
Assessment of Gallbladder Wall Vascularity from Laparoscopic
Images using Deep Learning
Constantinos Loukas
1a
and Dimitrios Schizas
2b
1
Medical Physics Lab, Medical School, National and Kapodistrian University of Athens, Mikras Asias 75 str.,
Athens, Greece
2
1
st
Department of Surgery, Laikon General Hospital, National and Kapodistrian University of Athens, Athens, Greece
Keywords: Surgery, Laparoscopic Cholecystectomy, Gallbladder, Vascularity, Classification, Cnn, Deep Learning.
Abstract: Despite the significant progress in content-based video analysis of surgical procedures, methods on analyzing
still images acquired during the operation are limited. In this paper we elaborate on a novel idea for computer
vision-based assessment of the vascularity of the gallbladder (GB) wall, using frames extracted from videos
of laparoscopic cholecystectomy. The motivation was based on the fact that the wall’s vascular pattern
provides an indirect indication of the GB condition (e.g. fat coverage, wall thickening, inflammation), which
in turn is usually related to the operation complexity. As the GB wall vascularity may appear irregular, in this
study we focus on the classification of rectangular sub-regions (patches). A convolutional neural network
(CNN) is proposed for patch classification based on two ground-truth annotation schemes: 3-classes (Low,
Medium and High vascularity) and 2-classes (Low vs. High). Moreover, we employed three popular classifiers
with a rich set of hand-crafted descriptors. The CNN achieved the best performance with accuracy: 98% and
83.1%, and mean F1-score: 98% and 80.4%, for 2-class and 3-class classification, respectively. The other
methods’ performance was lower by 2%-6% (2-classes) and 6%-17% (3 classes). Our results indicate that
CNN-based patch classification is promising for intraoperative assessment of the GB wall vascularity.
1 INTRODUCTION
Laparoscopic surgery is a widely used technique for
the treatment of various diseases of the
gastrointestinal tract. The operation is performed via
long-shaft tools and a laparoscopic camera, inserted
into the body through small incisions. Compared to
open surgery, LS requires the demonstration of
advanced psychomotor skills, mostly due to lack of
direct 3D vision, the limited working space, the
fulcrum effect and minimal force feedback. On the
other hand, LS offers several benefits for the patient
such as less postoperative pain, minimal blood loss,
faster recovery and better cosmetic results. In
addition, LS provides the opportunity to capture
video and image data via the laparoscopic camera,
providing valuable visual information. The data may
be used for various purposes, such as documentation,
archival, retrospective analysis of the procedure,
skills assessment and surgical training. If processed
online, they may also be used to provide context
a
https://orcid.org/0000-0001-7879-7329
b
https://orcid.org/0000-0002-7046-0112
specific information to the surgical staff for decision
for extra support and resource scheduling (Twinanda
et al., 2019).
In the literature various studies have proposed
computer vision techniques, mostly for surgical
workflow analysis and tool detection applications
(Loukas, 2018). Surgical workflow analysis aims to
segment the recorded video into the main phases of
the operation. In offline mode these techniques could
be utilized for video database indexing and retrieval
(Loukas and Georgiou, 2013), whereas when applied
online they could be utilized to improve staff
coordination and resource scheduling in the operating
room (Twinanda et al., 2017). Given the close relation
between surgical phases and the tool types employed,
recent works have proposed the joint detection of
tools and phases (Jin et al., 2019). Some approaches
also aim to detect and localize the tool tip for tool
motion analysis and skill assessment (Jin et al., 2018).
Other developments in video-based analysis of
surgical interventions include shot detection (Loukas
28
Loukas, C. and Schizas, D.
Assessment of Gallbladder Wall Vascularity from Laparoscopic Images using Deep Learning.
DOI: 10.5220/0008918800280036
In Proceedings of the 13th International Joint Conference on Biomedical Engineer ing Systems and Technologies (BIOSTEC 2020) - Volume 2: BIOIMAGING, pages 28-36
ISBN: 978-989-758-398-8; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

et al., 2016), keyframe extraction (Loukas et al.,
2018), event detection (Loukas and Georgiou, 2015)
and surgery classification (Twinanda et al., 2015).
Despite the significant progress in surgical video
analysis, studies related to the analysis of still images
captured intentionally during the operation are
limited. These images may be captured for various
purposes, such as: (a) for patient information, (b) as
supplementary material to the formal report, (c) for
future reference with regard to the condition of the
operated organ or the patient’s anatomy, (d) as
evidence of the procedure outcome, and (e) for
medical research. Although retrospective image
extraction from the video stream is technically
feasible, manual video browsing and image selection
is tedious and time consuming. Moreover, still images
provide an important supplement to the operational
video, depicting certain visual characteristics of the
operated organ and the patient’s anatomy
(Petscharnig and Schöffmann, 2018).
Based on the aforementioned remarks, in this
paper we investigate a novel concept for visual
assessment of the gallbladder (GB), the operated
organ in laparoscopic cholecystectomy (LC), based
on computer vision. In particular, we investigate
various image analysis and machine learning
techniques for assessment of the GB condition from
intraoperative images. LC is a widely used technique
for the treatment of GB diseases, with approximately
600,000 operations performed every year in the
United States (Pontarelli et al., 2019). The purpose of
the operation is the removal of the GB following
some preoperative indications, such as the presence
of cholecystitis, gallstones, etc. The procedure is
mainly divided into 7 phases, some of which are
either repeated or not required to be performed,
depending on the operation progress (Twinanda et al.,
2017). An important task at the initial stage of the
operation (‘preparation phase’), is when the surgeon
inspects the GB to assess its condition, such as the
thickness of its wall, presence of inflammation, fat
coverage, etc. Among the various parameters
assessed by the surgeon is the GB wall vascularity
pattern. For example, the vascular pattern may
become less visible when the GB wall is covered by
fat or due to wall thickening, potentially as a result of
cholecystitis. Hence, in this study we aim to
investigate the feasibility of a computer vision
approach to assess the vascular pattern of the GB
wall, using laparoscopic images extracted from the
operation’s video. Such a system could potentially be
utilized for the management and classification of
surgical image databases, as well as to support
surgical training (Loukas et al., 2011). For example,
it could be utilized to retrieve cases from a video
database where the GB appears similar to that in a
query image. This would help trainees retrieve and
review similar operations in terms of the GB
condition. Another application could be the
automated classification of LC operations. Various
useful metrics for the operations (e.g. mean/longest
duration, etc.) associated with a particular GB
condition could then be extracted for: management
decision support, resource scheduling and evaluation
purposes. The proposed system could also be used in
online fashion, to assess the complexity of the
operation or for recruitment of extra resources (e.g.
senior surgical staff). To date, the assessment based
on preoperative imaging (e.g. US, CT), cannot always
provide adequate evidence about the GB condition,
such as for example presence of an acute or chronic
cholecystitis. Visual assessment of the GB condition
during the initial stage of LC is an important factor
for early assessment of the operation progress or for
potential need of extra resources.
Figure 1 illustrates an overview of how such a
system could potentially be utilized in practice. Given
a database of surgical videos (a), an image of the GB
is captured during the preparation phase (b). Then,
regions of interest (ROIs) from the GB wall are
manually selected and annotated by an experienced
surgeon (c). The ROIs along their ground truth labels
are fed into a supervised learning algorithm to
develop a predictive model of the GB’s vascular
pattern (e.g. Low, Medium and High). In surgical
practice (d), the surgeon outlines a ROI on the GB
image. Using the predictive model, the GB is
classified based on the selected ROI (or aggregation
of ROIs).
2 METHODOLOGY
2.1 Dataset
To create the GB image dataset, 31 LC videos were
selected from the publicly available Cholec80 video
collection (Twinanda et al., 2017). From each video
we manually extracted various still-images (854 ×
480) of the GB. The images originated mostly from
the preparation phase of LC, during which the
surgeon approaches the endoscope towards the GB
for inspection. Besides, the GB is lift with the aid of
a grasper, providing an appropriate view of
the GB
body. From the videos we managed to extract 121
images (median 4) with a large and clear view of the
GB wall.
Assessment of Gallbladder Wall Vascularity from Laparoscopic Images using Deep Learning
29
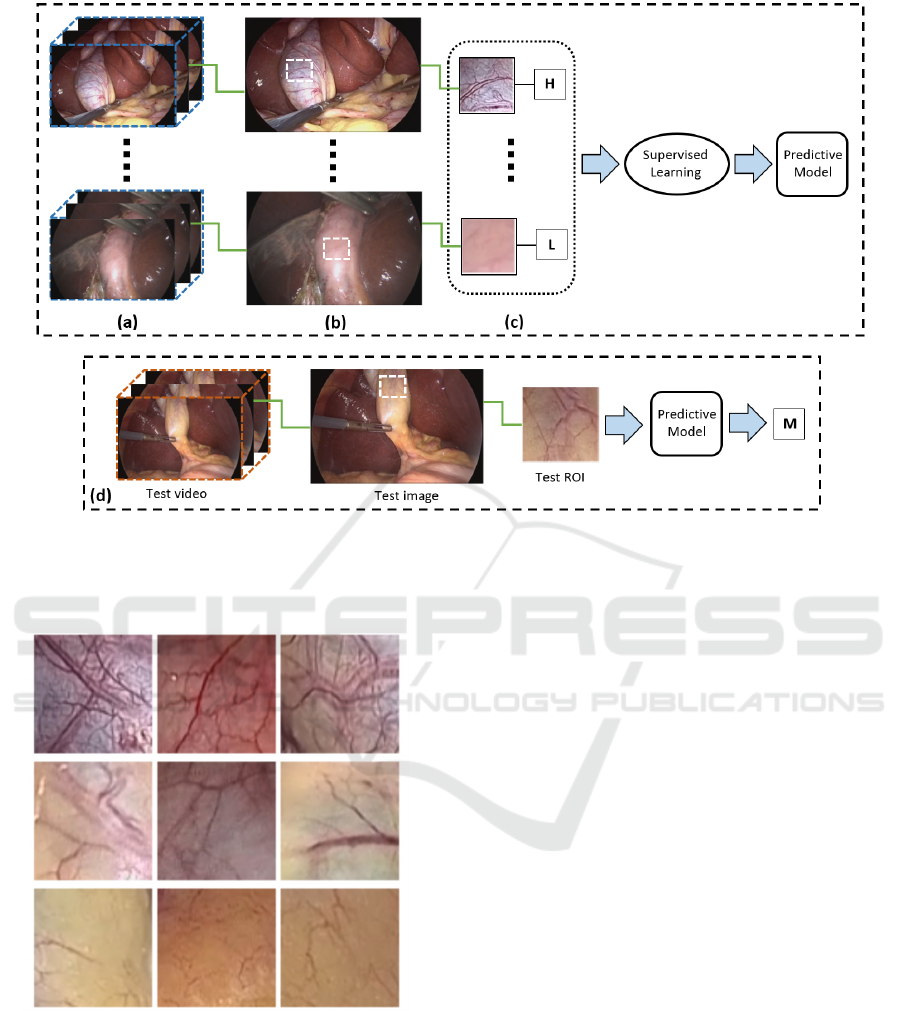
Figure 1: Graphical overview of the proposed system (see text for details). (a) Database of laparoscopic videos. (b) GB images
extracted from the videos. (c) ROIs along with annotations are provided by an expert surgeon. The ROIs are used to build a
predictive model based on supervised learning. (d) Use-case scenario: a sample ROI outlined on the GB image is classified
based on the predictive model.
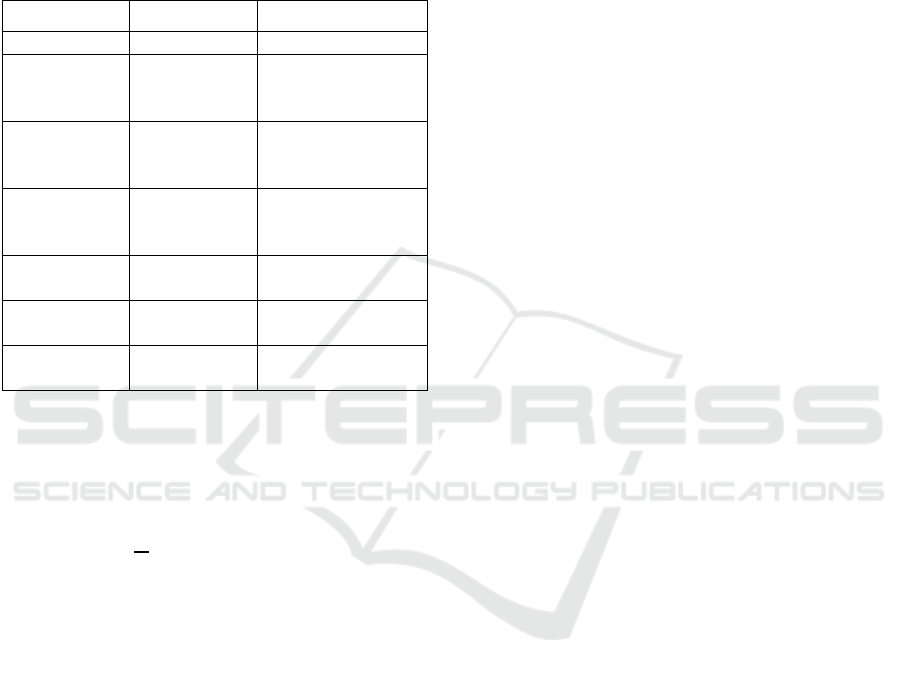
Figure 2: Example image-
p
atches with different vascula
r
patterns. From top row to bottom: High-, Medium- an
d
Low-degree of vascularity. All patches come from differen
t
GB images.
Then, rectangular patches with various patterns of
vascularity were selected from the GB wall (body and
fundus areas). GB regions with specular reflection
were excluded. At this point it should be noted that
the GB may not necessarily exhibit the same vascular
pattern across its wall. There may be regions of high
vascularity separated by regions covered with fat (i.e.
low vascularity), and vice versa. Hence, a significant
step towards the assessment of the entire GB wall is
to be able to classify the vascular pattern of individual
sub-regions (patches). After experimentation with
various patch sizes, a 70 × 70 size was selected
providing a compromise between distinct pattern of
vascularity and adequate resolution to perform
assessment. Figure 2 illustrates sample image patches
with various patterns of vascularity. Note the high
inter-class color variance and the color similarity
between the middle and bottom row patches.
From the GB images, a total number of 525 image
patches was selected. The patches were classified by
a faculty surgeon based on two schemes, as this is the
first time that such a classification is examined. The
first scheme employed a 3-class classification: low-
(L), medium- (M), and high-degree (H) of
vascularity. In particular, H denotes presence of
prominent superficial vessels, L denotes absence of
vessels or extensive fat coverage, and M denotes
moderate vascularity or/and fat coverage. Second, we
employed a 2-class classification so that all patches
classified as M, were reclassified as L or H,
depending on whether they are closer to one class or
BIOIMAGING 2020 - 7th International Conference on Bioimaging
30

the other. An overview of the surgeon’s annotations
for the two schemes is provided in Table 1.
Table 1: Ground-truth data statistics. Number of image-
patches per class for the two classification schemes.
L M H
3-class classification
188 133 204
2-class classification
262 - 263
2.2 Classification based on
Handcrafted Features
From each image patch we extracted a rich set of
color, edge, texture, and statistical features that
describe the image on a global level (Lux and
Marques, 2013),(Vallières et al., 2015). Color and
color-edge information was based on the improved
color coherence (Pass, et al., 1996), auto color
correlogram (Huang et al., 1997), color histogram,
and color edge magnitude/direction feature vectors.
To limit the number of colors, the RGB images were
first quantized to k = 32 colors based on a k-means
algorithm applied on the training set. This method
was preferred instead of the standard uniform color
quantization process, were the RGB color space is
divided into equal-sized partitions, as it was observed
that the colors of the images distribute across a
limited region of the entire RGB space. The improved
color coherence vector takes into account the size and
locations of the regions with a particular quantized
color, whereas the auto color correlogram counts how
often a quantized color finds itself in its immediate
neighborhood. The color edge magnitude and
direction histograms were based on the gradient of
each quantized color image plane using the Sobel
gradient operator.
Image texture and edge description was based on
the Histogram of oriented gradients (HOG) (Dalal
and Triggs, 2005), Tamura features (coarseness,
contrast and directionality) (Tamura et al., 1978), and
edge histogram descriptor (Vikhar and Karde, 2016),
extracted from the intensity component of the color
image.
Moreover, we extracted various statistical
features using the radiomics feature extraction
process, applied on the quantized color image
(Vallières et al., 2015). In particular, we extracted
global features such as variance, skewness and
kurtosis as well as statistical measures using higher-
order matrix-based texture types: GLCM (gray-level
co-occurrence matrix), GLRLM (gray-level run-
length matrix), GLSZM (gray-level size zone matrix)
and NGTDM (neighborhood gray-tone difference
matrix). Note that compared to the standard
calculation of these matrices on 2D images using 8-
heignboors connectivity, in our case each color image
is considered a 3D volume and thus the texture
matrices were determined by considering 26-
connected voxels (i.e. pixels were considered to be
neighbors in all 13 directions in three dimensions).
From each texture matrix various statistical features
were extracted such as: energy, contrast, etc.
(GLCM); short run emphasis, long run emphasis, etc.
(GLRLM); small zone emphasis, large zone
emphasis, etc. (GLSZM); complexity, strength, etc.
(NGTDM).
After feature extraction from each image patch,
we performed early fusion by combining all features
resulting in a feature vector with D = 900 dimensions.
Because the number of training image-patches was
less than D, a PCA for high-dimensional data
(Bishop, 2006) was applied leading to feature vectors
with dimensionality of 165 (accounting for ≥ 95% of
the overall variability).
For the classification step we employed 3
classifiers (Bishop, 2006): Support vector machines
(SVM), k-nearest neighbors (KNN) and Naïve Bayes
(NB). For SVM we employed the Gaussian kernel;
the hyperparameters to optimize were the box
constraint and kernel scale. For KNN the
hyperparameters were the distance function
(Euclidean, cosine and city block) and the number of
nearest neighbors. NB was based on a Gaussian
kernel and the hyperparameter was the kernel window
width. For each classifier the optimization was
performed on the validation set via a grid search (10
values for each measurable hyperparameter).
2.3 CNN based Classification
The proposed CNN model is shown in Table 2.
Overall we used 3 convolutional, 3 max-pooling and
3 fully connected (FC) layers. The convolution kernel
size was 3 × 3; the convolution filters were set to 16,
32, and 64. All convolutions had stride and padding
equal to 1. Each convolution layer was followed by
Batch Normalization (Ioffe and Szegedy, 2015) and a
rectified linear unit (ReLU) activation. For max-
pooling, the stride was 2 whereas the size of the first
and the other two layers was 2 × 2 and 3 × 3,
respectively. For the three FC layers, the number of
neurons was set to 512, 256 and C (number of
classes), respectively. The first two FC layers were
followed by ReLU activation, whereas the last FC by
a softmax function, which provided the class
probabilities. To prevent overfitting, a dropout layer
with probability p = 0.2 was used after each FC layer
(except the last one) (Srivastava et al., 2014). The
Assessment of Gallbladder Wall Vascularity from Laparoscopic Images using Deep Learning
31
weights of the convolutional filters and FC layers
were randomly initialized from a normal distribution
with zero mean and 0.01 standard deviation, whereas
all biases were set to zero.
Table 2: The employed CNN model. BN+ReLU denotes
Batch normalization followed by ReLU. ReLU+Drop
denotes ReLU followed by dropout with probability p.
C
denotes the number of classes (dataset dependent).
Layer Filter size Output W×H×K
Input - 70 × 70 × 3
Conv1 3 × 3 × 16 70 × 70 × 16
BN+ReLU - 70 × 70 × 16
Max-pool1 2 × 2 35 × 35 × 16
Conv2 3 × 3 × 32 35 × 35 × 32
BN+ReLU - 35 × 35 × 32
Max-pool2 3 × 3 17 × 17 × 32
Conv3 3 × 3 × 64 17 × 17 × 64
BN+ReLU - 17 × 17 × 64
Max-pool3 3 × 3 8 × 8 × 64
FC1 - 512
ReLU+Drop - 512
FC2 - 256
ReLU+Drop - 256
FC3 - C
Softmax - C
During training we adopted a batch size of 35 and
the Adam optimization with default parameters
(Kingma and Ba, 2014). The loss function was based
on the categorical cross-entropy:
1
,
log
(1)
where j ∊ {1,…,C} is the class index, y
i,j
∊ {0,1}
is the ground truth corresponding to class j and image
i, and φ(·) denotes the softmax output for the
activations of the FC3 layer. To reduce overfitting, an
L
2
regularization term (weight decay) with λ = 1e-4
was added to the loss function.
The model was trained either for 40 epochs or
until the loss on the validation set was larger than the
previously smallest loss for 5 evaluations. The
evaluation was performed every 10 iterations. The
initial learning rate was 0.001, which was dropped by
a factor of 0.1 after every 8 epochs. To increase the
dataset size and improve generalization of the model,
data augmentation was performed: horizontal flip,
vertical flip and rotation by ±30
o
and ±60
o
.
3 EXPERIMENTAL RESULTS
The image patch dataset was randomly split into
training (60%), validation (20%) and test set (20%),
based on a 5-fold cross validation. Specifically, the
image patches were randomly split into five parts of
equal size with the constraint to preserve the
frequency of each class among the folds. The
frequency of each class was also preserved among the
three sets in each fold. Each of the five folds was
selected as test-set and the other ones for training and
validation. The performance of the aforementioned
approaches was evaluated in terms of the following
metrics:
Acc = (TP + TN)/(P + N) (2)
Pre = TP/(TP + FP) (3)
Rec = TP/(TP + FN) (4)
F1 = 2 × Pre × Rec/(Pre + Rec) (5)
where Acc, Pre, Rec, F1 denote: Accuracy,
Precision, Recall, and F1-score, respectively; TP, TN,
FP, FN, P, N denote: true positives, true negatives,
false positives, false negatives, positives and
negatives, respectively. In addition, we computed the
area under curve (AUC) from the receiver operating
characteristic (ROC) plot (i.e. true positive rate =
TP/P vs. false positive rate = FP/N). The results are
presented as mean values across the 5 folds, unless
otherwise stated.
Table 3 shows the performance of the four
methods for the 2-class classification problem (i.e. L
vs. H). The performance metrics of all methods are ≥
91%. The CNN model achieves the best performance
across all metrics; its accuracy is 98% and a similar
value is observed for the mean Precision, Recall and
F1-score of the two classes (Table 4). SVM is the
second best method with ~2% lower performance.
KNN is ranked third (~94.4% mean performance) and
NB fourth (~92.2% mean performance). It is worth
noting that for all methods the Recall for the H class
is higher than that of the L class, which means that
high vascularity images are discriminated slightly
better. The Precision of the CNN for the two classes
is similar (~98%), so the model generates similar
false positives. For the other methods, the Precision
between the classes is mixed. Figure 3 shows the
normalized confusion matrices of the four methods.
The matrices were normalized after summation of the
raw confusion matrices of the test-folds. From NB to
CNN the performance rises with increasing on Recall
and decreasing on misclassification (false positives).
BIOIMAGING 2020 - 7th International Conference on Bioimaging
32
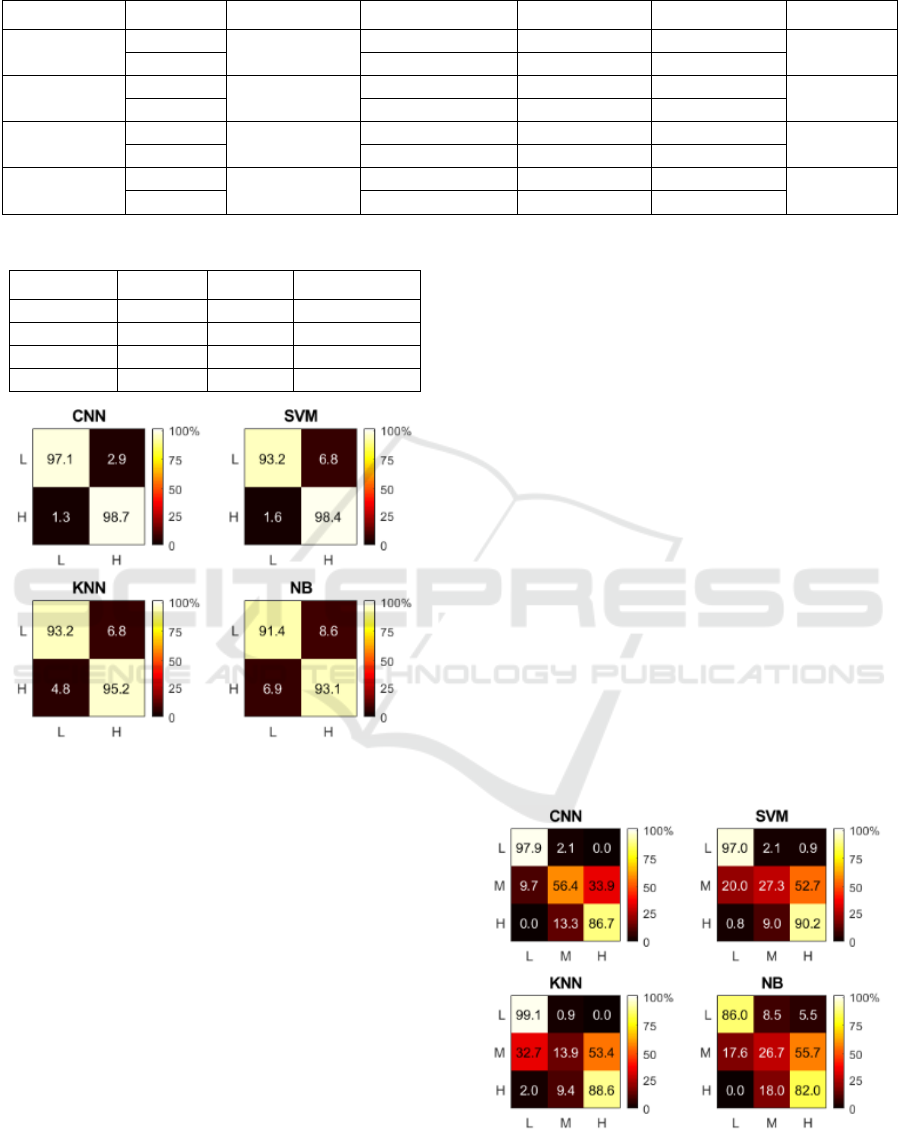
Table 3: Performance comparison for 2-class classification.
Table 4: Mean performance comparison for 2 classes.
Method Pre (%) Rec (%) F1 (%)
CNN 98.0 97.9 98.0
SVM 96.5 95.8 96.1
KNN 94.4 94.2 94.2
NB 92.3 92.2 92.2
Figure 3: Color-coded confusion matrices for 2-class
classification. The X and Y-axis represent predicted and
ground truth labels, respectively.
Table 5 shows the methods’ performance for the
ranking of the methods with respect to their
performance is the same as that in 2-class
classification (1
st
CNN, 2
nd
SVM, 3
rd
KNN and 4
th
NB), although in overall the methods’ performance is
lower, as expected. Specifically, the accuracy of the
CNN is 83.1% whereas the mean Precision, Recall
and F1-score of the three classes is 81.5%, 80.3% and
80.4%, respectively (Table 6).
From Table 5 it is observed that the Recall of the
L class is higher than that of the other two classes,
denoting better classification for the L class. The
same result is also valid for the Precision of the L
class, implying that misclassification of the other two
classes as L is lower. Moreover, for the L and H
classes the Recall is higher than Precision, denoting
that the methods generate less false negatives
compared to false positives. Among the three classes,
the best performance is yielded for the L class and the
worst for the M class. For the CNN model, the F1-
scores of the three classes are: 95.7% (L), 83.0% (H)
and 62.7% (M). It is worth noting that this
performance ranking is the same in all methods.
However, the performance of the SVM, KNN and NB
is much lower than CNN’s.
For the CNN model and the L class, it can be
noticed that compared to 2-class classification (Table
3), in 3-class classification (Table 5), the Precision
and F1 are only 4.6% and 2% lower respectively,
whereas the Recall is 0.8% higher. However, the
performance of the H class deteriorates considerably
(≥ 12%). Hence, it seems that the addition of the M
class has a negative impact on the classification of the
H class, mostly because samples between these two
classes are misclassified as one another. This may
also be noticed from Figure 4 that shows the
normalized confusion matrices. It is observed that: (a)
the CNN yields no confusion between L and H,
whereas in the other methods there is a slight
confusion, (b) the H class is more confused as M than
what the L class is, (c) the M class is mostly confused
Figure 4: Color-coded confusion matrices for 3-class
classification. The X and Y-axis represent predicted and
ground truth labels, respectively.
Method Class Acc (%) Pre (%) Rec (%) F1 (%) AUC
CNN
L
98.0
98.2 97.1 97.7
0.996
H 97.9 98.7 98.3
SVM
L
96.2
97.8 93.2 95.4
0.991
H 95.1 98.4 96.7
KNN
L
94.4
93.7 93.2 93.4
0.969
H 95.0 95.2 95.1
NB
L
92.4
91.0 91.4 91.1
0.977
H 93.7 93.1 93.3
Assessment of Gallbladder Wall Vascularity from Laparoscopic Images using Deep Learning
33
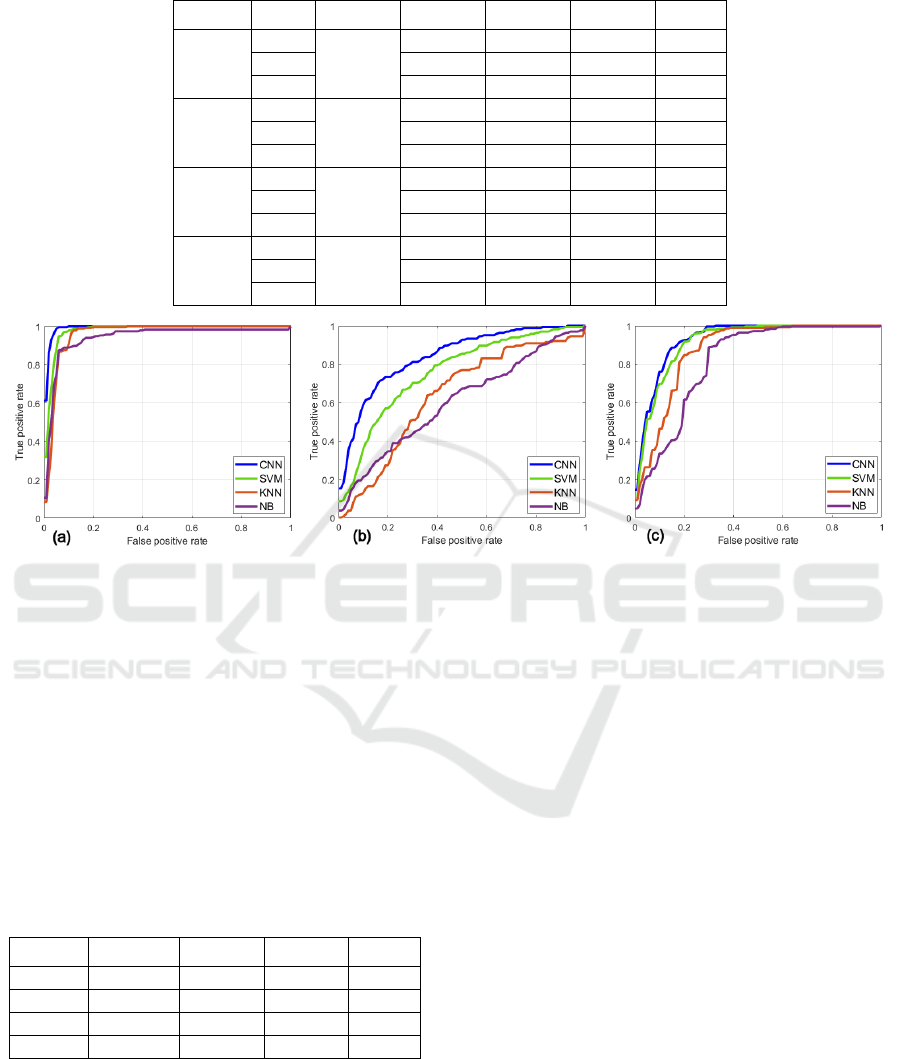
Table 5: Performance comparison for 3-class classification.
Method Class Acc (%) Pre (%) Rec (%) F1 (%) AUC
CNN
L
83.1
93.6 97.9 95.7
0.992
M 71.0 56.4 62.7
0.843
H 79.8 86.7 83.0
0.929
SVM
L
76.8
86.7 97.0 91.6
0.977
M 67.0 27.3 36.9
0.759
H 72.2 90.2 80.0
0.916
KNN
L
73.6
79.9 99.1 88.5
0.965
M 46.9 13.9 21.3
0.663
H 72.0 88.6 79.5
0.883
NB
L
69.5
87.3 86.0 86.4
0.947
M 38.5 26.7 31.1
0.605
H 67.3 82.0 73.6
0.845
Figure 5: The ROC curves for the: (a) L, (b) M and (c) H classes, respectively.
as H and this confusion is greater than all other
confusions, and (d) the confusion of the M class as H
is more than the confusion of H as M. Overall, CNN
outputs the fewest misclassifications across the three
classes.
Tables 5 and 6 also report the classes’ and class-
average AUC values, respectively. The mean ROC
curves for every class and method are depicted in
Figure 5. The CNN model yields the highest AUC
across all classes: 0.992 (L), 0.843 (M) and 0.929 (H),
something that may be also concluded from the ROC
curves.
Table 6: Mean performance comparison for 3 classes.
Method Pre (%) Rec (%) F1 (%)
AUC
CNN 81.5 80.3 80.4
0.921
SVM 75.3 71.5 69.5
0.884
KNN 66.3 67.2 63.1
0.837
NB 64.4 64.9 63.7
0.799
4 CONCLUSIONS
In this paper we present a novel idea for visual
assessment of the GB wall vascularity from
intraoperative LC images, based on machine learning.
To the best of our knowledge, the research work
presented in this paper is the first one that attempts to
investigate this application field. As described in the
Introduction, the vascular pattern of the GB wall
provides some clues about the GB condition, the
operation complexity, potentially need of extra
resources (e.g. advanced surgical skills) and generally
a means to characterize the operation. The
classification of the vascular pattern was based on
two alternative evaluations provided by an
experienced surgeon (2-class classification and 3-
class classification), as there is no established
consensus about the most appropriate vascularity
grading scheme from laparoscopic images. Our
results lead to the following conclusions.
First, the CNN model outperforms all other
methods both in 2-class and 3-class classification
(e.g. by ≥ 1.8% and ≥ 6.3% in accuracy, respectively).
Second, for all methods the best performance is
achieved in 2-class classification. Third, in 2-class
and 3-class classification, the CNN model yields a
very high performance for the L class (Pre ≥ 93.6%
and Rec ≥ 97.1%). Fourth, for the H class the
performance is very high in 2-class classification (Pre
= 97.9% and Rec = 98.7%), but lower in 3-class
BIOIMAGING 2020 - 7th International Conference on Bioimaging
34

classification (Pre = 79.8% and Rec = 86.7%). Fifth,
the inclusion of the M class mostly deteriorates the
performance of the H classification (e.g. for CNN:
13.3% confusion). Moreover, the M class is mostly
confused as H (33.9% for CNN), something that is
observed for all methods. Hence, the M class was the
most difficult one to recognize. A quantitative
measure of independent reviewers’ agreement on the
annotation of the M class would help evaluating the
difficulty of this task, or even whether the 3-class
classification scheme is indeed appropriate for our
application. In the future we aim to elaborate further
on this issue.
Given that this study investigates a novel
application in the area of computer assisted surgery,
there are still open issues for further research. First,
the results in this study are based on the ground truth
assessment
provided by a single expert. The
recruitment of additional experts is essential in order
to evaluate their level of agreement and most
importantly to establish the most appropriate
vascularity annotation scheme. Moreover, we aim to
expand our dataset by including more images from
additional LC operations. Second, the results are
based on classification of patches extracted from GB
images. Hence, it is important to extend the CNN
model to predict the vascular pattern of the entire GB
region in the laparoscopic image. A potential solution
would be to sequentially extract patches from a user-
specified GB region and then aggregate the CNN’s
patch predictions. The investigation of more
advanced CNN models, alternative loss functions to
penalize misclassifications of extreme classes, and
color preprocessing techniques for visual
enhancement of the GB wall vessels, are also major
topics of interest for future research work.
ACKNOWLEDGEMENTS
The author thanks Special Account for Research
Grants and National and Kapodistrian University of
Athens for funding to attend the meeting.
REFERENCES
Bishop, C.M., 2006. Pattern recognition and machine
learning. New York: Springer-Verlag New York, Inc.
Dalal, N. and Triggs, B., 2005. Histograms of Oriented
Gradients for Human Detection, In IEEE Computer
Society Conference on Computer Vision and Pattern
Recognition (CVPR’05), pp. 886–893.
Huang J. et al., 1997. Image indexing using color
correlograms, In Proceedings of IEEE Computer
Society Conference on Computer Vision and Pattern
Recognition. IEEE Comput. Soc, pp. 762–768.
Ioffe, S. and Szegedy, C., 2015. Batch Normalization:
accelerating deep network training by reducing internal
covariate shift, In Proceedings of the 32nd
International Conference on International Conference
on Machine Learning (ICML ’15), pp. 448–456.
Jin, A. et al., 2018. Tool detection and operative skill
assessment in surgical videos using region-based
convolutional neural networks, In IEEE Winter
Conference on Applications of Computer Vision
(WACV). Lake Tahoe, NV, USA, pp. 691–699.
Jin, Y. et al., 2019. Multi-task recurrent convolutional
network with correlation loss for surgical video
analysis. arXiv preprint. Available at:
http://arxiv.org/abs/1907.06099.
Kingma, D.P. and Ba, J, 2014. Adam: a method for
stochastic optimization. arXiv preprint. Available at:
http://arxiv.org/abs/1412.6980.
Loukas, C. et al., 2016. Shot boundary detection in
endoscopic surgery videos using a variational Bayesian
framework, International Journal of Computer Assisted
Radiology and Surgery, 11(11), pp. 1937–1949.
Loukas, C. et al., 2018. Keyframe extraction from
laparoscopic videos based on visual saliency detection,
Computer Methods and Programs in Biomedicine, 165,
pp. 13–23.
Loukas, C., 2018. Video content analysis of surgical
procedures, Surgical Endoscopy, 32(2), pp. 553–568.
Loukas, C. and Georgiou, E., 2013. Surgical workflow
analysis with Gaussian mixture multivariate
autoregressive (GMMAR) models: a simulation study,
Computer Aided Surgery, 18(3–4), pp. 47–62.
Loukas, C. and Georgiou, E., 2015. Smoke detection in
endoscopic surgery videos: a first step towards retrieval
of semantic events, International Journal of Medical
Robotics and Computer Assisted Surgery, 11(1), pp.
80–94.
Loukas, C. et al., 2011. The contribution of simulation
training in enhancing key components of laparoscopic
competence, The American Surgeon, 77(6), pp. 708-
715.
Lux, M. and Marques, O., 2013. Visual information
retrieval using Java and LIRE, Synthesis Lectures on
Information Concepts, Retrieval, and Services. Edited
by G. Marchionini. Morgan & Claypool.
Pass, G. et al., 1996. Comparing images using color
coherence vectors, In
Proceedings of the fourth ACM
international conference on Multimedia-
MULTIMEDIA ’96. New York, New York, USA: ACM
Press, pp. 65–73.
Petscharnig, S and Schöffmann, K., 2018. Binary
convolutional neural network features off-the-shelf for
image to video linking in endoscopic multimedia
databases, Multimedia Tools and Applications, 77(21),
pp. 28817–28842.
Assessment of Gallbladder Wall Vascularity from Laparoscopic Images using Deep Learning
35

Pontarelli, E.M. et al., 2019. Regional cost analysis for
laparoscopic cholecystectomy, Surgical Endoscopy,
33(7), pp. 2339–2344.
Srivastava, N. et al., 2014. Dropout: a simple way to prevent
neural networks from overfitting, Journal of Machine
Learning Research, 15(1), pp. 1929–1958.
Tamura, H., Mori, S. and Yamawaki, T., 1978. Textural
features corresponding to visual perception, IEEE
Transactions on Systems, Man, and Cybernetics, 8(6),
pp. 460–473.
Twinanda, A.P. et al., 2015. Classification approach for
automatic laparoscopic video database organization,
International Journal of Computer Assisted Radiology
and Surgery, 10(9), pp. 1449–1460.
Twinanda, A.P. et al., 2017. EndoNet: A deep architecture
for recognition tasks on laparoscopic videos, IEEE
Transactions on Medical Imaging, 36(1), pp. 86–97.
Twinanda, A.P. et al., 2019. RSDNet: learning to predict
remaining surgery duration from laparoscopic videos
without manual annotations, IEEE Transactions on
Medical Imaging, 38(4), pp. 1069–1078.
Vallières, M. et al., 2015. A radiomics model from joint
FDG-PET and MRI texture features for the prediction
of lung metastases in soft-tissue sarcomas of the
extremities, Physics in Medicine and Biology, 60(14),
pp. 5471–5496.
Vikhar, P. and Karde, P., 2016. Improved CBIR system
using Edge Histogram Descriptor (EHD) and Support
Vector Machine (SVM), In International Conference
on ICT in Business Industry & Government (ICTBIG).
IEEE, pp. 1–5.
BIOIMAGING 2020 - 7th International Conference on Bioimaging
36
