
Unsupervised Descriptive Text Mining for Knowledge Graph Learning
Giacomo Frisoni, Gianluca Moro
∗
and Antonella Carbonaro
Department of Computer Science and Engineering – DISI, University of Bologna,
Via dell’Universit
`
a 50, I-47522 Cesena, Italy
Keywords:
Text Mining, Knowledge Graphs, Unsupervised Learning, Semantic Web, Ontology Learning, Rare Diseases.
Abstract:
The use of knowledge graphs (KGs) in advanced applications is constantly growing, as a consequence of their
ability to model large collections of semantically interconnected data. The extraction of relational facts from
plain text is currently one of the main approaches for the construction and expansion of KGs. In this paper, we
introduce a novel unsupervised and automatic technique of KG learning from corpora of short unstructured
and unlabeled texts. Our approach is unique in that it starts from raw textual data and comes to: i) identify a set
of relevant domain-dependent terms; ii) extract aggregate and statistically significant semantic relationships
between terms, documents and classes; iii) represent the accurate probabilistic knowledge as a KG; iv) extend
and integrate the KG according to the Linked Open Data vision. The proposed solution is easily transferable
to many domains and languages as long as the data are available. As a case study, we demonstrate how it is
possible to automatically learn a KG representing the knowledge contained within the conversational messages
shared on social networks such as Facebook by patients with rare diseases, and the impact this can have on
creating resources aimed to capture the “voice of patients”.
1 INTRODUCTION
We are witnessing a continuous growth of unstruc-
tured textual content on the Web, especially in con-
texts such as social networks. It is increasingly diffi-
cult for humans to consume the information contained
in text documents of their interest at the same rate as
they are produced and accumulated over time. This
problem highlights the importance of automatic read-
ing and natural language understanding (NLU).
The automatic extraction of high-value semantic
knowledge directly from text corpora is a common
need. Depending on the domain, there are many ques-
tions to which we would like to find answers, avoiding
the manual reading of documents inside the starting
corpus. For example, we may want to analyze the ef-
fectiveness of a drug, the most difficult activities for
patients, the main causes of destructive accidents, or
the reasons behind negative reviews.
The identification of useful answers for the de-
scription of phenomena from unstructured texts is
called descriptive text mining. The knowledge from
which we want to derive phenomena explanations
can be seen as a set of aggregative, interpretable
and quantifiable semantic relationships between un-
∗
Contact author: gianluca.moro@unibo.it
bounded combinations of relevant concepts.
Learning this type of knowledge requires the over-
coming of several challenges. An automatic learn-
ing process is preferred to avoid the necessary guid-
ance of a human analyst, and domain independent so-
lutions are sought to extend their applicability. Un-
supervised and semantic approaches are preferable
because the data is mostly unlabeled and the mean-
ing of words and phrases can provide important in-
dications. Knowledge extraction must take place on
a global level, as an aggregation of the whole cor-
pus. The statistical significance quantification (e.g.,
p-value) of the identified relationships is necessary for
subsequent processing.
The vast majority of existing techniques only par-
tially solve the illustrated problem, which has recently
been addressed in (Frisoni et al., 2020) where we pro-
posed a descriptive text mining methodology for the
discovery of statistically significant evidences, here
referenced as DTM4ED. In particular, we allowed
the identification of scientific medical correlations di-
rectly from the patients’ posts, with an accuracy of
about 78%. The general output of DTM4ED is a flat
list of correlated and quantified sets of terms (e.g.,
<“dysphagia swallowing”: 86%; “alcohol acid re-
flux”: 81%>). However this approach has some lim-
itations: terms are not semantically tagged and there
316
Frisoni, G., Moro, G. and Carbonaro, A.
Unsupervised Descriptive Text Mining for Knowledge Graph Learning.
DOI: 10.5220/0010153603160324
In Proceedings of the 12th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2020) - Volume 1: KDIR, pages 316-324
ISBN: 978-989-758-474-9
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

are no links between different correlation sets. For in-
stance, our previous contribution does not infer that
(i) “alcohol” is a liquid food, (ii) “acid reflux” and
“GERD” are equivalent concepts in the context under
consideration, (iii) “dysphagia” and “acid reflux” are
both symptoms or diseases in their own right.
The limited expressiveness that characterizes
NLU models — such as the one just described — can
be overcome by integrating qualitative techniques and
moving towards “mixed AI”. Natural Language Pro-
cessing (NLP) and Semantic Web technologies are
increasingly combined together to achieve better re-
sults, and in the future it is expected that the joint
use of subsymbolic and symbolic AI will be central
(Patel and Jain, 2019). According to the 2019 Gart-
ner’s Hype Cycle
1
, knowledge graphs (KGs) are con-
sidered one of the most promising technologies of the
next decade. With their ability to represent informa-
tion through a collection of interlinked entities based
on semantics and meaning, KGs are a powerful tool
for the representation of knowledge, its integration
and inference based on automatic reasoning.
In this paper, we introduce a novel unsupervised
and automatic technique for knowledge graph learn-
ing, which extends the expressive power of the results
produced by DTM4ED. With the idea of combining
subsymbolic and symbolic AI, the research has two
main advantages in both directions. Firstly, it en-
riches the KG learning approaches known in litera-
ture, distinguishing itself thanks to the strengths in-
herited from the methodology on which it is built.
Secondly, it increases the expressive power, the in-
terpretability and the interrogability of the knowl-
edge extracted with descriptive text mining, offering
significant application advantages in many domains.
By tagging the terms and representing the correla-
tions between concepts with a KG, a non-expert user
can more easily understand the results of the analy-
sis (e.g., “poem” is a surgical technique), work hier-
archically with meta-terms and choose which kind of
correlations search without knowing the specific in-
stances (e.g., drug ↔ drug, drug ↔ symptom, “chest
pain” ↔ food).
The paper is organized as follows. Section 2 sum-
marizes the related works. Section 3 discusses how
DTM4ED can be implemented to enable the construc-
tion of KGs. Section 4 introduces and discusses our
KG learning method. Section 5 shows the application
of the contribution on a medical case study. Finally,
Section 6 sums up the work and presents possible fu-
ture developments.
1
https://www.gartner.com/smarterwithgartner/5-trends-
appear-on-the-gartner-hype-cycle-for-emerging-technolo
gies-2019/
2 RELATED WORK
Handcrafting big knowledge representations is an ex-
tremely intensive, non-scalable and time consuming
task. Ontology learning (OL) studies the mechanisms
to transform the creation and maintenance of ontolo-
gies into a semi or complete automatic process, and
the first works date back to several years ago (Maed-
che and Staab, 2001). OL techniques have been
widely investigated for various domain purposes and
application scenarios (Asim et al., 2018).
With the huge increase in magnitude and through-
put related to the generation of textual content, sev-
eral attempts have been made to bring some level
of automation in the process of ontology acquisi-
tion directly from unstructured text. As a conse-
quence, the development of OL currently goes hand in
hand with that of NLP and advanced machine learn-
ing approaches which are essential to extract knowl-
edge from text documents (Domeniconi et al., 2015).
Transfer learning to target domains with unlabeled
data is becoming increasingly necessary (Domeniconi
et al., 2014b; Domeniconi et al., 2014c; Domeniconi
et al., 2016b; Domeniconi et al., 2017; Pagliarani
et al., 2017; Moro et al., 2018), and is the most fre-
quent case for social messages (Domeniconi et al.,
2016b).
After the massive introduction of KGs by Google
(Singhal, 2012) for the representation of knowledge
extracted from the Web, today we are seeing a grow-
ing spread of these solutions, also for their poten-
tial in bringing common sense into neural networks
(Lin et al., 2019). In the community, many authors
claim that the real divide between ontologies and
knowledge graphs lies in the nature of data (Ehrlinger
and W
¨
oß, 2016). While ontologies are generally re-
garded as smaller hand-curated collections of asser-
tions with a focus on the schema and on the reso-
lution of domain-specific tasks, KGs are based on
facts, can have significant dimensions and are there-
fore the most suitable solution for the representation
of text-mined knowledge. Freebase (Bollacker et al.,
2008), DBpedia (Bizer et al., 2009), YAGO (Tanon
et al., 2020) and WordNet (Miller, 1998) are among
the most widely used KGs in NLP applications.
Knowledge graph learning (KGL) from text fol-
lows the same principles as OL from text, and it
is usually based on a multistep approach known as
learning layer cake (Buitelaar et al., 2005). The com-
plete model includes the extraction of terms and their
synonyms from the underlying text, the combination
of them to form concepts, the identification of taxo-
nomic and non-taxonomic relationships between the
found concepts, and finally the generation of rules.
Unsupervised Descriptive Text Mining for Knowledge Graph Learning
317

The works published in literature can be distinguished
on the basis of how they deal with the various stages.
A great categorization resulting from the review of
140 papers has been proposed in (Asim et al., 2018),
where the authors have observed better results from
hybrid linguistic-statistical approaches. In this sense,
Table 1 shows a summary of the most used techniques
for solving the sub-tasks that make up the ontology
and KG learning layer cake.
Table 1: Main linguistic and statistical techniques adopted
for the implementation of Ontology and Knowledge Graph
Learning Layer Cakes.
Techniques
Step
Linguistics Statistical
Preprocessing
Part of Speech Tagging
Dependency Parsing
Word Sense Disambiguation
Lemmatization
Term /
Synonym /
Concept
Extraction
Regular Expressions
Syntactic Analysis
Linguistic Filters
Subcategorization Frames
Seed Word Extraction
Named Entity Recognition
Named Entity Recognition
Term Weighting
C/NC value
Contrastive Analysis
Language Models
Clustering
Concept
Hierarchy
Regular Expressions
Dependency Analysis
Lexico Syntactic Pattern
WordNet-based
Term Subsumption
Formal Concept Analysis
Hierarchical Clustering
Relation
Extraction
Regular Expressions
Open Information Extraction
WordNet-based
Association Rule Mining
Bootstrapping
Logistic Regression
Open Information Extraction
Modern alternative methods, such as COMET
(Bosselut et al., 2019), automatically construct KGs
within deep learning models, but using labeled data.
Several tools for ontology learning are avail-
able (e.g., Text2Onto
2
, OntoGen
3
, GATE
4
). Typically
their main objective is not to automatically build an
ontology starting from a body of texts, but help user to
do it. From a representation perspective, a newly KG
Management System has been introduced by Grakn
5
,
also supporting hypergraphs, type systems and rea-
soning, but it is independent of the knowledge learn-
ing process. Promising results have been achieved by
FRED
6
and Ontotext
7
, but they are unable to meet
all the requirements demanded by a flexible KGL so-
lution. In fact, the methodologies currently used in
state-of-the-art OL and KGL often need human inter-
vention and large labeled datasets, lack explainability
due to the use of black box models, struggle to in-
tegrate new knowledge and evolve dynamically, are
trained to recognize a pre-established set of entities
and/or relationships without allowing semantic per-
2
http://neon-toolkit.org/wiki/1.x/Text2Onto.html
3
http://ontogen.ijs.si/
4
https://gate.ac.uk/
5
https://grakn.ai/
6
http://wit.istc.cnr.it/stlab-tools/fred
7
https://www.ontotext.com/
sonalization, do not consider uncertainty, are based
on solutions strongly dependent on domain, or inef-
fective in non-general contexts.
The contribution described in this paper wants to
differentiate itself by proposing an unsupervised KGL
approach from unstructured text based on DTM4ED
(Frisoni et al., 2020), handling unlabeled data, con-
cepts without a predefined schema, semantic mod-
eling of both terms and documents, statistical quan-
tification, interpretability, and support for execution
without man-in-the-analysis.
3 DESCRIPTIVE TEXT MINING
Section 3.1 briefly summarizes DTM4ED, and Sec-
tion 3.2 shows how it can be empowered to produce
results for the construction of a KG.
3.1 Methodology
DTM4ED recognizes semantic correlations between
terms and documents, and can be used to provide a
description of a phenomenon of interest. Assuming
that a phenomenon can be represented by a distribu-
tion of documents having a certain class, the last goal
is addressed by searching for a set of relevant terms
representative of the document distribution itself.
The solution consists of various modules whose
implementation can be adapted to the specific prob-
lem under consideration: quality preprocessing (to
improve the quality of the text documents contained
within the starting corpus), document classification
(to recognize the phenomenon to be investigated),
analysis preprocessing (to prepare the data for the
analysis), term weighting (to identify the significance
of each term in each document, defining also the
vocabulary), and language modeling (to bring out
semantic similarities between terms and documents
within a low-dimensional latent vector space).
The composition of the phenomenon description
is carried out incrementally and is based on the con-
struction of a query (i.e., an artificial document). Af-
ter identifying a first term or considering a starting
query, at each step the query is folded into latent
space. From here, we search for new terms semanti-
cally close to the query and with greater significance,
choosing the one that — if combined with the current
description — continues to be representative of the
phenomenon. The degree of correlation between the
query and the phenomenon is indicated by the p-value
resulting from the application of the chi-squared (χ
2
)
statistical hypothesis test, together with R-precision.
KDIR 2020 - 12th International Conference on Knowledge Discovery and Information Retrieval
318

The process ends when it is no longer possible to en-
rich the query and remain below the pre-established
p-value threshold.
3.2 Methodology Definition for
Knowledge Graph Learning
3.2.1 Entity Tagging
In a basic version, DTM4ED works with plain and flat
terms. The information content of documents within
the corpus can be significantly extended through the
use of pre-trained Named Entity Recognition (NER)
and Named Entity Linking (NEL) systems.
Named Entity Recognition. A NER system al-
lows the unsupervised detection and classification of
the entities mentioned in unstructured text into pre-
defined categories (e.g., drug, symptom, food, place).
Advanced solutions are capable of handling several
hundreds of very fine-grained types, also organized
in a hierarchical taxonomy. Many recent works use
contextualized learning and Freebase-based type hi-
erarchies (Ren et al., 2016; Li et al., 2020).
Named Entity Linking. NEL is the task of aligning
a textual mention of a named-entity to an appropri-
ate entry in a target knowledge base (KB), annotating
it with a unique identity and link (Rao et al., 2013).
It can be seen as an instance of an extremely fine-
grained NER, where the types are the actual entries of
an entity database (e.g., Wikipedia pages) or Linked
Open Data resources (e.g., Wikidata, DBpedia, Free-
base, YAGO). NEL is typically carried out following
a NER phase, because entity linking identifies specific
entities assuming that the correct mentions have al-
ready been previously recognized. NEL systems can
also perform a normalization operation in assigning
the same identifier to two or more synonyms (e.g.,
“soda water”, “fizzy water” → “carbonated water”).
Since NEL requires selecting the proper concept from
a restricted set of candidates, it is also called Named
Entity Disambiguation (NED).
Promising results come from the joint learning of
NER and NEL (Martins et al., 2019).
Within DTM4ED, NER and NEL systems can
be applied in the early steps of quality preprocess-
ing. Information on recognized entities can be re-
ported directly within the textual content of the doc-
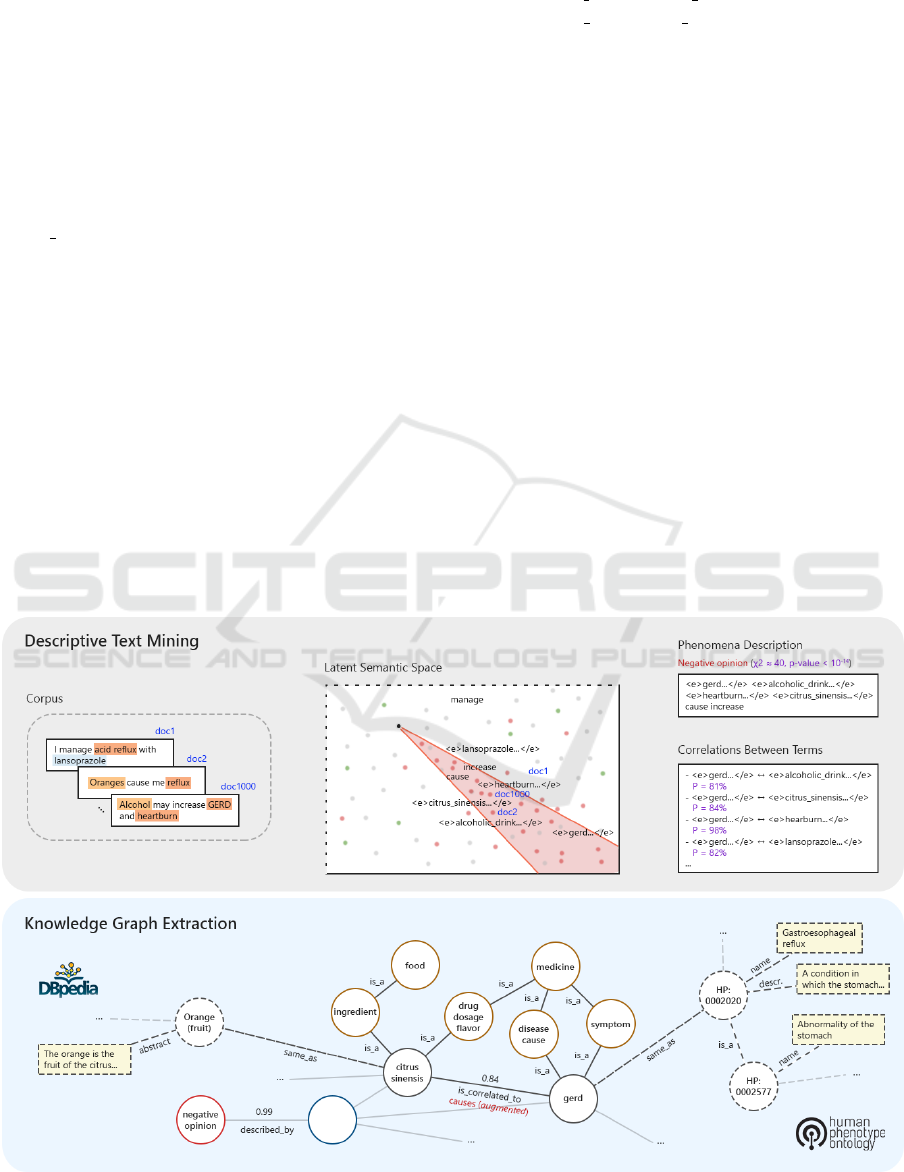
uments, carrying out an entity tagging phase (Figure
1). By adopting a word-level unigram tokenization or
by making use of support data structures, it is pos-
sible to preserve the information associated with the
labeled terms for all the rest of the analysis.
Even if not recognized as entities, the considera-
tion of terms deemed to be relevant after term weight-
Figure 1: Entity tagging on a sample document.
ing continues to be fundamental. We refer to these
terms as standard terms, which can be verbs, adjec-
tives or domain-specific concepts without a corre-
sponding entity on other KBs.
3.2.2 Correlation between Terms
By calculating the correlation between each pair of
terms, it is possible to extract a very useful knowl-
edge for tasks such as query auto-completion and KG
extraction. For each term, we can keep track of the
top N terms related to it, in descending order and with
a positive correlation above a certain threshold. The
inverse correlations can be significant or not depend-
ing on the language model chosen. Correlations can
be expressed as probabilities. Some language models
already represent correlations in this way, like pLSA
(Hofmann, 2013) which discovers the underlying se-
mantic structure of the data in a probabilistic way.
Alternatively, we suggest two main ways to calculate
them: (i) cosine similarities remapped from [−1,1] to
[0,1]; (ii) p-values deriving from chi-squared tests be-
tween terms, observing and estimating the number of
times the two terms appear together, do not appear or
appear alternately in the latent semantic space.
4 KNOWLEDGE GRAPH
LEARNING
Here we illustrate how the knowledge obtained using
the methodology defined in Section 3 can be modeled
with a KG, discussing also the resulting advantages.
Figure 2 summarizes the process.
4.1 Learning Layer Cake
Preprocessing. This step is directly included in
DTM4ED. It generally concerns encoding and sym-
bols normalization, URL removal, word lengthening
fixing, entity tagging, lemmatization, stemming, doc-
ument classification, case-folding, special characters
and stopwords removal, tokenization, term-document
matrix, term weighting, feature selection and lan-
guage modeling.
Term/Concept Extraction. After entity tagging, the
terms are distinguished between standard and entity
Unsupervised Descriptive Text Mining for Knowledge Graph Learning
319
ones. In the field of ontology or KG learning, the def-
inition of term can be compared to that of standard
term in the case of descriptive text mining. Similarly,
a concept corresponds to an entity recognized by a
NER system and possibly identified by a NEL sys-
tem, for which id, types and optional links to external
KBs are known. However, the definition of concept
can also be extended to the standard terms that have
passed the feature selection phase, significant docu-
ments and classes representing phenomena.
Concept Hierarchy. A hierarchy of concepts formed
by is a relationships is easily derivable from the re-
sults of the NER on the taxonomy of interest.
Relations. In addition to hierarchical relationships,
we can model the significant correlations between the
terms that emerged in the low-rank latent space with
a global analysis, i.e. considering all the documents
within the corpus (Section 3.2.2). By looking at se-
mantic relationships with high probability, we can
represent the set of bonds that each concept (typed or
non-typed) has with the others. The extracted knowl-
edge can therefore be mapped into RDF triples (sub-
ject, predicate, and object). As for entities, by using
rules or by training a classifier (Onuki et al., 2019),
it is also possible to extract or predict more specific
relations than the general correlation ones. For ex-
ample, in the case of a strong correlation between a
medical treatment and a specialized center, the rela-
tion type has correlated term could actually cor-
respond to is performed at. A further type of rela-
tion is that between a phenomenon and its description
(i.e., a set of representative terms), modelable with
blank nodes. The correlations extracted with text min-
ing techniques are not certain, but stochastic. These
probability values can be used to label the relations
represented within an ontology or a KG, and to en-
able probabilistic reasoning.
4.2 Knowledge Integration
Linking entity mentions to existing KBs is core to Se-
mantic Web population (Gangemi, 2013; Carbonaro
et al., 2018). Starting from the results returned by the
NEL system, it is possible to derive references to rep-
resentations of the same concepts on different KBs.
This allows us to automatically integrate our own on-
tology or KG with existing resources, and to signif-
icantly extend the knowledge representation accord-
ing to the Linked Open Data (LOD) vision and, in
general, with any kind of medical ontologies (e.g. ge-
nomic ones (Domeniconi et al., 2014a; Domeniconi
et al., 2016a)).
Figure 2: High-level process for the automatic extraction of knowledge from a corpus of text documents. The examples refer
to a medical domain, with DBpedia and HPO as integrated knowledge bases.
KDIR 2020 - 12th International Conference on Knowledge Discovery and Information Retrieval
320

4.3 Expressive Power, Interrogability
and Interpretability
Compared to DTM4ED, the output is no longer a se-
quence or a flat set of clusters made up of correlated
unlabeled terms, but a KG. The inclusion of terms
recognized as entities within a hierarchical taxonomy
means that the correlations are interconnected, and no
longer independent of each other. The consequence is
the increase in expressive power.
The typing introduced by the NER system
also represents an important step towards inter-
pretability, allowing a non-expert user to better
understand what a certain term represents (e.g.,
“poem” is a /medicine/surgical technique, “gemelli”
is a /medicine/hospital). The modularity of the lan-
guage model allows us to avoid the use of neural so-
lutions (Allen et al., 2019), and to have greater inter-
pretability and reliability also in the KGL process.
From the greater expressive power, new forms of
interrogability arise. If a user wants to investigate
all the significant correlations between two or more
types of entities (e.g., drug ↔ symptom, symptom
↔ food), he is no longer forced to check them in-
dividually and to know all the terms related to the in-
stances of the types considered (e.g., <“lansoprazole
gerd”: ?>, <“aspirin headache”: ?>, <“gerd alco-
hol”: ?>, . ..). Now it is possible to manage queries
concerning the meta-levels of the concept hierarchy.
Similarly to what happens with a Prolog inference
engine based on the unification theory, correlations
between ground concepts can be obtained by substi-
tution. Meta-level correlations can also involve only
one unbounded term (“chest pain” ↔ drug), and can
be filtered also within DTM4ED.
5 CASE STUDY AND
EXPERIMENTS
To assess the effectiveness of the method described
above, we performed some experiments on the same
case study and dataset introduced in (Frisoni et al.,
2020), as a natural continuation of the research.
5.1 Dataset
The case study belongs to the medical field and con-
cerns the online patient communities. With the aim of
extracting and representing the knowledge contained
in the conversational messages shared on social net-
works by patients with a rare disease, it is focused on
an Italian Facebook Group dedicated to Esophageal
Achalasia (ORPHA: 930).
The dataset consists of 6,917 posts and 61,692
first-level comments, published between 21/02/2009
and 05/08/2019. It contains experiences, questions
and suggestions of about 2,000 users, shared in
the private Facebook Group
8
managed by AMAE
Onlus
910
, the main Italian patient organization for
Esophageal Achalasia. Collaborating with the orga-
nization, the data were downloaded anonymously and
considering only the textual messages.
As suggested in the original paper, DTM4ED was
implemented with R and Python, adopting Latent Se-
mantic Analysis (LSA) (Landauer and Dumais, 1997)
as an algebraic language model capable of respond-
ing to the need for explainability that is typical of the
health sector.
5.2 Entity Management
Considering the heterogeneity and specificity of the
entity types linked to the topics of interest from the
patient’s point of view (e.g., symptoms, drugs, foods,
places), it is necessary to adopt highly comprehensive
NER and NEL systems. To this end, we selected Text-
Razor
11
, which is able to identify 36,584,406 entities
(model version: 2020-03) organized in thousands of
different taxonomic types in both Freebase and DB-
Pedia. Moreover, it supports entity disambiguation
and linking to Wikipedia, DBPedia, Wikidata, Free-
base, and PermID
12
. Using this commercial NLP sys-
tem, we obtained data for 15,687 and 73,095 entities
in posts and comments, respectively.
We introduced a new taxonomy for the case study,
on which we mapped the Freebase and DBpedia types
(e.g., DBpediaType ∈ {Beer ∨ Vodka ∨ Wine} →
Type = / f ood/beverage/alcoholic
beverage). This
reconciliation phase avoids the maintenance of two
distinct typizations with different levels of granular-
ity, and allows the filtering of only the entities labeled
with a type of interest.
5.3 Knowledge Graph
The construction of the KG was carried out starting
from the results associated with the highest evalua-
tion score obtained with DTM4ED, which led to the
achievement of 77.68% accuracy and 78.9% precision
on a set of 224 gold standard medical correlations
with a confidence threshold 1 − pvalue ≥ 0.8.
8
https://www.facebook.com/groups/36705181245/
9
http://www.amae.it/
10
https://www.orpha.net/consor/cgi-bin/SupportGroup S
earch.php?lng=EN&data id=106412
11
https://www.textrazor.com/
12
https://permid.org/
Unsupervised Descriptive Text Mining for Knowledge Graph Learning
321
For the realization of the knowledge graph, we
selected Prot
´
eg
´
e
13
and Apache Jena
14
. Prot
´
eg
´
e was
used to build a starting skeleton OWL ontology, mod-
eling the taxonomy chosen for the case study as class
hierarchy, and the relations as object and data prop-
erties. From this base, the combination of Apache
Jena and RServe
15
allowed the invocation of the R
analyzes from Java code through socket server con-
nection, and the automatic population of the KG from
the results obtained. Following the learning layer
cake proposed in Section 4, we represented the stan-
dard and entity terms, the hierarchical links of the
latter with the defined taxonomy, the owl:sameAs
connections to existing representations on DBpedia
and Freebase, and the relationships of statistical ev-
idence between concepts. We also considered the pa-
tients’ opinion as the phenomenon to be described
(i.e, documentClass = opinionClass ∈ {pos ∨ neg ∨
neutral}), and we tagged each correlation with the p-
value associated with the dependency on the various
classes. We conducted experiments with generic (i.e.,
non-augmented) relationships, indicating probabilis-
tic correlations between concepts by additional class
nodes and taking the OBAN model (Sarntivijai et al.,
2016) as reference. As design choice, we considered
terms as individuals (with OWL2 punning
16
for link-
ing to external KBs), representing concept hierarchy
and correlations as classes, relations as object proper-
ties, and correlation probabilities as data properties.
5.4 Experiments
To test the greater expressive power introduced with
KGs and the new meta-level queries discussed in
Section 4.3, we used the Semantic Query-enhanced
Web Rule Language (SQWRL) (O’Connor and Das,
2009). SQWRL enables powerful queries on OWL re-
sources, using a high-level abstract syntax for the rep-
resentation of First Order Logic (FOL) and Horn-like
rules. We performed several SQWRL queries with
Pellet
17
reasoner on the KG learned from the textual
corpus. The higher expressive power of the queries
allows to search by related concepts, rather than by
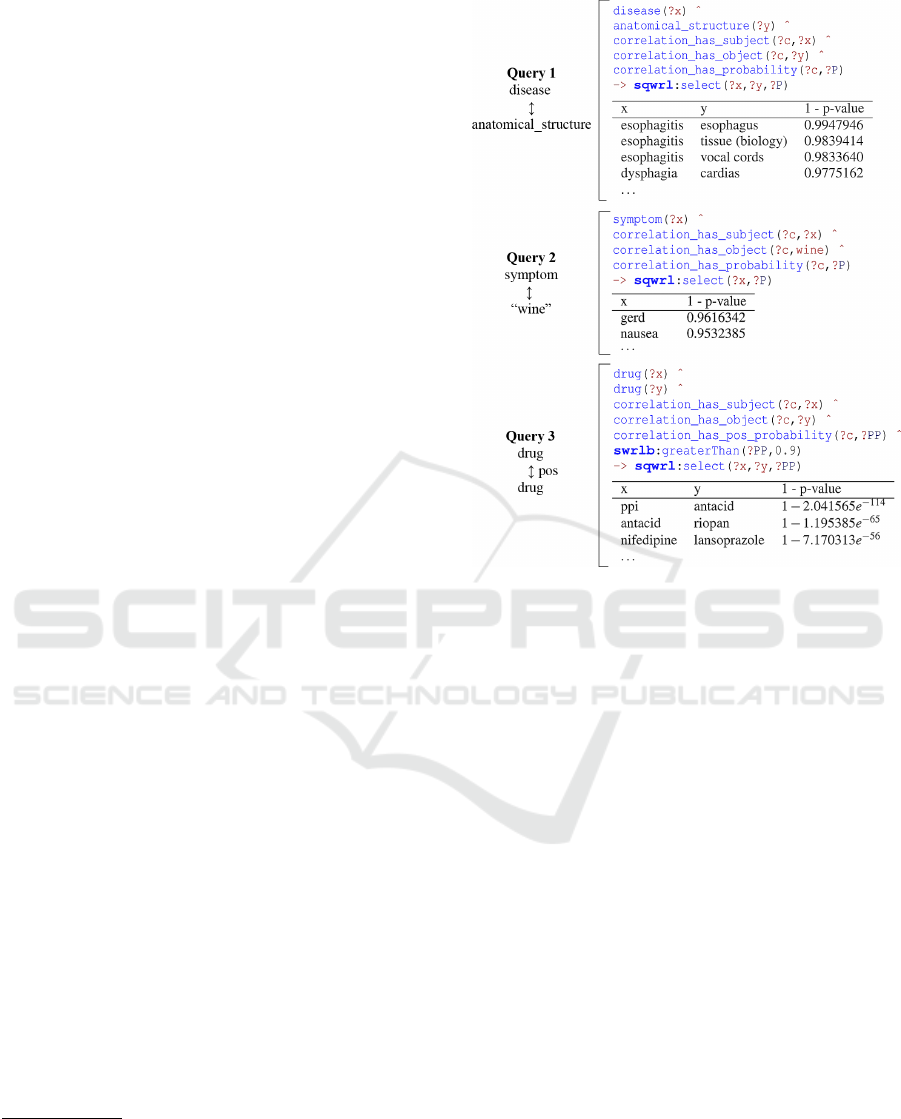
only related words. Figure 3 shows some key exam-
ples of queries related to the case study, previously
not possible with DTM4ED.
13
https://protege.stanford.edu/
14
https://jena.apache.org/
15
https://cran.r-project.org/web/packages/Rserve/
16
Technique focused on creating an individual with the
same IRI as a class.
17
https://github.com/stardog-union/pellet
Figure 3: SQRL queries on the KG based on Esophageal
Achalasia. Query 1 searches for unbounded correlations be-
tween diseases and anatomical structures; Query 2 searches
for correlations between symptoms (unbounded) and wine
(bounded); Query 3 searches for all pairs of related drugs
linked to a positive patients’ opinion.
6 CONCLUSIONS
We proposed an unsupervised and automatic tech-
nique of knowledge graph learning from corpora of
short unstructured and unlabeled texts. By extend-
ing a modular descriptive text mining methodology
previously introduced by us, we demonstrated how it
can be effectively applied to the task in question and
how KGs allow a significant increase in expressive
power, interrogability, and interpretability relatively
the extracted knowledge. We conducted experiments
as part of a case study focused on Esophageal Acha-
lasia, with the aim of building a KG directly from the
social messages shared by patients within the ever-
increasing number of online communities. SQWRL
queries were executed on the KG learned from the
textual corpus and their results show the potential
deriving from the new meta-level knowledge intro-
duced in the system. The contribution is not based
on dictionaries, and can be applied on other diseases
or completely different domains and languages (Ric-
cucci et al., 2007; Carbonaro, 2012).
KDIR 2020 - 12th International Conference on Knowledge Discovery and Information Retrieval
322

The work is extendable in several directions, such
as the embedding of the KG learning process within
neural networks, the semantic enrichment through
classifier systems for relation augmentation, and the
use of probabilistic reasoning. We also plan to carry
out a comprehensive validation of our contribution.
REFERENCES
Allen, C., Balazevic, I., and Hospedales, T. M. (2019). On
understanding knowledge graph representation. arXiv
preprint arXiv:1909.11611.
Asim, M. N., Wasim, M., Khan, M. U. G., et al. (2018).
A survey of ontology learning techniques and appli-
cations. Database, 2018.
Bizer, C., Lehmann, J., Kobilarov, G., et al. (2009). DB-
pedia - A crystallization point for the Web of Data.
Journal of web semantics, 7(3):154–165.
Bollacker, K., Evans, C., Paritosh, P., et al. (2008). Free-
base: a collaboratively created graph database for
structuring human knowledge. In Proceedings of
the 2008 ACM SIGMOD international conference on
Management of data, pages 1247–1250.
Bosselut, A., Rashkin, H., Sap, M., et al. (2019). Comet:
Commonsense transformers for automatic knowledge
graph construction. arXiv preprint arXiv:1906.05317.
Buitelaar, P., Cimiano, P., and Magnini, B. (2005). Ontology
learning from text: methods, evaluation and applica-
tions, volume 123. IOS press.
Carbonaro, A. (2012). Interlinking e-learning resources
and the web of data for improving student experience.
Journal of e-Learning and Knowledge Society, 8(2).
Carbonaro, A., Piccinini, F., and Reda, R. (2018). Integrat-
ing heterogeneous data of healthcare devices to enable
domain data management. Journal of e-Learning and
Knowledge Society.
Domeniconi, G., Masseroli, M., Moro, G., and Pinoli, P.
(2014a). Discovering new gene functionalities from
random perturbations of known gene ontological an-
notations. In KDIR 2014 - Proceedings of the In-
ternational Conference on Knowledge Discovery and
Information Retrieval, Rome, Italy, pages 107–116.
SciTePress.
Domeniconi, G., Masseroli, M., Moro, G., and Pinoli, P.
(2016a). Cross-organism learning method to discover
new gene functionalities. Computer Methods and Pro-
grams in Biomedicine, 126:20–34.
Domeniconi, G., Moro, G., Pagliarani, A., and Pasolini, R.
(2017). On deep learning in cross-domain sentiment
classification. In Proceedings of the 9th International
Joint Conference on Knowledge Discovery, Knowl-
edge Engineering and Knowledge Management - (Vol-
ume 1), Funchal, Madeira, Portugal, 2017, pages 50–
60. SciTePress.
Domeniconi, G., Moro, G., Pasolini, R., and Sartori, C.
(2014b). Cross-domain text classification through it-
erative refining of target categories representations. In
Fred, A. L. N. and Filipe, J., editors, KDIR 2014 - Pro-
ceedings of the International Conference on Knowl-
edge Discovery and Information Retrieval, Rome,
Italy, 21 - 24 October, 2014, pages 31–42. SciTePress.
Domeniconi, G., Moro, G., Pasolini, R., and Sartori, C.
(2014c). Iterative refining of category profiles for
nearest centroid cross-domain text classification. In
International Joint Conference on Knowledge Discov-
ery, Knowledge Engineering, and Knowledge Man-
agement, pages 50–67. Springer.
Domeniconi, G., Moro, G., Pasolini, R., and Sartori, C.
(2015). A Study on Term Weighting for Text Cate-
gorization: A Novel Supervised Variant of tf. idf. In
DATA, pages 26–37.
Domeniconi, G., Semertzidis, K., Lopez, V., Daly, E. M.,
Kotoulas, S., et al. (2016b). A novel method for
unsupervised and supervised conversational message
thread detection. In DATA, pages 43–54.
Ehrlinger, L. and W
¨
oß, W. (2016). Towards a Definition
of Knowledge Graphs. SEMANTiCS (Posters, Demos,
SuCCESS), 48.
Frisoni, G., Moro., G., and Carbonaro., A. (2020). Learn-
ing interpretable and statistically significant knowl-
edge from unlabeled corpora of social text messages:
A novel methodology of descriptive text mining. In
Proceedings of the 9th International Conference on
Data Science, Technology and Applications - Volume
1: DATA,, pages 121–132. INSTICC, SciTePress.
Gangemi, A. (2013). A comparison of knowledge extrac-
tion tools for the semantic web. In Cimiano, P., Cor-
cho, O., et al., editors, The Semantic Web: Seman-
tics and Big Data, pages 351–366, Berlin, Heidelberg.
Springer Berlin Heidelberg.
Hofmann, T. (2013). Probabilistic latent semantic analysis.
arXiv preprint arXiv:1301.6705.
Landauer, T. K. and Dumais, S. T. (1997). A solution to
Plato’s problem: The latent semantic analysis theory
of acquisition, induction, and representation of knowl-
edge. Psychological review, 104(2):211.
Li, J., Sun, A., Han, J., et al. (2020). A survey on deep learn-
ing for named entity recognition. IEEE Transactions
on Knowledge and Data Engineering.
Lin, B. Y., Chen, X., Chen, J., et al. (2019). Kagnet:
Knowledge-aware graph networks for commonsense
reasoning. arXiv preprint arXiv:1909.02151.
Maedche, A. and Staab, S. (2001). Ontology learning for
the semantic web. IEEE Intelligent systems, 16(2):72–
79.
Martins, P. H., Marinho, Z., and Martins, A. F. (2019). Joint
learning of named entity recognition and entity link-
ing. arXiv preprint arXiv:1907.08243.
Miller, G. A. (1998). WordNet: An electronic lexical
database. MIT press.
Moro, G., Pagliarani, A., Pasolini, R., and Sartori, C.
(2018). Cross-domain & in-domain sentiment anal-
ysis with memory-based deep neural networks. In
KDIR, pages 125–136.
O’Connor, M. J. and Das, A. K. (2009). Sqwrl: a query
language for owl. In OWLED, volume 529.
Unsupervised Descriptive Text Mining for Knowledge Graph Learning
323

Onuki, Y., Murata, T., Nukui, S., et al. (2019). Relation pre-
diction in knowledge graph by multi-label deep neural
network. Applied Network Science, 4(1):20.
Pagliarani, A., Moro, G., Pasolini, R., and Domeniconi, G.
(2017). Transfer learning in sentiment classification
with deep neural networks. In 9th International Joint
Conference, IC3K 2017, Funchal, Madeira, Portugal,
2017, Revised Selected Papers, volume 976 of Com-
munications in Computer and Information Science,
pages 3–25. Springer.
Patel, A. and Jain, S. (2019). Present and future of semantic
web technologies: a research statement. International
Journal of Computers and Applications, pages 1–10.
Rao, D., McNamee, P., and Dredze, M. (2013). Entity link-
ing: Finding extracted entities in a knowledge base. In
Multi-source, multilingual information extraction and
summarization, pages 93–115. Springer.
Ren, X., He, W., Qu, M., et al. (2016). Afet: Automatic
fine-grained entity typing by hierarchical partial-label
embedding. In Proceedings of the 2016 Conference on
Empirical Methods in Natural Language Processing,
pages 1369–1378.
Riccucci, S., Carbonaro, A., and Casadei, G. (2007).
Knowledge acquisition in intelligent tutoring sys-
tem: A data mining approach. In Lecture Notes in
Computer Science, volume 4827, pages 1195–1205,
Berlin, Heidelberg. Springer Berlin Heidelberg.
Sarntivijai, S., Vasant, D., Jupp, S., et al. (2016). Linking
rare and common disease: mapping clinical disease-
phenotypes to ontologies in therapeutic target valida-
tion. Journal of biomedical semantics, 7(1):1–11.
Singhal, A. (2012). Introducing the knowledge graph:
thing, not strings. Official Blog of Google. http:
//goo.gl/zivFV.
Tanon, T. P., Weikum, G., and Suchanek, F. (2020). Yago 4:
A reason-able knowledge base. In European Semantic
Web Conference, pages 583–596. Springer.
KDIR 2020 - 12th International Conference on Knowledge Discovery and Information Retrieval
324
