
Expression of HSA-MIR-155-5P and mRNA Suppressor of Cytokine
Signalling 1 (SOCS1) on Plasma at Early-stage and Late-stage of
Nasopharyngeal Carcinoma
Recita Indraswary
1a
, Sofia Mubarika Haryana
2b
Agus Surono
3c
1
Department of Molecular Medicine and Oral Biology, Faculty of Dentistry, Universitas Islam Sultan Agung, Semarang,
Indonesia
2
Post-Doctoral Program, Faculty of Medicine, Universitas Gadjah Mada, Yogyakarta, Indonesia
3
Department of Ear, Nose, Throat, Faculty of Medicine, Universitas Gadjah Mada, Yogyakarta, Indonesia
Keywords: Plasma, Nasopharyngeal carcinoma, Hsa-miR-155-5p, mRNA SOCS1
Abstract: Nasopharyngeal carcinoma (NPC) is a head and neck tumor with high prevalence and recurrence rate in Asia.
Accurate therapy based on carcinoma pathogenesis at molecular level is urgently needed. Overexpression of
hsa-miR-155-5p has been identified in various carcinomas include NPC. In previous in-silico research, hsa-
miR-155-5p are known to target mRNA SOCS1, as a transduction signal suppressor for gene transcription
activator.
This study aimed to analyze different expressions of hsa-miR-155-5p and mRNA SOCS1 on plasma
patients at an early and late stage of nasopharynx carcinoma. Hsa-miR-155-5p and mRNA SOCS1 were
isolated from blood sample plasma using miRCURY RNA Isolation Kit-Biofluid. cDNA synthesized using
cDNA Synthesis kit II, 8-64 rxns running by PCR thermal cycler (Bio-Rad c1000), and Real-time qPCR (Bio-
Rad CFX 96) for Hsa-miR-155-5p. mRNA SOCS1 was analyzed by One-Step qRT-PCR using KAPA™
SYBR® kit. Hsa-miR-155-5p expressions change was upregulated 1.13-fold (p value=0.713) on the late-stage
compared to the early stage, while for mRNA SOCS1 downregulated 1.1-fold (p value=0.891) on late-stage
compared with early stage. Hsa-mir-155-5p was overexpressed on late-stage nasopharynx carcinoma and
aligned with mRNA SOCS1 downregulated expression, compared to early stage. Deregulation of -miR-155-
5p dan mRNA SOCS1 may play important role in NPC progressivity.
1 INTRODUCTION
Nasopharyngeal carcinoma (NPC) is a head and neck
tumor with a high prevalence and recurrence in Asia,
commonly in men (Wah et al., 2014; Wildeman et al.,
2013). A better understanding of the molecular basis
of tumorigenesis does improve clinical outcomes and
useful to develop early detection, for more accurate
prognosis, and developing cancer individualized
therapy (Kong et al., 2012). miRNAs offer great
potential as biomarkers for cancer detection because
of their remarkable stability in blood and their
characteristic expression in different diseases
MicroRNAs (miRNAs) are a family of small
non-coding RNA molecules with 20–23 nucleotides
a
https://orcid.org/0000-0003-1871-5333
b
https://orcid.org/0000-0001-7205-625X
c
https://orcid.org/0000-0003-3363-4563
in length. It has function to negatively regulate
protein-coding genes at the post-transcriptional level
by mRNA uncapping and deadenylation. This process
will lead to increased mRNA turnover and decreased
target gene expression (Abba et al., 2014; Calin &
Croce, 2006; Koturbash et al., 2011; Lu et al., 2014;
Vishwamitra et al., 2012). Deregulation of
microRNAs (miRNAs) is indicated in several
conditions such as inflammation, cancer development
and tumor progression (Calin & Croce, 2006; Y.
Huang et al., 2013; X. Liu et al., 2009; Sotiropoulou
et al., 2009; Tang et al., 2012; H. Zheng et al., 2013;
S.-R. Zheng et al., 2012).
Hsa-miR-155-5p is prominent in cancer biology.
Among microRNAs that have been linked to cancer,
it is the most commonly overexpressed in human
Indraswary, R., Haryana, S. and Surono, A.
Expression of HSA-MIR-155-5P and mRNA Suppressor of Cytokine Signalling 1 (SOCS1) on Plasma at Early-stage and Late-stage of Nasopharyngeal Carcinoma.
DOI: 10.5220/0010488000750080
In Proceedings of the 1st Jenderal Soedirman International Medical Conference in conjunction with the 5th Annual Scientific Meeting (Temilnas) Consortium of Biomedical Science Indonesia
(JIMC 2020), pages 75-80
ISBN: 978-989-758-499-2
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
75

malignancies (Du et al., 2011; Jiang, Zhang, Lu, He,
Li, Gu, et al., 2010; Palma et al., 2014; Zhang et al.,
2013). Those miRNAs that lead to tumorigenesis and
cancer are classified as oncomiRs. These oncomiRs
are not only therapeutic targets but also important
biomarkers for cancer detection and management (Du
et al., 2011).
Cytokines activate multiple intracellular
signaling pathways to produce their physiological
effects. One of the most studied pathways is involving
the receptor-associated janus kinases (JAKs) and the
latent cytoplasmic transcription factors signal
transducers and activators of transcription (STATs).
Suppressors of Cytokine Signalling1 (SOCS1) is a
negative regulator for STAT3. Activation of STAT3
will induce transcription of several genes which has a
role in oncogeneses such as cell proliferation,
differentiation, invasion, and angiogenesis (Y. Huang
et al., 2013; Jiang, Zhang, Lu, He, Li, Gu, et al.,
2010). Studies have proved that hsa-miR-155-5p
target mRNA SOCS1(L. Liu et al., 2013). Even
ribonuclease was found in human plasma and
miRNAs have proven stable in blood plasma covering
by lipid complex or lipoproteins like an apoptotic
body, micro vesicle, or exosomes (Du, 2012; Kim et
al., 2012; H. Zheng et al., 2013). This makes miRNAs
has exciting prospect as powerful and minimal-
invasive cancer biomarkers using plasma sample.
Deregulation expressions of Hsa-miR-155-5p and
mRNA SOCS1 can monitor disease progression and
treatments. This research aimed to analyze difference
expressions of hsa-miR-155-5p and mRNA SOCS1
on plasma at early and late stage of NPC.
2 MATERIALS AND METHODS
First, the insilico study was done by bioinformatic
analysis through non-profit database of microRNA-
based on notation sequences, such as miRbase,
miRTarbase, Diana miR-Path and microRNAMap.
The result shows that mRNA SOCS1 is one of hsa-
miR-155-5p with mRNAs targets. Hsa-miR-155-5p
binding site complementary occurs in along 401
nucleotides 3' untranslated region (UTR), at 15-34
nucleotides withal Minimum Free Energy (MFE) -
15,50 kj/mol, or 218-242 nucleotides withal MFE -
15,90 kj/mol, and also possible binding at nucleotides
404-425 withal MFE -9,13 kj/mol (figure1).
This initial study included 37 nasopharyngeal
carcinoma patients of early stage and late stage (Table
1). samples were obtained from Venous blood. Hsa-
miR-155-5p and mRNA SOCS1 were isolated from
blood sample plasma using miRCURY RNA
Isolation Kit-Biofluid. cDNA was synthesized using
cDNA Synthesis kit II, 8-64 rxns running by PCR
thermal cycler (Bio-Rad c1000), and Real-time qPCR
(Bio-Rad CFX 96) for Hsa-miR-155-5p. mRNA
SOCS1 was analyzed by One-Step qRT-PCR using
KAPA™ SYBR® kit. The Ct value for each product
was determined. MiR-16 was used as a reference gene
for has-miR-155-5p quantification, and mRNA Beta
Actin was used as a reference for mRNA SOCS1.
Relative quantification for transcript accumulation
was performed according to the comparative Livak
Method: 2
-∆∆Cq
.
Figure 1: hsa-miR-155-5p several binding site on 3’UTR
region mRNA SOCS1.
Table 1: Characteristics of samples.
Characteristics n
(
%
)
Sta
g
es
Earl
y
6
(
16
)
Late 31 (84)
Sex
Men 27 (73)
Women 10 (27)
Age
20-30 5
(
13.5
)
31-40 10
(
27
)
41-50 11
(
30
)
51-60 6 (16)
61-71 5 (13.5)
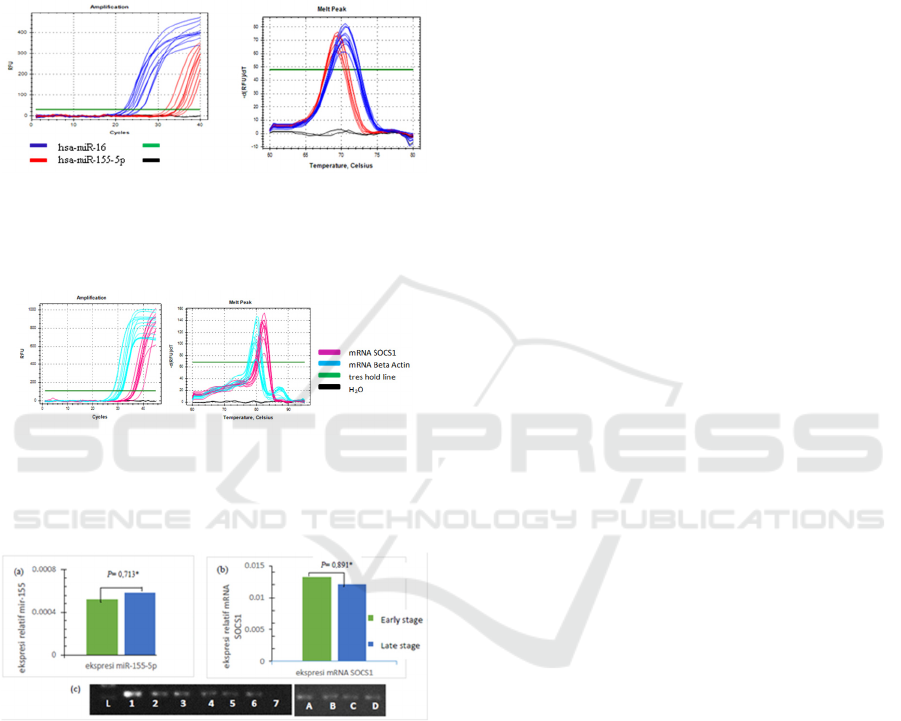
3 RESULTS
Quantitative real-time PCR was performed to
evaluate hsa-miR-155-5p and mRNA SOCS1
expressions in 37 NPC patients. From qRT-PCR
examination, the amplification of hsa-miR-155-5p
using hsa-miR-16 as a reference gene on samples
were occurring (figure 2a). No shift in peak melt
curve was validating product specificity (figure 2b).
Specific results mRNA SOCS1 obtained using
the kit KAPA ™ SYBR® One-Step qRT-PCR (figure
3), where the optimal primer concentration at 10 pmol
with annealing temperature 59.4
o
C.
Hsa-miR-155-5p and mRNA SOC1 expressions
data quantifying were anlyzed statistically to
determine whether the data were normally
TGACCGGCAGCGCCC
G
CCGTGCACGCAGCATTAACTGGGATGCCGTGTTATTTT
GTTATTACTTGCCTGGAACCATGTGGGTACCCTCCCCGGCCTGGGTTGGAGGGA
GCGGATGGGTGTAGGGGCGAGGCGCCTCCCGCCCTCGGCTGGAGACGAGGCCGC
AGACCCCTTCTCACCTCTTGAGGGGGTCCTCCCCCTCCTGGTGCTCCCTCTGGG
TCCCCCTGGTTGTTGTAGCAGCTTAACTGTATCTGGAGCCAGGACCTGAACTCG
CACCTCCTACCTCTTCATGTTTACATATACCCAGTATCTTTGCACAAACCAGGG
GTTGGGGGAGGGTCTCTGGCTTTATTTTTCTGCTGTGCAGAATCCTATTTTATA
T
TTTTTAAAGTCAGTTTAGGTAATAAACTTTATTATGAAAGTTTTTTTTTT
JIMC 2020 - 1’s t Jenderal Soedirman International Medical Conference (JIMC) in conjunction with the Annual Scientific Meeting
(Temilnas) Consortium of Biomedical Science Indonesia (KIBI )
76
distributed. Shapiro-Wilk test results indicate that the
data quantifying of expression of hsa-miR-155-5p
and mRNA SOCS1 both early-stage and late-stage
normally distributed, Furthermore, the value of ΔΔCq
hsa-miR-155-5p with hsa-miR-16 (table 2) and
mRNA SOCS1 with Beta-actin mRNA (Table 3)
analyzed used Livak method; Fold change = 2
-∆∆Cq
.
Figure 2: (a) hsa-miR-155-5p and miR-16 amplification
curves on plasma NPC, analyzed by Bio-Rad CFX
ManagerTM Software. (b) hsa-miR-155-5p and hsa-miR-
16 melt peak.
Figure 3: (a) mRNA SOCS1 and mRNA Beta-actin
amplification curves done with one-step qRT-PCR using
KAPA ™ SYBR® One-Step qRT-PCR kit. (b) mRNA
SOCS1 and mRNA. Beta-actin Melt peak.
Figure 4. (a) Hsa-miR-155-5p expressions increased 1.13-
fold. (b) mRNA SOCS1 expressions decreased 1.1-fold.
Plasma samples of early-stage NPC compared with late-
stage NPC (c) mRNA SOCS1 electrophoresis result on
Agarose gel. L = Ladder (100bp), 1-3 = SOCS1 NPC early
stage, 4-6 = SOCS1 NPC late-stage, 7 = negative control.
A and B = β-actin early-stage NPC, C dan D = β-actin late-
stage NPC.
Bio-Rad CFX Manager™ Software was used to
analyzed qRT-PCR result. The results showed that
hsa-miR-155-5p expressions fold change was
upregulated 1.13 on the late-stage compared with
early-stage (figure 4a). For mRNA SOCS1, it was
downregulated 1.1-fold on late stage compared with
early stage (figure 4b). qRT-PCR results were
subsequently confirmed by electrophoresis. This was
done using 2% Agarose gel electrophoresis, running
by 100 volts for 30 minutes to determine the
specificity and consistency of SOCS1 results of qRT-
PCR (figure 4c).
4 DISCUSSION
This study was aimed to analyze different expressions
of hsa-miR-155-5p and mRNA SOCS1 on plasma
patients at an early and late stage of NPC. Livak
method result showed that hsa-miR-155-5p were
overexpressed in late-stage NPC plasma compared
with early-stage NPC.
Overexpression’s of hsa-miR-155-5p possibly
aligned with NPC progressions. Hsa-miR-155-5p is a
micro RNA that has been shown to be associated with
the progression of carcinoma in human (Li et al.,
2012; L. Liu et al., 2013).
MiR-155 overexpression is
also correlated with poor prognosis in cancer (H.
Zheng et al., 2013). It also had been proven that miR-
155 expressions in breast cancer patients serum were
associated with clinical stages (Rawlings et al., 2004).
mRNA SOCS1 expressions downregulated in
the NPC plasma late stage compared to the early-
stage. SOCS1 mRNA may be targeted by hsa-miR-
155-5p. Other studies also proved there is a negative
correlation between the expression level of miR-155
and SOCS1 (C. Huang et al., 2013). The results
showed expression of SOCS1 decreased by 1.1-fold
in the NPC plasma late stage compared to the early
stage. Downregulated of SOCS1 may increase
STAT3 expression. SOCS1 is a negative feedback
regulator on the JAK / STAT pathway and these
pathways stimulate cells to proliferate, migrate, or
apoptosis (Chen et al., 2013).
STAT3 plays an important role in carcinoma
invasion by inducing inflammatory pro-oncogenic
pathways and tumor cell immunity, including nuclear
factor-kappaB (NF-kappaB) and interleukin-6 (IL-6)
-GP130-JAK pathway. SOCS1 acts as a negative
regulator of this pathway by inhibiting STAT3
phosphorylation (Jiang, Zhang, Lu, He, Li, & Gu,
2010). The antiproliferative mechanism is carried by
SOCS1 through inhibition of JAK2 kinase activity
Expression of HSA-MIR-155-5P and mRNA Suppressor of Cytokine Signalling 1 (SOCS1) on Plasma at Early-stage and Late-stage of
Nasopharyngeal Carcinoma
77
Table 2. Hsa-miR-155-5p and miR-16 expressions in plasma NPC early-stage and late-stage.
NPC Plasma Targets Mean (∆Cq) ±
Deviation
∆∆Cq Fold change
(2
-∆∆Cq
)
Early stage Hsa-miR-155-5p 10.92 ± 2.26 -0.175 1.13
Hsa-miR-16
Late sta
g
e Hsa-miR-155-5
p
10.74 ± 1.89
Hsa-miR-16
Table 3. mRNA SOCS1 and mRNA Beta-actin expressions in plasma early stage and late stage of NPC.
NPC plasma Targets Mean (∆Cq) ±
Deviation
∆∆Cq 2
-∆∆Cq
Fold change
-1/2
-∆∆Cq
Earl
y
sta
g
e mRNA SOCS1 6.24± 2.15 0.14 0.91 -1.102
mRNA BA
Late sta
g
e mRNA SOCS1 6.38± 2.42
mRNA BA
which prohibit the activation (phosphorylation) of
STAT3 (Yoshimura et al., 2012).
There have been many studies proving that
STAT3 is an oncogene in various malignancies. Hsa-
miR-155-5p overexpressed in Human Laryngeal
Squamous Cell Carcinoma late stage compared to
early-stage followed by downregulated of mRNA
SOCS1 expressions and overexpression of STAT3 in
late stage. Other studies have shown a decrease of
SOCS1 expressions in colonic adenocarcinoma
progression and SOCS1 was found very low
expressed on the late stage (David et al., 2014).
Until now the NPC classifications are based on
TNM (Tumor, Node, Metastasis) as defined by the
American Joint Committee on Cancer (AJCC) (Lee et
al., 2004). On the late stage, TNM has a higher value
than the early stage, reflecting the progression of
cancer cell growth exceeded from the early stage.
Hsa-miR-155-5p has an important role in tumor
progression related to its ability to modulate
epithelial-to-mesenchymal transition (EMT)
(Bouyssou et al., 2014). EMT is a cell program, where
epithelial cells transformed into mesenchymal cells
characterized by loss of polarity, adhesion loss,
motility, and increased potential ability. Cells able to
move as metastasis incidence of malignancy.
Knockdown on miR-155 resulted in the increase of
SOCS1 and STAT3 decreases in malignancy. This is
consistent with the results obtained, in which the hsa-
miR-155-5p expressions increased in the late-stage
followed by SOCS1 mRNA expression decreased.
5 CONCLUSIONS
The identification of carcinoma molecular
mechanisms is substantial to obtain successful
therapy. Plasma samples are feasible as media in NPC
molecular examinations as shown by the results
similarity in pattern with the tissues itself. Hsa-miR-
155-5p and mRNA SOCS1 were proven stable in
NPC blood plasma and able to be quantitatively
analysed. This research shows that hsa-mir-155-5p
was overexpressed on late-stage nasopharynx
carcinoma, aligned with mRNA SOCS1
downregulated expression, compared to the early
stage. Therefore, deregulation of -miR-155-5p dan
mRNA SOCS1 are believed to have a significant role
in NPC progressivity.
ACKNOWLEDGEMENTS
The authors thank the DIKTI funding research for
providing financial support of this study. The authors
thank to Universitas Gadjah Mada, Universitas Islam
Sultan Agung, Prof. Dr. dr. Teguh Aryandono Sp.B
(K) Onk, Dr. Med. dr. Indwiani Astuti, Prof. Dr.
Mustafa, Apt., Kes, dr. Ahmad Hamim Sadewa,
Ph.D., Drs. Zulaela, Dipl. Med. Stat., M.Si, Dr. dr.
Cita Herawati, Risky Oktriani, S. Si, M. Biotech, dr.
Sumadi Lukman A., Ph.D., dr. Zulrachman,
Nihayatus Saadah, S. Si, and the entire members of
the study group microRNA (genomiR) for their
support to this research.
JIMC 2020 - 1’s t Jenderal Soedirman International Medical Conference (JIMC) in conjunction with the Annual Scientific Meeting
(Temilnas) Consortium of Biomedical Science Indonesia (KIBI )
78

REFERENCES
Abba, M., Patil, N., & Allgayer, H. (2014). MicroRNAs in
the Regulation of MMPs and Metastasis. 625–645.
https://doi.org/10.3390/cancers6020625
Bouyssou, J. M. C., Manier, S., Huynh, D., Issa, S.,
Roccaro, A. M., & Ghobrial, I. M. (2014). Regulation
of microRNAs in cancer metastasis. Biochimica et
Biophysica Acta, 1845(2), 255–265.
https://doi.org/10.1016/j.bbcan.2014.02.002
Calin, G. a, & Croce, C. M. (2006). MicroRNA signatures
in human cancers. Nature Reviews. Cancer, 6(11), 857–
866. https://doi.org/10.1038/nrc1997
Chen, Y., Lan, Q., Zheng, T., Zhao, N., Holford, T. R.,
Lerro, C., Dai, M., Huang, H., Liang, J., Ma, S.,
Leaderer, B., Boyle, P., Chanock, S., Rothman, N., &
Zhang, Y. (2013). Polymorphisms in JAK/STAT
Signaling Pathway Genes and Risk of Non-Hodgkin
Lymphoma. Leuk Res, 37(9), 1120–1124.
https://doi.org/10.1016/j.leukres.2013.05.003.Polymor
phisms
David, M., Naudin, C., Letourneur, M., Polrot, M., Renoir,
J.-M., Lazar, V., Dessen, P., Roche, S., Bertoglio, J., &
Pierre, J. (2014). Suppressor of cytokine signaling 1
modulates invasion and metastatic potential of
colorectal cancer cells. Molecular Oncology, 8(5), 942–
955. https://doi.org/10.1016/j.molonc.2014.03.014
Du, Z. (2012). Biomarkers in Nasopharyngeal Carcinoma.
Karolinska Institutet, Stockholm, Sweden.
Du, Z., Hu, L., Wang, H., Yan, L., Zeng, Y., & Shao, J.
(2011). Upregulation of MiR-155 in Nasopharyngeal
Carcinoma is Partly Driven by LMP1 and LMP2A and
Downregulates a Negative Prognostic Marker
JMJD1A. 6(4).
https://doi.org/10.1371/journal.pone.0019137
Huang, C., Li, H., Wu, W., Jiang, T., & Qiu, Z. (2013).
Regulation of miR-155 affects pancreatic cancer cell
invasiveness and migration by modulating the STAT3
signaling pathway through SOCS1. Oncology Reports,
30(3), 1223–1230.
https://doi.org/10.3892/or.2013.2576
Huang, Y., Yang, Y. B., Zhang, X. H., Yu, X. L., Wang, Z.
Bin, & Cheng, X. C. (2013). MicroRNA-21 gene and
cancer. Med Oncol, 30(376).
https://doi.org/10.1007/s12032-012-0376-8
Jiang, S., Zhang, H.-W., Lu, M.-H., He, X.-H., Li, Y., Gu,
H., Liu, M.-F., & Wang, E.-D. (2010). MicroRNA-155
functions as an OncomiR in breast cancer by targeting
the suppressor of cytokine signaling 1 gene. Cancer
Research, 70(8), 3119–3127.
https://doi.org/10.1158/0008-5472.CAN-09-4250
Jiang, S., Zhang, H., Lu, M., He, X., Li, Y., & Gu, H.
(2010). MicroRNA-155 Functions as an OncomiR in
Breast Cancer by Targeting the Suppressor of Cytokine
Signaling 1 Gene. 6, 3119–3128.
https://doi.org/10.1158/0008-5472.CAN-09-4250
Kim, D.-J., Linnstaedt, S., Palma, J., Park, J. C., Ntrivalas,
E., Kwak-Kim, J. Y. H., Gilman-Sachs, A., Beaman,
K., Hastings, M. L., Martin, J. N., & Duelli, D. M.
(2012). Plasma components affect accuracy of
circulating cancer-related microRNA quantitation. The
Journal of Molecular Diagnostics : JMD, 14(1), 71–80.
https://doi.org/10.1016/j.jmoldx.2011.09.002
Kong, Y. W., Ferland-McCollough, D., Jackson, T. J., &
Bushell, M. (2012). microRNAs in cancer
management. The Lancet Oncology, 13(6), e249-58.
https://doi.org/10.1016/S1470-2045(12)70073-6
Koturbash, I., Zemp, F. J., Pogribny, I., & Kovalchuk, O.
(2011). Small molecules with big effects: the role of the
microRNAome in cancer and carcinogenesis. Mutation
Research, 722(2), 94–105.
https://doi.org/10.1016/j.mrgentox.2010.05.006
Lee, A. W. M., Au, J. S. K., Teo, P. M. L., Leung, T. W.,
Chua, D. T. T., Sze, W. M., Zee, B. C. Y., Oncology,
C., Hospital, Q. M., & Kong, H. (2004). Staging of
Nasopharyngeal Carcinoma : Suggestions for
Improving the Current UICC / AJCC Staging System 1.
Clin Oncol (R Coll Radiol), 269–276.
https://doi.org/10.1016/j.clon.2004.01.008
Li, S., Chen, T., Zhong, Z., Wang, Y., Li, Y., & Zhao, X.
(2012). microRNA-155 silencing inhibits proliferation
and migration and induces apoptosis by upregulating
BACH1 in renal cancer cells. Molecular Medicine
Reports, 5(4), 949–954.
https://doi.org/10.3892/mmr.2012.779
Liu, J., Mao, Q., Liu, Y., Hao, X., Zhang, S., & Zhang, J.
(2013). Analysis of miR-205 and miR-155 expression
in the blood of breast cancer patients. Chin J Cancer
Res, 25(1), 46–54. https://doi.org/10.3978/j.issn.1000-
9604.2012.11.04
Liu, L., Li, W., Wei, X., Cui, Q., Lou, W., Wang, G., Hu,
X., & Qian, C. (2013). Potent antitumor activity of
oncolytic adenovirus-mediated SOCS1 for
hepatocellular carcinoma. Gene Therapy, 20(1), 84–92.
https://doi.org/10.1038/gt.2012.4
Liu, X., Chen, Z., Yu, J., Xia, J., & Zhou, X. (2009).
MicroRNA Profiling and Head and Neck Cancer. 2009.
https://doi.org/10.1155/2009/837514
Lu, J., Xu, X., Liu, X., Peng, Y., Zhang, B., Wang, L., Luo,
H., Peng, X., Li, G., Tian, W., He, M., & Li, X. (2014).
Predictive value of miR-9 as a potential biomarker for
nasopharyngeal carcinoma metastasis. British Journal
of Cancer, 110(2), 392–398.
https://doi.org/10.1038/bjc.2013.751
Palma, C. A., Sheikha, D. Al, Lim, T. K., Bryant, A., Vu,
T. T., Jayaswal, V., & Ma, D. D. F. (2014). MicroRNA-
155 as an inducer of apoptosis and cell differentiation
in Acute Myeloid Leukaemia. 1–15.
Rawlings, J. S., Rosler, K. M., & Harrison, D. a. (2004).
The JAK/STAT signaling pathway. Journal of Cell
Science, 117(Pt 8), 1281–1283.
https://doi.org/10.1242/jcs.00963
Sotiropoulou, G., Pampalakis, G., Lianidou, E. V. I., &
Mourelatos, Z. (2009). Emerging roles of microRNAs
as molecular switches in the integrated circuit of the
cancer cell. 1443–1461.
https://doi.org/10.1261/rna.1534709.RNA
Tang, J., Ahmad, A., & Sarkar, F. H. (2012). The Role of
MicroRNAs in Breast Cancer Migration, Invasion and
Metastasis. International Journal of Molecular
Expression of HSA-MIR-155-5P and mRNA Suppressor of Cytokine Signalling 1 (SOCS1) on Plasma at Early-stage and Late-stage of
Nasopharyngeal Carcinoma
79

Sciences, 13(10), 13414–13437.
https://doi.org/10.3390/ijms131013414
Vishwamitra, D., Li, Y., Wilson, D., Manshouri, R., Curry,
C. V, Shi, B., Tang, X. M., Sheehan, A. M., Wistuba, I.
I., Shi, P., & Amin, H. M. (2012). MicroRNA 96 is a
post-transcriptional suppressor of anaplastic lymphoma
kinase expression. The American Journal of Pathology,
180(5), 1772–1780.
https://doi.org/10.1016/j.ajpath.2012.01.008
Wah, S., Ling, Y., Man, C., Shin, P., Ming, V., Lau, Y.,
Zhang, G., & Wai, K. (2014). Etiological factors of
nasopharyngeal carcinoma. Oral Oncology, 50(5),
330–338.
https://doi.org/10.1016/j.oraloncology.2014.02.006
Wildeman, M. a, Fles, R., Herdini, C., Indrasari, R. S.,
Vincent, A. D., Tjokronagoro, M., Stoker, S.,
Kurnianda, J., Karakullukcu, B., Taroeno-Hariadi, K.
W., Hamming-Vrieze, O., Middeldorp, J. M.,
Hariwiyanto, B., Haryana, S. M., & Tan, I. B. (2013).
Primary treatment results of Nasopharyngeal
Carcinoma (NPC) in Yogyakarta, Indonesia. PloS One,
8(5), e63706.
https://doi.org/10.1371/journal.pone.0063706
Yoshimura, A., Suzuki, M., Sakaguchi, R., Hanada, T., &
Yasukawa, H. (2012). SOCS, Inflammation, and
Autoimmunity. Frontiers in Immunology, 3(March),
20. https://doi.org/10.3389/fimmu.2012.00020
Zhang, C., Zhao, J., & Deng, H. (2013). MiR-155 promotes
proliferation of human breast cancer MCF-7 cells
through targeting tumor protein 53-induced nuclear
protein 1. Journal of Biomedical Science, 20(1), 1.
https://doi.org/10.1186/1423-0127-20-79
Zheng, H., Zhang, L., Zhao, Y., Yang, D., Song, F., Wen,
Y., & Hao, Q. (2013). Plasma miRNAs as Diagnostic
and Prognostic Biomarkers for Ovarian Cancer. 8(11).
https://doi.org/10.1371/journal.pone.0077853
Zheng, S.-R., Guo, G.-L., Zhang, W., Huang, G.-L., Hu, X.-
Q., Zhu, J., Huang, Q.-D., You, J., & Zhang, X.-H.
(2012). Clinical significance of miR-155 expression in
breast cancer and effects of miR-155 ASO on cell
viability and apoptosis. Oncology Reports, 27(4),
1149–1155. https://doi.org/10.3892/or.2012.1634
JIMC 2020 - 1’s t Jenderal Soedirman International Medical Conference (JIMC) in conjunction with the Annual Scientific Meeting
(Temilnas) Consortium of Biomedical Science Indonesia (KIBI )
80
