
Immunohistochemistry of KRAS Protein in Colorectal Cancer
Dody Novrial
1a
, Kamal Agung Wijayana
2b
and Hanif Kun Cahyani
2c
1
Department of Anatomical Pathology, Faculty of Medicine,Universitas Jenderal Soedirman, Purwokerto, Indonesia
2
Department of Surgery, Faculty of Medicine, Universitas Jenderal Soedirman Purwokerto, Indonesia
3
Faculty of Medicine, Universitas Jenderal Soedirman Purwokerto, Indonesia
Keywords: Ras gene aberration, KRAS mutation, Colorectal cancer.
Abstract: PCR assays are used for the detection of KRAS mutations in colorectal cancer (CRC). However, there are
some disadvantages in the clinical application such as the high-cost value and specific codon properties of
this method. Our study aimed to evaluate the effectivity of immunohistochemistry (IHC) in assessing ras
gene aberration in CRC before PCR testing. Paraffin-embedded tissue samples from 45 CRC patients were
stained immunohistochemically using anti-KRAS protein polyclonal antibody. KRAS protein expression was
assessed and correlated with clinicopathological features. A chi-square test was performed to evaluate the
results statistically. KRAS protein positivity was observed in 31.1% of cases. The positivity was correlated
with female predominance (p=0.03). No significant correlation between KRAS protein expression with age,
tumor topography, lymph node metastases, TNM staging, and tumor differentiation. In conclusion, IHC was
regarded as cost- effective, non-specific for codons, and may complement PCR in the detection of KRAS gene
mutation.
1 INTRODUCTION
Colorectal cancer (CRC) is one of the most common
cancer worldwide and is the second leading cause of
cancer deaths (Jemal et al., 2011). Sporadic CRC
frequently arises through the activation of oncogenes
such as KRAS and BRAF as well as inactivation of
tumor suppressor and mismatch repair genes
(Raskov, Pommergaard, Burcharth, & Rosenberg,
2014). KRAS mutation is one of the first alterations
that occur in colorectal tumorigenesis (Rajagopalan et
al., 2002). Mutation of KRAS occurs approximately in
14%-50% of CRC and frequently detected in codon
12 and codon 13 (Sammoud et al., 2012).
Anti-epidermal growth factor receptor (EGFR)
antibody such as cetuximab and panitumumab has
been approved for CRC treatment. It engaged with the
extracellular domain of EGFR, blocks ligand binding,
and leads to inhibition of downstream RAS-RAF-
MEK-ERK signaling pathway (Porru, Pompili,
Caruso, Biroccio, & Leonetti, 2018). However, this
therapy is not recommended for use in patients with
a
https://orcid.org/0000-0002-3807-852X
b
https://orcid.org/0000-0002-2870-5358
c
https://orcid.org/0000-0002-4333-8701
KRAS mutation because of anti-EGFR antibody
resistance (De Roock et al., 2010). Therefore, KRAS
status becomes an important biomarker for patient
selection.
PCR is an established assay for KRAS mutation
detection since this analysis has favorable sensitivity
even in samples with low tumor cells (Cree, 2016).
However, there are some weaknesses of PCR assays
in the clinical setting such as high-cost value and
specific codon properties of this method.
Immunohistochemistry (IHC) has been suggested as
the prior screening method before genetic testing
(Wan Juhari et al., 2015). It has been a part of routine
service in most of the pathology laboratories which
not as expensive as molecular detection screening.
In the detection of MMR defects, IHC showed
high sensitivity and specificity compared to
molecular MSI-testing (Shia, 2008). However, in the
screening of ras gene aberration, several previous
studies revealed conflicting results (Elsabah & Adel,
2013; Piton, Borrini, Bolognese, Lamy, & Sabourin,
2015)
.
In this study, we evaluated the
Novrial, D., Wijayana, K. and Cahyani, H.
Immunohistochemistry of KRAS Protein in Colorectal Cancer.
DOI: 10.5220/0010487500470051
In Proceedings of the 1st Jenderal Soedirman International Medical Conference in conjunction with the 5th Annual Scientific Meeting (Temilnas) Consortium of Biomedical Science Indonesia
(JIMC 2020), pages 47-51
ISBN: 978-989-758-499-2
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
47
immunoexpression of KRAS proteins in CRC. We
discuss the result of KRAS immunohistochemistry
and its
relationship with some of the
clinicopathological features.
2 MATERIALS AND METHODS
This study was approved by the Research Ethics
Committee of Faculty of Medicine Universitas
Jenderal Soedirman. A total of 45 Formalin-fixed
paraffin-embedded (FPPE) tissue blocks of CRC
patients underwent colectomy were collected from
Margono Soekarjo Hospital and private pathology
laboratory from January to December 2019.
FPPE tissue blocks were subjected to staining by
immunohistochemical assays. Sections of FPPE
tissue blocks (4 µm) were transferred to positively
charged slides. Then, they were subjected to
deparaffinized, rehydration, and blocking with
hydrogen peroxide and antigen retrieval (Dako target
retrieval solution, citrate buffer pH 6.0) at 100
0
C for
10 minutes. After a short rinsed in phosphate-
buffered saline (PBS), slides then incubated overnight
at room temperature with primary antibody against
KRAS oncoprotein (orb53139 KRAS polyclonal
antibody: dilution 1:100, UK). Slides were washed
with PBS, then incubated for 30 minutes with labeled
secondary antibody. Product visualization (Dako)
was performed with diaminobenzidine substrate as
the chromogen. The slides were counterstained with
Mayer’s hematoxylin and washed once each with
distilled water and PBS. Finally, slides dehydrated
with ethanol, cleared in xylene, and mounted under a
coverslip.
Slides were evaluated under a light microscope by
pathologists subjectively. Sample with no primary
antibody added was used as a negative control. The
cytoplasmic expression of KRAS protein was assessed
using the previously established criteria of Allred et
al (Allred, Harvey, Berardo, & Clark, 1998). We
considered positive expression if the score of more
than 2 in tumor cells.
The corresponding clinical and pathological data
including sex, age, tumor topography, lymph node
metastases, TNM staging, and tumor differentiation
were obtained from a review of patient records.
Association between clinicopathological parameters
and KRAS protein immunostaining was statistically
examined by the Chi-square test, and p-value < 0.05
was considered significant.
3 RESULTS
Table 1 shows the clinicopathological features of
CRC cases. More than half of the patients were male.
The mean age of the patients was 52.2±12.5 years,
and most of them were >40 years old age. The
majority of the CRC were located on the distal colon,
and most of them were in stage 3 with well/moderate
differentiation.
Table 1: Association between KRAS protein expression
and clinicopathological features of CRC.
Features KRAS protein
expression
p
Positive
n (%)
Negative
n (%)
Sex 0.03
Male 5(35.7) 22(71)
Female 9(64.3) 9(29)
Age 0.63
≤40 3(21.4) 7(22.6)
>40 11(78.6) 24(77.4)
Topography 0.62
Proximal colon 5(35.7) 11(35.5)
Distal colon 9(64.3) 20(64.5)
Lymph Node
Status
0.15
Positive 7(50) 9(29)
Negative 7(50) 22(71)
Staging 0.15
Stage 2 7(50) 9(29)
Stage 3 7(50) 22(71)
Differentiation 0.69
Well and
moderate
14(100) 30(96.8)
Poor 0(0) 1(3.2)
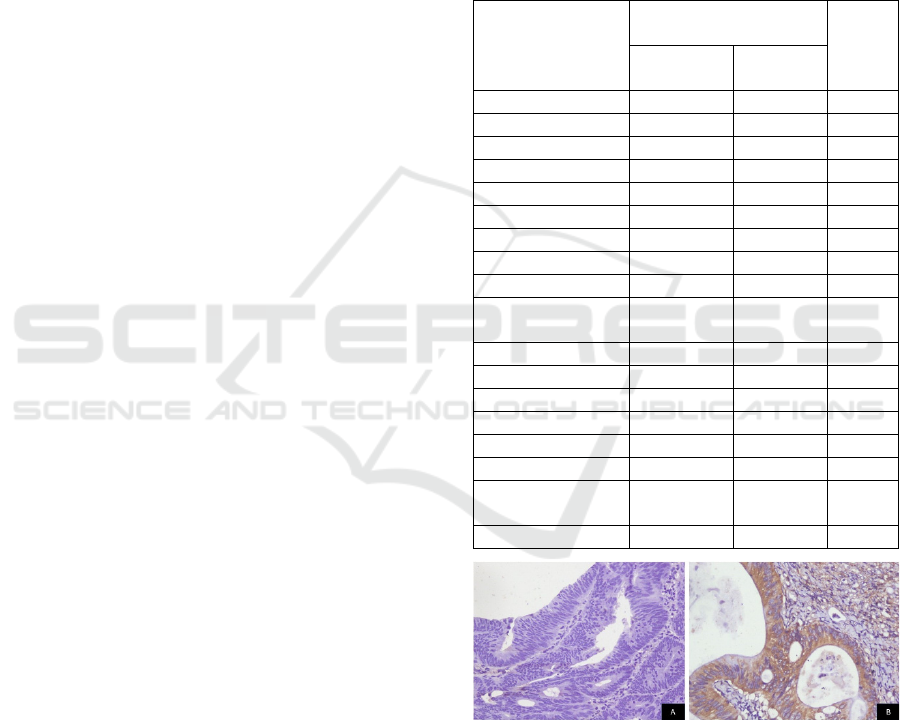
Figure 1: Representative photomicrographs of KRAS
immunostaining in CRC tissues (A. Negative expression;
B. Positive expression; 400x magnification).
Positive KRAS protein expression was found in
14(31.1%) of cases (Figure 1). We found a significant
association between positive KRAS protein
expression and female predominance. Nevertheless,
our study did not reveal any statistical association
between KRAS protein expression with age,
JIMC 2020 - 1’s t Jenderal Soedirman International Medical Conference (JIMC) in conjunction with the Annual Scientific Meeting
(Temilnas) Consortium of Biomedical Science Indonesia (KIBI )
48

topography, lymph node status, tumor staging, and
tumor differentiation.
4 DISCUSSION
Mutation of RAS proto-oncogenes is frequently
found in human cancer. In colorectal carcinogenesis,
mutation of KRAS leads to uncontrolled proliferation
and malignant transformation. Nowadays, the
determination of the KRAS mutation is mandatory for
treatment with anti-EGFR therapy in patients with
CRC. KRAS and BRAF mutations play a pivotal role
in colorectal carcinogenesis and are related to the
main resistance to anti-EGFR therapy (Larki et al.,
2017). Unfortunately, mutation of KRAS has only
been detected commonly in codon 12,13, and 61 (Lee
et al., 2017). Therefore, expanding RAS testing in
CRC to analyze more mutations may better predict
benefit from anti-EGFR therapy.
However, increase testing volume will increase
testing costs which may have economic implications.
Previous studies revealed that single standard KRAS
exon 2 testing was more than threefold costly than
single antibody IHC assay (Kircher, Mohindra, &
Nimeiri, 2015; Muirhead, Aoun, Powell, Juncker, &
Mollerup, 2010). Thus, the morphological study
followed by protein detection using IHC appears to
be an alternative to molecular screening. IHC of
KRAS might be useful as a prognostic and predictive
marker in CRC. KRAS positive protein expression
was associated with the disease aggressiveness of
CRC. There was a significantly reduced relapse-free
survival (RPS) in rectal cancer patients with KRAS
positive protein expression (Kanik, Gajjar, & Ghosh,
2018).
In the present study, we detected KRAS protein
expression in 31.1% of CRC cases. This result was
close to study in Europe (33%) (Piton et al., 2015) but
lower than study in Egypt (42.3%) (Elsabah & Adel,
2013). This KRAS IHC positivity was also close to a
study that revealed KRAS mutation in 32.8% of CRC
cases previously (Liu, Jakubowski, & Hunt, 2011).
However, other studies found mutation positivity
range between 14% to 50% (Sammoud et al., 2012).
These differences were reported because of several
weaknesses of mutation analysis such as codon
specific mutation sites, ethnic variations (Zhang et al.,
2015), diet, and lifestyle factors (Hughes, Simons,
van den Brandt, van Engeland, & Weijenberg, 2017).
Our data demonstrated the predominance of
KRAS protein expression in female CRC patients.
This finding was in line with several previous studies
that reported a correlation between mutated KRAS
and the female gender in CRC (Li et al., 2015; Tong
et al., 2014). However, other reports using IHC assays
did not demonstrate such a relationship (Kanik et al.,
2018; Sammoud et al., 2012). A possible explanation
for our result is likely complex and multifactorial
including lifestyle factors and the composition of gut
microbiota which varies among gender (Kostic et al.,
2011). Female sex hormones are related to colorectal
carcinogenesis by their effects on the production of
bile acid, bowel transit time, and bacterial
fermentation (Sammoud et al., 2012).
In this recent study, we did not find an association
between KRAS protein expression and the age of
patients. This is in keeping with some previous
reports both using IHC or molecular testing (Elsabah
& Adel, 2013; Rosty et al., 2013). However, another
study reported that ras p21 IHC overexpression was
relatively related to the advanced age of patients
(Sammoud et al., 2012). This difference result might
be caused by different classification of the patient's
age.
Besides, we did not discover any significant
relationship between KRAS protein expression and
other clinicopathological parameters such as
topography, lymph node status, stage, and
differentiation of the tumors. Our result was in
agreement with previous studies using either IHC
(Elsabah & Adel, 2013; Piton et al., 2015) or PCR
assays (Sammoud et al., 2012). Nevertheless, other
reports detected an association between mutant KRAS
with mucinous subtype and greater differentiation of
CRC (Zhang et al., 2015). Unfortunately, in our
study, we only had one sample of poorly
differentiated CRC, and showing negative KRAS
protein staining.
5 CONCLUSIONS
The prevalence of positive KRAS protein in CRC was
relatively the same as the prevalence of KRAS
mutation. However, the use of polyclonal antibody
which not specific to detect KRAS mutant became one
of the limitations of KRAS IHC assays. Therefore, the
development of a monoclonal antibody designed
against the mutated KRAS domain is necessary. This
could greatly assist the screening of CRC patients for
anti-EGFR therapies. In the future, IHC could
become a promising tool in diagnostic and prognostic
decisions.
Immunohistochemistry of KRAS Protein in Colorectal Cancer
49

REFERENCES
Allred, D. C., Harvey, J. M., Berardo, M., & Clark, G. M.
(1998). Prognostic and predictive factors in breast
cancer by immunohistochemical analysis. Mod Pathol,
11(2), 155-168.
Cree, I. A. (2016). Diagnostic RAS mutation analysis by
polymerase chain reaction (PCR). Biomol Detect
Quantif, 8, 29-32. doi:10.1016/j.bdq.2016.05.001
De Roock, W., Jonker, D. J., Di Nicolantonio, F., Sartore-
Bianchi, A., Tu, D., Siena, S., . . . Tejpar, S. (2010).
Association of KRAS p.G13D mutation with outcome
in patients with chemotherapy-refractory metastatic
colorectal cancer treated with cetuximab. JAMA,
304(16), 1812-1820. doi:10.1001/jama.2010.1535
Elsabah, M. T., & Adel, I. (2013). Immunohistochemical
assay for detection of K-ras protein expression in
metastatic colorectal cancer. J Egypt Natl Canc Inst,
25(1), 51-56. doi:10.1016/j.jnci.2013.01.003
Hughes, L. A. E., Simons, C., van den Brandt, P. A., van
Engeland, M., & Weijenberg, M. P. (2017). Lifestyle,
Diet, and Colorectal Cancer Risk According to
(Epi)genetic Instability: Current Evidence and Future
Directions of Molecular Pathological Epidemiology.
Curr Colorectal Cancer Rep, 13(6), 455-469.
doi:10.1007/s11888-017-0395-0
Jemal, A., Bray, F., Center, M. M., Ferlay, J., Ward, E., &
Forman, D. (2011). Global cancer statistics. CA Cancer
J Clin, 61(2), 69-90. doi:10.3322/caac.20107
Kanik, P., Gajjar, K., & Ghosh, N. (2018).
Immunohistochemical Localization of KRAS and
BRAF and its Clinical Utility in Patients with
Colorectal Cancer. Colorectal Cancer: Open Access,
04(01). doi:10.21767/2471-9943.100051
Kircher, S. M., Mohindra, N., & Nimeiri, H. (2015). Cost
estimates and economic implications of expanded RAS
testing in metastatic colorectal cancer. Oncologist,
20(1), 14-18. doi:10.1634/theoncologist.2014-0252
Kostic, A. D., Gevers, D., Pedamallu, C. S., Michaud, M.,
Duke, F., Earl, A. M., . . . Meyerson, M. (2011).
Genomic analysis identifies association of
Fusobacterium with colorectal carcinoma. Genome
Research, 22(2), 292-298. doi:10.1101/gr.126573.111
Larki, P., Gharib, E., Yaghoob Taleghani, M., Khorshidi,
F., Nazemalhosseini-Mojarad, E., & Asadzadeh
Aghdaei, H. (2017). Coexistence of KRAS and BRAF
Mutations in Colorectal Cancer: A Case Report
Supporting The Concept of Tumoral Heterogeneity.
Cell J, 19(Suppl 1), 113-117.
doi:10.22074/cellj.2017.5123
Lee, S. H., Chung, A. M., Lee, A., Oh, W. J., Choi, Y. J.,
Lee, Y.-S., & Jung, E. S. (2017). KRAS Mutation Test
in Korean Patients with Colorectal Carcinomas: A
Methodological Comparison between Sanger
Sequencing and a Real-Time PCR-Based Assay.
Journal of Pathology and Translational Medicine,
51(1), 24-31. doi:10.4132/jptm.2016.10.03
Li, W., Qiu, T., Zhi, W., Shi, S., Zou, S., Ling, Y., . . . Lu,
N. (2015). Colorectal carcinomas with KRAS codon 12
mutation are associated with more advanced tumor
stages. BMC Cancer, 15, 340. doi:10.1186/s12885-
015-1345-3
Liu, X., Jakubowski, M., & Hunt, J. L. (2011). KRAS gene
mutation in colorectal cancer is correlated with
increased proliferation and spontaneous apoptosis. Am
J Clin Pathol, 135(2), 245-252.
doi:10.1309/AJCP7FO2VAXIVSTP
Muirhead, D., Aoun, P., Powell, M., Juncker, F., &
Mollerup, J. (2010). Pathology economic model tool: a
novel approach to workflow and budget cost analysis in
an anatomic pathology laboratory. Arch Pathol Lab
Med, 134(8), 1164-1169. doi:10.1043/2000-0401-OA.1
Piton, N., Borrini, F., Bolognese, A., Lamy, A., & Sabourin,
J. C. (2015). KRAS and BRAF Mutation Detection: Is
Immunohistochemistry a Possible Alternative to
Molecular Biology in Colorectal Cancer?
Gastroenterol Res Pract, 2015, 753903.
doi:10.1155/2015/753903
Porru, M., Pompili, L., Caruso, C., Biroccio, A., & Leonetti,
C. (2018). Targeting KRAS in metastatic colorectal
cancer: current strategies and emerging opportunities. J
Exp Clin Cancer Res, 37(1), 57. doi:10.1186/s13046-
018-0719-1
Rajagopalan, H., Bardelli, A., Lengauer, C., Kinzler, K. W.,
Vogelstein, B., & Velculescu, V. E. (2002).
Tumorigenesis: RAF/RAS oncogenes and mismatch-
repair status. Nature, 418(6901), 934.
doi:10.1038/418934a
Raskov, H., Pommergaard, H. C., Burcharth, J., &
Rosenberg, J. (2014). Colorectal carcinogenesis--
update and perspectives. World J Gastroenterol,
20(48), 18151-18164. doi:10.3748/wjg.v20.i48.18151
Rosty, C., Young, J. P., Walsh, M. D., Clendenning, M.,
Walters, R. J., Pearson, S., . . . Buchanan, D. D. (2013).
Colorectal carcinomas with KRAS mutation are
associated with distinctive morphological and
molecular features. Modern Pathology, 26(6), 825-834.
doi:10.1038/modpathol.2012.240
Sammoud, S., Khiari, M., Semeh, A., Amine, L., Ines, C.,
Amira, A., . . . Saadia, B. (2012). Relationship between
expression of ras p21 oncoprotein and mutation status
of the K-ras gene in sporadic colorectal cancer patients
in Tunisia. Appl Immunohistochem Mol Morphol,
20(2), 146-152. doi:10.1097/PAI.0b013e3182240de1
Shia, J. (2008). Immunohistochemistry versus
microsatellite instability testing for screening colorectal
cancer patients at risk for hereditary nonpolyposis
colorectal cancer syndrome. Part I. The utility of
immunohistochemistry. J Mol Diagn, 10(4), 293-300.
doi:10.2353/jmoldx.2008.080031
Tong, J. H., Lung, R. W., Sin, F. M., Law, P. P., Kang, W.,
Chan, A. W., . . . To, K. F. (2014). Characterization of
rare transforming KRAS mutations in sporadic
colorectal cancer. Cancer Biol Ther, 15(6), 768-776.
doi:10.4161/cbt.28550
Wan Juhari, W. K., Wan Abdul Rahman, W. F., Mohd
Sidek, A. S., Abu Hassan, M. R., Ahmad Amin
Noordin, K. B., Zakaria, A. D., . . . Zilfalil, B. A.
(2015). Analysis of Hereditary Nonpolyposis
Colorectal Cancer in Malay Cohorts using
JIMC 2020 - 1’s t Jenderal Soedirman International Medical Conference (JIMC) in conjunction with the Annual Scientific Meeting
(Temilnas) Consortium of Biomedical Science Indonesia (KIBI )
50

Immunohistochemical Screening. Asian Pac J Cancer
Prev, 16(9), 3767-3771.
doi:10.7314/apjcp.2015.16.9.3767
Zhang, J., Zheng, J., Yang, Y., Lu, J., Gao, J., Lu, T., . . .
Liu, T. (2015). Molecular spectrum of KRAS, NRAS,
BRAF and PIK3CA mutations in Chinese colorectal
cancer patients: analysis of 1,110 cases. Sci Rep, 5,
18678. doi:10.1038/srep18678
Immunohistochemistry of KRAS Protein in Colorectal Cancer
51
