Interventions. John Wiley & Sons, Chichester, UK, 2
edition.
Marshall, C. (2019). The systematic review toolbox. http://
systematicreviewtools.com/index.php. [Last accessed
26. May 2019].
Marshall, I. J., Kuiper, J., Banner, E., and Wallace, B. C.
(2017). Automating biomedical evidence synthesis:
RobotReviewer. Proceedings of the Conference of
the Association for Computational Linguistics (ACL),
2017:7–12.
Mikolov, T., Chen, K., Corrado, G., and Dean, J. (2013).
Efficient estimation of word representations in vector
space.
Niu, Y., Hirst, G., Mcarthur, G., and Rodriguez-Gianolli, P.
(2003). Answering clinical questions with role iden-
tification. Proceedings of the ACL 2003 workshop on
Natural language processing in biomedicine -.
Nye, B., Li, J. J., Patel, R., Yang, Y., Marshall, I., Nenkova,
A., and Wallace, B. (2018). A corpus with multi-level
annotations of patients, interventions and outcomes to
support language processing for medical literature. In
Proceedings of the 56th Annual Meeting of the Associ-
ation for Computational Linguistics (Volume 1: Long
Papers), pages 197–207, Melbourne, Australia. Asso-
ciation for Computational Linguistics.
Nye, B., Li, J. J., Patel, R., Yang, Y., Marshall, I., Nenkova,
A., and Wallace, B. (2019). Ebm-nlp leaderboard.
https://ebm-nlp.herokuapp.com/. [Last accessed 16.
October 2019].
Ouzzani, M., Hammady, H., Fedorowicz, Z., and Elma-
garmid, A. (2016). Rayyan—a web and mobile app
for systematic reviews. Systematic Reviews, 5(1).
Rajpurkar, P., Zhang, J., Lopyrev, K., and Liang, P. (2016).
Squad: 100,000+ questions for machine comprehen-
sion of text.
Schmidt, L., Shokraneh, F., Steinhausen, K., and Adams,
C. E. (2019). Introducing raptor: Revman parsing tool
for reviewers. Systematic Reviews, 8(1).
Schmidt, S., Stange, R., Lischka, E., Kiehntopf, M., Deufel,
T., Loth, D., and Uhlemann, C. (2010). Unkon-
trollierte klinische studie zur wirksamkeit ambulanten
heilfastens bei patienten mit arthrose. Forschende
Komplement
¨
armedizin / Research in Complementary
Medicine, 17(2):87–94.
Shokraneh, F. and Adams, C. E. (2019). Study-based regis-
ters reduce waste in systematic reviewing: discussion
and case report. Systematic Reviews, 8(1):129.
Data Mining in Clinical Trial Text: Transformers for Classification and Question Answering Tasks
93

Vong, W. and Then, P. H. H. (2015). Information seeking
features of a pico-based medical question-answering
system. In 2015 9th International Conference on IT
in Asia (CITA), pages 1–7.
Wolf, T., Debut, L., Sanh, V., Chaumond, J., Delangue, C.,
Moi, A., Cistac, P., Rault, T., Louf, R., Funtowicz,
M., and Brew, J. (2019). Huggingface’s transformers:
State-of-the-art natural language processing. ArXiv,
abs/1910.03771.
Xiao, H. (2018). bert-as-service. https://github.com/
hanxiao/bert-as-service.
APPENDIX
Availability of the Code and Data
Scripts and supplementary material, as well as fur-
ther illustrations are available from https://github.
com/L-ENA/HealthINF2020. Training data for sen-
tence classification and question answering are freely
available from the cited sources.
Additionally, the Cochrane Schizophrenia Group
extracted, annotated and made available data from
studies included in over 200 systematic reviews. This
aims at supporting the development of methods for
reviewing tasks, and to increase the re-use of their
data. These data include risk-of-bias assessment, re-
sults including all clean and published outcome data
extracted by reviewers, data on PICOs, methods, and
identifiers such as PubMed ID and a link to their
study-based register. Additionally, a senior reviewer
recently carried out a manual analysis of all 33,000
outcome names in these reviews, parsed and allocated
to 15,000 unique outcomes in eight main categories
(Schmidt et al., 2019).
HEALTHINF 2020 - 13th International Conference on Health Informatics
94
