
A Deep Transfer Learning Framework for Pneumonia Detection from
Chest X-ray Images
Kh Tohidul Islam
a
, Sudanthi Wijewickrema
b
, Aaron Collins
c
and Stephen O’Leary
d
Department of Surgery (Otolaryngology), Faculty of Medicine, Dentistry and Health Sciences, University of Melbourne,
Melbourne, Victoria 3010, Australia
Keywords:
Pneumonia Detection using X-ray Images, Deep Learning, Transfer Learning, Feature Extraction, Artificial
Neural Networks.
Abstract:
Pneumonia occurs when the lungs are infected by a bacterial, viral, or fungal infection. Globally, it is the
largest solo infectious disease causing child mortality. Early diagnosis and treatment of this disease are critical
to avoid death, especially in infants. Traditionally, pneumonia diagnosis was performed by expert radiologists
and/or doctors by analysing X-ray images of the chest. Automated diagnostic methods have been developed
in recent years as an alternative to expert diagnosis. Deep learning-based image processing has been shown to
be effective in automated diagnosis of pneumonia. However, deep leaning typically requires a large number
of labelled samples for training, which is time consuming and expensive to obtain in medical applications as
it requires the input of human experts. Transfer learning, where a model pretrained for a task on an existing
labelled database is adapted to be reused for a different but related task, is a common workaround to this issue.
Here, we explore the use of deep transfer learning to diagnose pneumonia using X-ray images of the chest. We
demonstrate that using two individual pretrained models as feature extractors and training an artificial neural
network on these features is an effective way to diagnose pneumonia. We also show through experiments that
the proposed method outperforms similar existing methods with respect to accuracy and time.
1 INTRODUCTION
Pneumonia is a serious lung infection, caused by
viruses, bacteria, or fungi (Banu, 2019). Nearly half a
billion people are affected by pneumonia globally per
year, resulting in approximately four million deaths
(Lodha et al., 2013). However, it is a treatable dis-
ease, if diagnosed and treated early. According to
the World Health Organization, chest X-ray imaging
is currently the best available approach for pneumo-
nia diagnosis (Organization, 2001; Chen et al., 2019).
Chest X-rays are typically examined by trained medi-
cal practitioners (Wang and Xia, 2018). This not only
requires expert knowledge but is also time intensive
and expensive (Siddiqi, 2019). Moreover, due to the
complex nature of chest X-ray images, it remains a
challenging task for an expert to interpret the images.
Automated frameworks for pneumonia detection us-
ing chest X-rays have been introduced as effective al-
a
https://orcid.org/0000-0003-2172-7041
b
https://orcid.org/0000-0001-8015-8577
c
https://orcid.org/0000-0002-5943-8467
d
https://orcid.org/0000-0001-6926-2103
ternatives to expert-based diagnosis.
Machine learning, which has been successfully
applied in many fields of medical image processing
(Qin et al., 2018), is a viable solution for pneumonia
diagnosis using chest X-rays. Traditionally, machine
learning methods required the generation of hand-
crafted image features as their input. In contrast, deep
learning techniques can be taught to learn the ideal
features for a given task as part of the training pro-
cess. In recent years, deep learning frameworks have
achieved remarkable success in numerous image pro-
cessing applications (Razzak et al., 2017; Fourcade
and Khonsari, 2019). Most of these models were orig-
inally trained and tested on a well-known large scale
natural image database called ImageNet (Deng et al.,
2009). This database contains millions of labeled im-
ages from thousands of categories, and offers a reli-
able opportunity for researchers to evaluate the per-
formance of their deep learning models.
However, despite the high accuracy levels
achieved by deep learning models, a large depository
of labelled images is required to train them. Obtaining
labelled medical images is expensive and time con-
286
Islam, K., Wijewickrema, S., Collins, A. and O’Leary, S.
A Deep Transfer Learning Framework for Pneumonia Detection from Chest X-ray Images.
DOI: 10.5220/0008927002860293
In Proceedings of the 15th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2020) - Volume 5: VISAPP, pages
286-293
ISBN: 978-989-758-402-2; ISSN: 2184-4321
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
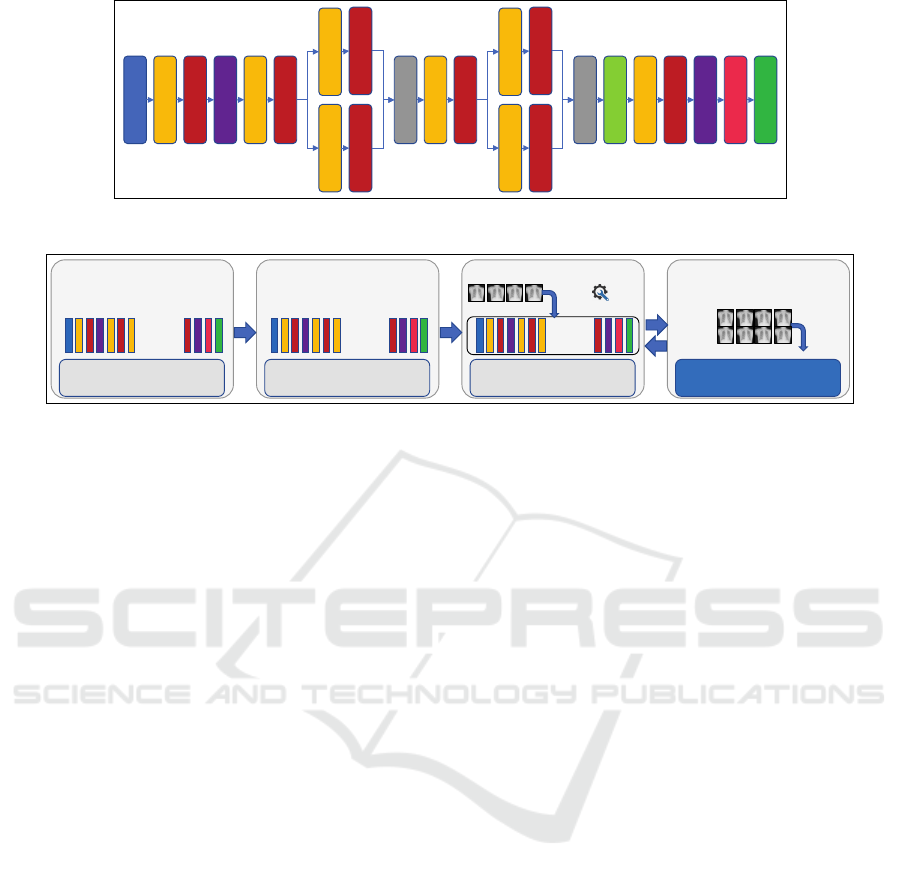
Input
Convolution
Activation
Pooling
Convolution
Activation
Convolution
Activation
Convolution
Activation
Combination
Convolution
Activation
Convolution
Activation
Convolution
Activation
Combination
Dropout
Convolution
Activation
Pooling
Softmax
Classification
Figure 1: An example of a pretrained model used for image classification (adopted from Iandola et al. (Iandola et al., 2016)).
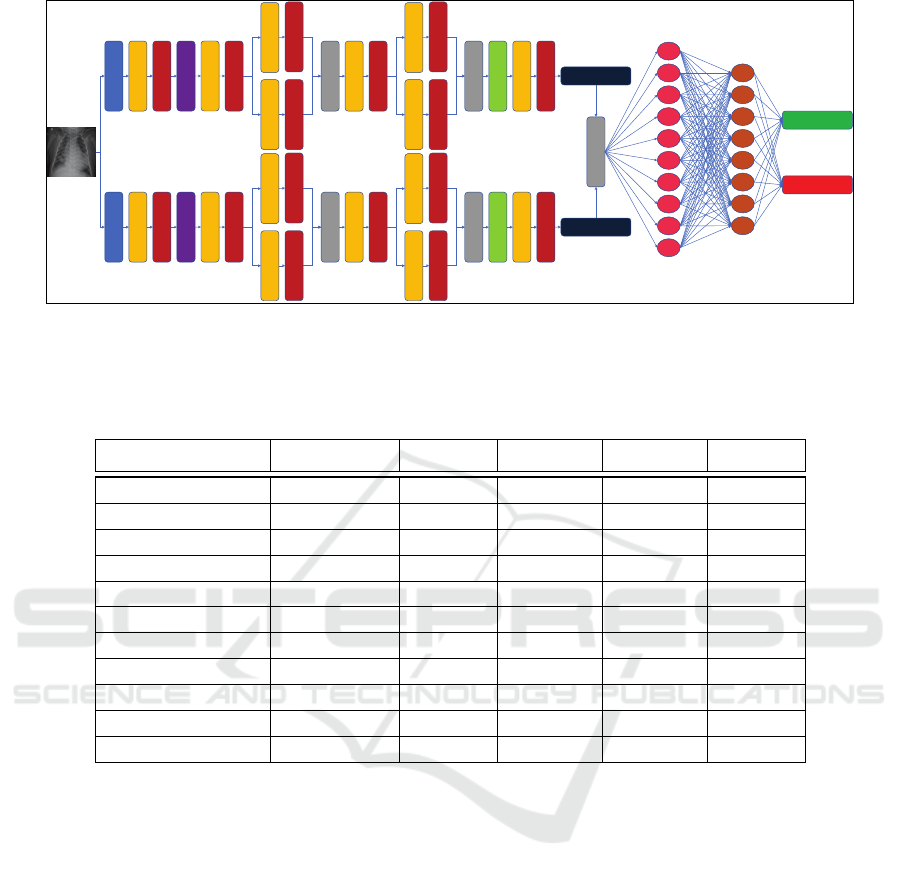
LOAD PRE-TRAINED MODEL
MILLIONS OF IMAGES
WITH 1000s OF CLASSES
…
EARLY
LAYERS
FINAL
LAYERS
REPLACE THE FINAL LAYERS
LEARN FASTER WITH
FEWER(N) CLASSES
…
REPLACE
LAYERS
RE-TRAIN THE MODEL
HUNDREDS OF IMAGES
WITH N CLASSES
…
TRAINING OPTIONS
TRAINING IMAGES
INTERPRET AND ASSESS
MODEL ACCURACY
TRAINED MODEL
TRAINING IMAGES
Figure 2: Overview of the deep transfer learning approach. Typically this involves changing the final layers of the pretrained
model and re-training the network to suit the task under consideration.
suming. Also, there are privacy concerns that have to
be considered when using/sharing patient data. In ad-
dition, training of deep learning models from scratch
requires high computational processing power and
is highly time-consuming. Deep transfer learning,
where a model that has been previously trained to per-
form one specific task is adapted and reused for an-
other similar task, is an alternative solution that over-
comes these issues. This approach has proven its ef-
fectiveness where only a limited amount of data is
available and computational power is of primary con-
cern (Yosinski et al., 2014). Typically, in deep transfer
learning, pretrained models can either be retrained for
a particular task using a limited number of domain-
specific images, or used as feature extractors in con-
junction with another classifier. An example of a pre-
trained deep neural network is shown in Figure 1.
In this paper, we use the concepts of deep transfer
learning to develop a framework for the diagnosis of
pneumonia. To this end, we explore the performance
of different deep neural network architectures as fea-
ture extractors when used in tandem with traditional
classification methods. We show through experimen-
tal results that the proposed method outperforms sim-
ilar existing methods. An overview of the deep trans-
fer learning approach is shown in Figure 2.
2 RELATED WORK
Medical imaging plays a vital role in disease diagno-
sis and the clinical decision-making process. Many
computer applications now assist medical profession-
als to provide fast and accurate diagnoses. Deep
learning models and deep transfer learning frame-
works are proven state-of-the-art techniques in the
field of medical image processing (Litjens et al.,
2017; Shie et al., 2015; Abidin et al., 2018). These
achievements have influenced the use of deep transfer
learning for pneumonia detection using chest X-ray
images (Kermany et al., 2018; Rajpurkar et al., 2017;
Liang and Zheng, 2019).
For example, Wang and Xia (Wang and Xia,
2018) proposed a deep transfer learning model for
diagnosing multiple thorax diseases, including pneu-
monia, using chest radiography by using a modi-
fied ResNet architecture (He et al., 2016). They
named their transfer learning model ChestNet and
compared their findings with three other deep learn-
ing models. They measured their performance us-
ing the area under the curve (AUC). They achieved
an average of AUC = 0.7810 per-class. However,
they used a high-powered computer configuration
(NVIDIA
R
TITAN Xp GPU×4, 128 gigabyte phys-
ical memory, 120 gigabyte solid state drive, Intel
R
Xeon
R
E5-2678V3×2) with 20 hours of training in
the CAFFE (Jia et al., 2014) deep learning framework
to achieve this performance.
Inspired by the original deep learning model of
DenseNet (Huang et al., 2017), a 121-layer deep
transfer learning model called CheXNet was pro-
posed by Rajpurkar et al. (Rajpurkar et al., 2017) for
pneumonia diagnosis from chest X-ray images. Apart
from pneumonia classification, they also demon-
strated that their model can determine the most symp-
tomatic areas of pneumonia in a chest X-ray. They
A Deep Transfer Learning Framework for Pneumonia Detection from Chest X-ray Images
287

compared their performance with that of radiologists
and demonstrated that the model can perform better
than an average radiologist’s performance for pneu-
monia diagnosis (F1 = 0.435) (Huang et al., 2017).
A transfer learning approach for pneumonia di-
agnosis was introduced by Kermany et al. (Kermany
et al., 2018) from chest X-ray images by modifying
the architecture of the original Inception-v3 model
(Szegedy et al., 2016). They achieved more than
90% classification accuracy on a publicly available
database. In addition to pneumonia classification,
they also demonstrated that the model can success-
fully perform tasks such as diabetic retinopathy diag-
nosis from optical coherence tomography images.
Antin et al. (Antin et al., 2017) proposed a ma-
chine learning model for pneumonia diagnosis from
chest X-ray images. First they used a logistic re-
gression model instead of a classifier as they theo-
rised it would be less memory-intensive than a clas-
sification model which treats each pixel of an im-
age a feature. From their initial findings, they con-
cluded that a logistic regression model does not work
well on their database of chest X-ray images (AUC
= 0.604). For this reason they moved to deep trans-
fer learning as a classification model which was in-
spired by CheXNet (Rajpurkar et al., 2017) as well
as the original DenseNet (Huang et al., 2017). They
employed Google
R
Cloud service, SciKit-Learn (Pe-
dregosa et al., 2011), and PyTorch (Paszke et al.,
2017) to implement their model. However, their clas-
sification results were not much improved (AUC =
0.609) compared to that of the logistic regression
model.
Recently, a deep residual network based trans-
fer learning method for pneumonia diagnosis from
chest X-rays has been introduced by Liang and Zheng
(Liang and Zheng, 2019). Initially, their network
of 2 dense layers and 49 convolutional layers was
trained on the publicly available ChestXray14 dataset
(Wang et al., 2017). Then, this trained network
was used as a deep transfer model and retrained on
the database provided by Kermany et al. (Kermany
et al., 2018). They compared the performance of
their method with four other deep learning archi-
tectures, VGG-16 (Simonyan and Zisserman, 2014),
DenseNet-201 (Huang et al., 2017), Xception (Chol-
let, 2017), and Inception-v3 (Szegedy et al., 2016).
They achieved an accuracy of 90.50% with their
method and 74.20%, 81.90%, 85.30%, 87.80% with
VGG-16, DenseNet-201, Xception, and Inception-v3
architectures respectively.
With the introduction of new deep learning archi-
tectures, the choice of models on which to build trans-
fer learning methods for complex classification tasks
has increased. In this paper, we explore their use as
deep transfer learning models for the task of pneumo-
nia diagnosis using chest X-rays.
3 OVERVIEW OF THE METHOD
Typically, deep learning models used in classification,
although they have their own complex network archi-
tectures, share a common trait. These networks de-
tect higher-level features towards the deeper layers
and lower-level features in the initial layers. There-
fore, once a deep neural network has been trained to
perform a classification task, it can also be used to ex-
tract high-level features learned for that task from its
deeper layers. These feature extractors can then be
used in conjunction with classification techniques to
perform the classification.
In this paper, we first retrained and compared
the performance of deep learning models to identify
which is most appropriate for the task of pneumo-
nia diagnosis from chest X-ray images. Then, we se-
lected the two best networks with respect to the per-
formance metrics of accuracy and sensitivity to use as
feature extractors. Next, we concatenated the two sets
of features and used them as input to traditional clas-
sifiers to determine the one best suited for pneumonia
diagnosis. In doing so, we developed a system that
combines the advantages of both deep learning and
traditional classification methods. Figure 3 shows an
overview of this method.
The database we used for the training and testing
of the proposed method was originally provided by
Kermany et al. (Kermany et al., 2018) and is pub-
licly available for research purposes. The database
contains 2D grayscale images of chest X-rays where
the average image dimensions are 1000 ×3500 pixels.
Each image is classified into one of two classes: nor-
mal or pneumonia. The database is divided into train-
ing and testing sets with the training set containing
1349 normal and 3883 pneumonia samples and the
testing set containing 234 normal and 390 pneumonia
samples. The pneumonia class in both training and
testing sets is subdivided into the classes of bacteria
or virus. Note that only the first level of classification
(normal or pneumonia) is used in our method.
To obtain an unbiased and complete view of clas-
sification results, we used classification accuracy, sen-
sitivity, specificity, and precision as performance met-
rics (Powers, 2011). For the development, train-
ing, and testing of methods, we used the MATLAB
R
academic framework, including the Deep Learning
Toolbox
TM
. A HP
R
Z6 G4 Workstation model com-
puter powered by Intel
R
Xeno
R
Silver 4108 CPU
VISAPP 2020 - 15th International Conference on Computer Vision Theory and Applications
288
Input
Convolution
Activation
Pooling
Convolution
Activation
Convolution
Activation
Convolution
Activation
Combination
Convolution
Activation
Convolution
Activation
Convolution
Activation
Combination
Dropout
Convolution
Activation
Concatenate
Input
Convolution
Activation
Pooling
Convolution
Activation
Convolution
Activation
Convolution
Activation
Combination
Convolution
Activation
Convolution
Activation
Convolution
Activation
Combination
Dropout
Convolution
Activation
F
1
F
2
F
3
F
4
…F
n
F
1
F
2
F
3
F
4
…F
n
Normal
Pneumonia
n
1
n
2
n
3
n
4
n
5
n
6
n
7
n
8
n
9
n
n
n
1
n
2
n
3
n
4
n
5
n
6
n
7
n
n
Figure 3: Overview of the proposed method for pneumonia detection from chest X-ray images. The image is input to two
different networks for feature extraction. Features extracted from those two networks are then concatenated and used as the
input to an artificial neural network for classification as either normal or pneumonia.
Table 1: Performance of the deep learning models retrained on the chest X-ray image database in identifying pneumonia
(Phase-1). The best results per metric are highlighted in bold.
Model Name Training Time Accuracy Sensitivity Specificity Precision
AlexNet 03:47:37 0.9311 0.8547 0.9769 0.9569
VGG-16 04:39:21 0.8814 0.6966 0.9923 0.9819
VGG-19 05:06:10 0.9359 0.8462 0.9897 0.9802
SqueezeNet 03:40:20 0.9471 0.9188 0.9641 0.9389
GoogLeNet 03:56:54 0.9327 0.8632 0.9744 0.9528
Inception-v3 04:09:09 0.9776 0.9615 0.9872 0.9783
DenseNet-201 23:47:54 0.9631 0.9359 0.9795 0.9648
ResNet-18 03:59:46 0.9022 0.7436 0.9974 0.9943
ResNet-50 04:29:44 0.9599 0.9017 0.9949 0.9906
ResNet-101 09:19:02 0.9359 0.8333 0.9974 0.9949
Inception-ResNet-v2 31:36:25 0.8958 0.7479 0.9846 0.9669
(1.80 GHZ) with 16 GB of physical memory and
5 GB of graphics memory (NVIDIA
R
QuADro
R
P2000 GPU) was used. The operating system used
was 64-bit Microsoft
R
Windows
R
10 Education.
4 SELECTION OF A FEATURE
EXTRACTOR
For the first step of our implementation, we used 11
pretrained models, trained on the ImageNet database
(Deng et al., 2009). The models considered were:
AlexNet (Krizhevsky et al., 2012), VGG-16 (Si-
monyan and Zisserman, 2014), VGG-19 (Simonyan
and Zisserman, 2014), SqueezeNet (Iandola et al.,
2016), GoogLeNet (Szegedy et al., 2015), Inception-
v3 (Szegedy et al., 2016), DenseNet-201 (Huang
et al., 2017), ResNet-18 (He et al., 2016), ResNet-
50 (He et al., 2016), ResNet-101 (He et al., 2016),
and Inception-ResNet-v2 (Szegedy et al., 2017).
We retrained all the pretrained models on the chest
X-ray image database (Kermany et al., 2018) us-
ing the same training configurations (stochastic gradi-
ent descent (Robbins and Monro, 1951) as the train-
ing optimizer with an initial learning rate of 0.0003,
maximum epochs of five, and maximum iterations of
5230). To avoid memory crashes, we used a five batch
minimum and to avoid network over-fitting, we used
a validation frequency of five iterations. We call this
Phase-1 of our network selection process. Table 1
shows the pretrained model selection results with re-
spect to normal and pneumonia classification.
We selected the highest performing models in
Phase-1 to be used in the next phase (Phase-2). For
example, we selected Inception-v3, DenseNet-201
and ResNet-50 as the three best models with re-
spect to accuracy. Likewise, we selected ResNet-18,
ResNet-50, and ResNet-101 as the three best mod-
els with regard to precision. We further retrained
A Deep Transfer Learning Framework for Pneumonia Detection from Chest X-ray Images
289
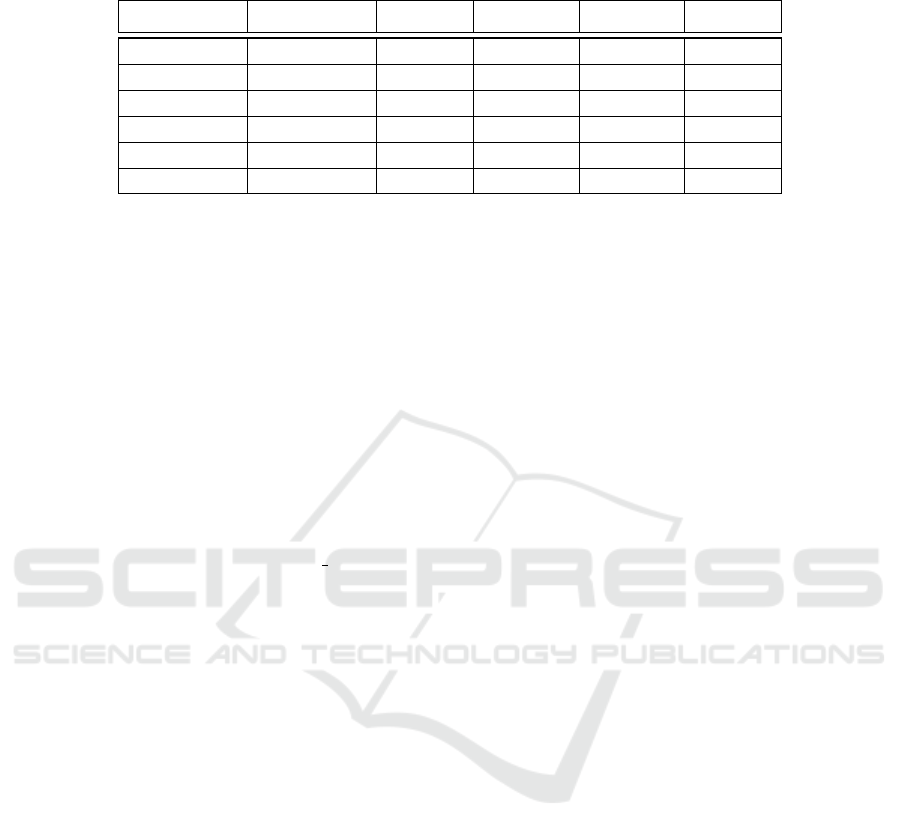
Table 2: Performance of the selected models after retraining with modified training parameters: initial learning rate and
maximum number of epochs (phase-2). The best results per metric are given in bold.
Model Name Training Time Accuracy Sensitivity Specificity Precision
SqueezeNet 08:03:11 0.9519 0.9145 0.9744 0.9554
Inception-v3 17:08:26 0.9744 0.9402 0.9949 0.9910
DenseNet-201 52:16:37 0.8446 0.5897 0.9974 0.9928
ResNet-18 08:36:19 0.9071 0.7564 0.9974 0.9944
ResNet-50 10:48:07 0.8798 0.6795 1.0000 1.0000
ResNet-101 29:29:54 0.8462 0.5897 1.0000 1.0000
them aiming to improve the performance of the se-
lected models. We decreased our initial learning rate
of 0.0003 to 0.0001 as Goodfellow et al. (Goodfel-
low et al., 2016) stated that when the learning rate is
high, rather than decreasing the training error gradi-
ent, it may possibly increase. Furthermore, we also
increased the maximum number of epochs (five to
ten) to train the selected models with more iterations
(maximum 10460). The results of this phase of train-
ing are shown in Table 2.
Then, we selected the models that performed best
with respect to accuracy and sensitivity (SqueezeNet
and Inception-v3) to be used as feature extractors.
We used the intermediate layers of the two net-
works (“fire9-expand3x3” and “conv2d 9” layers for
SqueezeNet and Inception-v3 respectively) to extract
features.
5 SELECTION OF A CLASSIFIER
In order to select an appropriate classifier, we com-
pared the performance of a few traditional classifica-
tion methods using the features extracted from the se-
lected deep neural networks as inputs. The classifiers
used were: support vector machines (SVM), k-nearest
neighbors (KNN), stacked auto-encoders (SAE), and
artificial neural networks (ANN) (Altman, 1992). To
train the SVM we used a linear kernel function with
the auto kernel scale parameter and box constraint
values set to 1. The KNN we used was a Fine KNN
model with Euclidean distance as the distance func-
tion and the number of neighbors set to 1. For the
SAE training, we used two encoders and the number
of neurons of the hidden layers of the first and sec-
ond encoders were 100 and 50 respectively. We also
used L2 regularizations of 0.004 and 0.002 for the two
encoders in that order. They were trained with a max-
imum of 100 epochs. The ANN had 200 neurons in
one hidden layer and it was trained using scaled con-
jugate gradient back-propagation (Møller, 1993).
We tested the performance of these classifiers with
the features of the two selected networks individually,
and also with concatenated features from both net-
works. We also explored the behaviour of each of the
classifiers when used in conjunction with feature ex-
tractors of different levels of retraining: no retraining
(original), retrained on chest X-ray images (Phase-1),
and retrained on chest X-ray images with adjusted
training parameters (Phase-2). For the training and
testing of the methods, we used four-fold cross valida-
tion using the combined images of training and testing
datasets of the chest X-ray image database of (Ker-
many et al., 2018). We calculated the average per-
formance of the four folds for each classifier for the
different combinations of features and retrain levels.
Table 3 shows the results of this comparison.
We observed that using the retrained (phase-2)
SqueezeNet and Inception-v3 models as feature ex-
tractors and concatenating the resulting features to be
used as input to an ANN classifier was the most effec-
tive way of achieving the highest levels of accuracy
and sensitivity. As such, we selected this combina-
tion of feature extractor and classifier as our preferred
method of pneumonia diagnosis.
6 PERFORMANCE RESULTS
We compared the performance of our method (us-
ing the combined features obtained from retrained
SqueezeNet and Inception-v3 networks and training
an ANN on these) with other similar existing meth-
ods that have been introduced for pneumonia diag-
nosis using chest X-rays (as discussed in Section
2). In order to make unbiased comparisons, we re-
implemented the existing methods in our system and
trained and tested them on the same database. The
results of the comparisons are shown in Table 4.
From the results, we can conclude that the pro-
posed method outperforms other similar existing
methods in every comparison metric except speci-
ficity where Wang and Xia (Wang and Xia, 2018)
is slightly better. The training time of the proposed
VISAPP 2020 - 15th International Conference on Computer Vision Theory and Applications
290

Table 3: Performance of classification methods using pretrained deep learning models as input. The results compare the
performance with respect to features extracted from the original models as well as the retrained models resulting from Phase-
1 and Phase-2. These are the average performance results obtained from four-fold cross validation. The best results per metric
are highlighted in bold.
Retrain
Level
Feature
Extractor
Feature
Extraction Time
Classifier
Training
Time
Accuracy Sensitivity Specificity Precision
Original
SqueezeNet 00:00:53
SVM 00:06:53 0.8461 0.6345 0.9539 0.9368
KNN 00:06:32 0.8726 0.6383 0.9651 0.9673
SAE 00:02:25 0.9164 0.6582 0.9358 0.9100
ANN 00:05:59 0.8542 0.6197 0.9949 0.9864
Inception-v3 00:02:19
SVM 00:04:43 0.8632 0.6528 0.9528 0.9374
KNN 00:04:10 0.8480 0.6253 0.9543 0.9365
SAE 00:02:05 0.9070 0.6274 0.9284 0.9005
ANN 00:01:21 0.8542 0.6197 0.9949 0.9864
SqueezeNet
+
Inception-v3
00:03:12
SVM 00:51:35 0.8835 0.9662 0.9617 0.9587
KNN 00:55:08 0.9053 0.6592 0.9544 0.9082
SAE 00:18:07 0.9551 0.9184 0.6832 0.9203
ANN 00:04:12 0.9602 0.9671 0.9705 0.9657
Phase-1
SqueezeNet 00:00:53
SVM 00:05:53 0.8594 0.6583 0.9541 0.9470
KNN 00:06:24 0.8763 0.6479 0.9666 0.9732
SAE 00:02:20 0.9137 0.6724 0.9405 0.9137
ANN 00:09:48 0.8622 0.6325 1.0000 1.0000
Inception-v3 00:02:17
SVM 00:04:45 0.8652 0.6533 0.9540 0.9382
KNN 00:04:12 0.8483 0.6258 0.9548 0.9378
SAE 00:02:10 0.9105 0.6270 0.9290 0.9071
ANN 00:01:30 0.9215 0.7906 1.0000 1.0000
SqueezeNet
+
Inception-v3
00:03:10
SVM 00:52:41 0.9154 0.9607 0.9714 0.9571
KNN 00:54:51 0.9135 0.6829 0.9621 0.8874
SAE 00:18:45 0.9782 0.9748 0.9824 0.9835
ANN 00:06:41 0.9847 0.9857 0.9834 0.9871
Phase-2
SqueezeNet 00:02:05
SVM 00:05:50 0.8891 0.6784 0.9645 0.9570
KNN 00:06:20 0.8793 0.6682 0.9695 0.9820
SAE 00:02:18 0.9317 0.6784 0.9490 0.9363
ANN 00:10:42 0.8830 0.6923 0.9974 0.9939
Inception-v3 00:02:11
SVM 00:04:40 0.8852 0.6596 0.9734 0.9593
KNN 00:04:10 0.8687 0.6754 0.9641 0.9577
SAE 00:02:11 0.9255 0.6380 0.9392 0.9281
ANN 00:01:01 0.9247 0.0753 1.0000 1.0000
SqueezeNet
+
Inception-v3
00:04:16
SVM 00:52:18 0.9768 0.9798 0.9739 0.9740
KNN 00:57:25 0.9773 0.9773 0.9773 0.9773
SAE 00:19:33 0.9815 0.9815 0.9832 0.9831
ANN 00:06:49 0.9899 0.9880 0.9918 0.9918
method is much lower than that of the other meth-
ods. This indicates that using a pretrained deep learn-
ing model as a feature extractor in tandem with a tra-
ditional classifier is effective in pneumonia detection
from X-ray images. As such, we were successful in
combining the advantages of both deep and traditional
learning. Furthermore, by concatenating the features
of the deep neural networks that showed the best ac-
curacy and sensitivity, we were able to improve the
classification performance. Figure 4 shows some ex-
amples of classification using this method, along with
the corresponding confidence levels.
A Deep Transfer Learning Framework for Pneumonia Detection from Chest X-ray Images
291
Table 4: Performance comparison with other similar methods. The best performance results are shown in bold.
Methods Training Time Accuracy Sensitivity Specificity Precision
(Liang and Zheng, 2019) 04:35:18 0.9050 0.9670 0.9549 0.8910
(Wang and Xia, 2018) 04:29:44 0.9599 0.9017 0.9949 0.9906
(Kermany et al., 2018) 04:09:09 0.9776 0.9615 0.9872 0.9783
(Rajpurkar et al., 2017) 23:47:54 0.9631 0.9359 0.9795 0.9648
Proposed 00:06:49 0.9899 0.9880 0.9918 0.9918
7 CONCLUSIONS
In this paper, we investigated the use of pretrained
deep neural networks as feature extractors along with
traditional classification methods to perform pneumo-
nia classification from X-ray images. We retrained the
pretrained networks on chest X-ray images, and se-
lected the two networks that provided the best levels
of accuracy and sensitivity to use as feature extractors.
We showed that by using the concatenated features
of these networks as inputs, the performance of tra-
ditional classification methods can be improved. Fi-
nally, we showed that this method outperformed sim-
ilar existing methods of pneumonia diagnosis. In fu-
ture work, we will test this method on other databases
and also compare its performance with that of human
experts.
ACKNOWLEDGEMENTS
This research was funded by the University of Mel-
bourne through a Melbourne Research Scholarship
(MRS) awarded to Kh Tohidul Islam in support of his
Doctor of Philosophy degree. The funders had no role
in study design, data collection and analysis, decision
to publish, or preparation of the manuscript. Also,
the authors would like to thank Dr. Jared Panario
a
b
c
d
e
f
g
h
i
j
Figure 4: Examples of chest X-ray images classified using
our method with corresponding confidence levels: (a) Nor-
mal, 100%, (b) Normal, 100%, (c) Pneumonia, 100%, (d)
Pneumonia, 100%, (e) Pneumonia, 91.10%, (f) Pneumonia,
99.90%, (g) Pneumonia, 100%, (h) Pneumonia, 100%, (i)
Pneumonia, 100%, and (j) Normal, 99.00%.
and Tayla Razmovski of the Department of Surgery
(Otolaryngology), University of Melbourne, Victoria,
Australia for their suggestions and clarifications.
REFERENCES
Abidin, A. Z., Deng, B., DSouza, A. M., Nagarajan, M. B.,
Coan, P., and Wism
¨
uller, A. (2018). Deep trans-
fer learning for characterizing chondrocyte patterns in
phase contrast X-Ray computed tomography images
of the human patellar cartilage. Computers in Biology
and Medicine, 95:24–33.
Altman, N. S. (1992). An introduction to kernel and nearest-
neighbor nonparametric regression. The American
Statistician, 46(3):175–185.
Antin, B., Kravitz, J., and Martayan, E. (2017). Detecting
Pneumonia in Chest X-Rays with Supervised Learn-
ing.
Banu, B. (2019). Pneumonia. In Reference Module in
Biomedical Sciences. Elsevier.
Chen, X., Chen, Y., Liu, H., Goldmacher, G., Roberts, C.,
Maria, D., and Ou, W. (2019). Pin92 pediatric bac-
terial pneumonia classification through chest x-rays
using transfer learning. Value in Health, 22:S209 –
S210. ISPOR 2019: Rapid. Disruptive. Innovative: A
New Era in HEOR.
Chollet, F. (2017). Xception: Deep learning with depthwise
separable convolutions. In 2017 IEEE Conference
on Computer Vision and Pattern Recognition (CVPR),
pages 1800–1807.
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., and Fei-Fei,
L. (2009). ImageNet: A large-scale hierarchical im-
age database. In 2009 IEEE Conference on Computer
Vision and Pattern Recognition. IEEE.
Fourcade, A. and Khonsari, R. (2019). Deep learning in
medical image analysis: A third eye for doctors. Jour-
nal of Stomatology, Oral and Maxillofacial Surgery,
120(4):279 – 288. 55th SFSCMFCO Congress.
Goodfellow, I., Bengio, Y., and Courville, A. (2016). Deep
Learning. MIT Press.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep Resid-
ual Learning for Image Recognition. In 2016 IEEE
Conference on Computer Vision and Pattern Recogni-
tion (CVPR). IEEE.
Huang, G., Liu, Z., van der Maaten, L., and Weinberger,
K. Q. (2017). Densely Connected Convolutional Net-
VISAPP 2020 - 15th International Conference on Computer Vision Theory and Applications
292

works. In 2017 IEEE Conference on Computer Vision
and Pattern Recognition (CVPR). IEEE.
Iandola, F. N., Han, S., Moskewicz, M. W., Ashraf, K.,
Dally, W. J., and Keutzer, K. (2016). Squeezenet:
Alexnet-level accuracy with 50x fewer parame-
ters and <0.5 MB model size. arXiv preprint
arXiv:1602.07360.
Jia, Y., Shelhamer, E., Donahue, J., Karayev, S., Long, J.,
Girshick, R., Guadarrama, S., and Darrell, T. (2014).
Caffe: Convolutional Architecture for Fast Feature
Embedding. arXiv preprint arXiv:1408.5093.
Kermany, D. S., Goldbaum, M., Cai, W., Valentim, C. C.,
Liang, H., Baxter, S. L., McKeown, A., Yang, G., Wu,
X., Yan, F., Dong, J., Prasadha, M. K., Pei, J., Ting,
M. Y., Zhu, J., Li, C., Hewett, S., Dong, J., Ziyar,
I., Shi, A., Zhang, R., Zheng, L., Hou, R., Shi, W.,
Fu, X., Duan, Y., Huu, V. A., Wen, C., Zhang, E. D.,
Zhang, C. L., Li, O., Wang, X., Singer, M. A., Sun, X.,
Xu, J., Tafreshi, A., Lewis, M. A., Xia, H., and Zhang,
K. (2018). Identifying Medical Diagnoses and Treat-
able Diseases by Image-Based Deep Learning. Cell,
172(5):1122–1131.e9.
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2012). Im-
ageNet Classification with Deep Convolutional Neu-
ral Networks. In Advances in Neural Information Pro-
cessing Systems 25, pages 1097–1105. Curran Asso-
ciates, Inc.
Liang, G. and Zheng, L. (2019). A transfer learning method
with deep residual network for pediatric pneumo-
nia diagnosis. Computer Methods and Programs in
Biomedicine.
Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A.,
Ciompi, F., Ghafoorian, M., van der Laak, J. A., van
Ginneken, B., and S
´
anchez, C. I. (2017). A survey
on deep learning in medical image analysis. Medical
Image Analysis, 42:60–88.
Lodha, R., Kabra, S., and Pandey, R. (2013). Antibiotics for
communityacquired pneumonia in children. Cochrane
Database of Systematic Reviews.
Møller, M. F. (1993). A scaled conjugate gradient algo-
rithm for fast supervised learning. Neural Networks,
6(4):525–533.
Organization, W. H. (2001). Standardization of interpreta-
tion of chest radiographs for the diagnosis of pneumo-
nia in children. Last Accessed: 14 May 2019.
Paszke, A., Gross, S., Chintala, S., Chanan, G., Yang, E.,
DeVito, Z., Lin, Z., Desmaison, A., Antiga, L., and
Lerer, A. (2017). Automatic differentiation in Py-
Torch. In NIPS-W.
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V.,
Thirion, B., Grisel, O., Blondel, M., Prettenhofer, P.,
Weiss, R., Dubourg, V., Vanderplas, J., Passos, A.,
Cournapeau, D., Brucher, M., Perrot, M., and Duch-
esnay, E. (2011). Scikit-learn: Machine Learning
in Python. Journal of Machine Learning Research,
12:2825–2830.
Powers, D. M. (2011). Evaluation: from precision, recall
and f-measure to roc, informedness, markedness and
correlation. Journal of Machine Learning Technolo-
gies, 2(1):37 – 63.
Qin, C., Yao, D., Shi, Y., and Song, Z. (2018). Computer-
aided detection in chest radiography based on artifi-
cial intelligence: a survey. BioMedical Engineering
OnLine, 17(1).
Rajpurkar, P., Irvin, J., Zhu, K., Yang, B., Mehta, H., Duan,
T., Ding, D., Bagul, A., Langlotz, C., Shpanskaya, K.,
et al. (2017). CheXNet: Radiologist-Level Pneumo-
nia Detection on Chest X-Rays with Deep Learning.
arXiv preprint arXiv:1711.05225.
Razzak, M. I., Naz, S., and Zaib, A. (2017). Deep Learning
for Medical Image Processing: Overview, Challenges
and the Future. In Lecture Notes in Computational Vi-
sion and Biomechanics, pages 323–350. Springer In-
ternational Publishing.
Robbins, H. and Monro, S. (1951). A Stochastic Approx-
imation Method. The Annals of Mathematical Statis-
tics, 22(3):400–407.
Shie, C.-K., Chuang, C.-H., Chou, C.-N., Wu, M.-H., and
Chang, E. Y. (2015). Transfer representation learn-
ing for medical image analysis. In 2015 37th Annual
International Conference of the IEEE Engineering in
Medicine and Biology Society (EMBC). IEEE.
Siddiqi, R. (2019). Automated pneumonia diagnosis using
a customized sequential convolutional neural network.
In Proceedings of the 2019 3rd International Confer-
ence on Deep Learning Technologies - ICDLT 2019.
ACM Press.
Simonyan, K. and Zisserman, A. (2014). Very Deep Con-
volutional Networks for Large-Scale Image Recogni-
tion. arXiv preprint arXiv:1409.1556.
Szegedy, C., Ioffe, S., Vanhoucke, V., and Alemi, A. (2017).
Inception-v4, Inception-ResNet and the Impact of
Residual Connections on Learning. In 2017 31st AAAI
Conference on Artificial Intelligence (AAAI-17).
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S.,
Anguelov, D., Erhan, D., Vanhoucke, V., and Rabi-
novich, A. (2015). Going deeper with convolutions. In
2015 IEEE Conference on Computer Vision and Pat-
tern Recognition (CVPR). IEEE.
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., and Wojna,
Z. (2016). Rethinking the Inception Architecture for
Computer Vision. In 2016 IEEE Conference on Com-
puter Vision and Pattern Recognition (CVPR). IEEE.
Wang, H. and Xia, Y. (2018). ChestNet: A Deep Neural
Network for Classification of Thoracic Diseases on
Chest Radiography. arXiv preprint arXiv:1807.03058.
Wang, X., Peng, Y., Lu, L., Lu, Z., Bagheri, M., and Sum-
mers, R. M. (2017). Chestx-ray8: Hospital-scale chest
x-ray database and benchmarks on weakly-supervised
classification and localization of common thorax dis-
eases. In 2017 IEEE Conference on Computer Vision
and Pattern Recognition (CVPR), pages 3462–3471.
Yosinski, J., Clune, J., Bengio, Y., and Lipson, H. (2014).
How transferable are features in deep neural net-
works? In Advances in Neural Information Process-
ing Systems 27, pages 3320–3328. Curran Associates,
Inc.
A Deep Transfer Learning Framework for Pneumonia Detection from Chest X-ray Images
293
