
CODO: An Ontology for Collection and Analysis of Covid-19 Data
Biswanath Dutta
1,*
and Michael DeBellis
2,
1
Indian Statistical Institute, Bangalore, India
2
Semantic Web Consultant, San Francisco, CA, U.S.A
Keywords: Domain Ontology, Ontology Engineering, COVID-19 Ontology, Novel Coronavirus Ontology, Disease,
Ontology Sharing and Reuse, Semantic Web.
Abstract: The COVID-19 Ontology for cases and patient information (CODO) provides a model for the collection and
analysis of data about the COVID-19 pandemic. The ontology provides a standards-based open source model
that facilitates the integration of data from heterogenous data sources. The ontology was designed by
analysing disparate COVID-19 data sources such as datasets, literature, services, etc. The ontology follows
the best practices for vocabularies by re-using concepts from other leading vocabularies and by using the
W3C standards RDF, OWL, SWRL, and SPARQL. The ontology already has one independent user and has
incorporated real world data from the government of India.
1 INTRODUCTION
The COVID-19 pandemic is a worldwide crisis
jeopardizing the health of everyone on the planet. One
of the tools to combat the pandemic is the collection
and analysis of data using FAIR principles.
1
Organizing data with technology based on FAIR
principles can provide open, federated data sources
that will provide healthcare workers with the critical
information required to track and eventually control
the growth of the pandemic. The COviD-19 Ontology
for cases and patient information (CODO) is a first
step at utilizing knowledge graph technology to help
combat the pandemic.
There are other initiatives that took a similar
approach (discussed in section 2.1). However, CODO
is unique in its scope and design approach. The main
goals of CODO are to:
1. Serve as an explicit ontology for use by data
and service providers to publish COVID-19
data using FAIR principles.
2. Develop and offer distributed, heterogenous,
semantic services and applications (e.g.,
decision support system, advanced
analytics).
*
https://sites.google.com/site/dutta2005/home
https://www.michaeldebellis.com/
1
https://www.go-fair.org/fair-principles/
3. Provide a standards-based reusable
vocabulary for the use of various
organizations (e.g., government agencies,
hospitals, academic researchers, data
publishers, news agencies, etc.) to annotate
and describe COVID-19 information.
The design of CODO has primarily been motivated
by the various COVID-19 data projection websites.
For example:
https://covid19.who.int/
https://www.isibang.ac.in/~athreya/incovid
19/
https://www.mygov.in/covid-19/
https://coronavirus.maryland.gov/
These sites show static presentations of COVID-19
cases, patient travel history, the relationships between
patients, etc. However, these kinds of static data and
visual representations need to be manually processed.
The search and visualization capabilities are typically
hard coded and impossible for users to customize
beyond the parameters defined in the software. More
importantly, the data is tightly coupled with specific
software to view it.
76
Dutta, B. and DeBellis, M.
CODO: An Ontology for Collection and Analysis of Covid-19 Data.
DOI: 10.5220/0010112500760085
In Proceedings of the 12th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2020) - Volume 2: KEOD, pages 76-85
ISBN: 978-989-758-474-9
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

With the development of the CODO ontology, we
aim at supporting the organization and representation
of COVID-19 case data on a daily basis, so that the
produced data can be queried and retrieved
semantically, and can also be taken as an input to
carry out advanced analytics (e.g., trend study,
growth projection). CODO also aims to facilitate the
representation of patient data, the relationships
between patients, between patient and locations,
changes over time, etc. This network data can support
the behaviour analysis of the disease, possible route
of disease spreading, various factors of disease
transmission, etc.
The CODO ontology will also help policymakers.
For example, in analysing how infrastructure was
utilized and where infrastructure could have been
utilized more effectively. Thus, CODO will help deal
with the current pandemic as well as provide a tool to
prepare for future potential crises.
The main contributions described in this paper are:
(i) Describe the CODO ontology. How it was
developed, how it relates to similar projects, how the
ontology can currently be leveraged to support
analysis of COVID-19 data and plans for future work.
(ii) Illustrate the process of automatic data integration
to the ontology.
(iii) Provide examples of how CODO has already
been utilized to analyse data about the pandemic.
The rest of the article is organized as follows:
section 2 describes the background that motivated
development of CODO. Specifically, a survey of
related work, an overview of FAIR principles and
how knowledge graphs can be utilized to provide
technology that implements these principles. Section
3 describes the methodology used to design the
CODO ontology. Section 4 describes the CODO
ontology highlighting some of the significant aspects
of it. Section 5 evaluates the CODO ontology by
automatically loading data on the pandemic and by
describing SPARQL queries that can analyse the data.
Finally, section 6 concludes the paper and discusses
next steps.
2 BACKGROUND
In this section we describe related work that we
surveyed before developing CODO. We also describe
2
https://bioportal.bioontology.org/ontologies/COVID19
3
https://github.com/oeg-upm/drugs4covid19-kg
4
http://covid19.squirrel.link/ontology/
the FAIR principles that were a driving rationale for
our decision to use knowledge graph technology.
2.1 Related Work
Dealing with a global pandemic is a knowledge
intensive process. As a result there have been several
ontologies developed related to the COVID-19
pandemic. Before developing CODO we did a survey
to determine if we could re-use an existing ontology.
We found nine relevant ontologies. However, none of
them were in the same space as what we needed: to
provide a semantic layer on top of case data from
India and the world. We briefly describe some of the
other COVID-19 ontologies in this section. Currently,
we have not found publications for any of them
except for the CIDO ontology (He et al., 2020).
The CIDO ontology (Ontology of Coronavirus
Infectious Disease) is part of the OBO Foundry
Ontology Library. CIDO is focused on analysing
Covid-19 from a medical standpoint. E.g., similarity
to other viruses, common symptoms, drugs that have
been attempted to treat the virus, etc.
COVID-19 Surveillance Ontology
2
is an
application ontology designed to support surveillance
in primary care. The main goal of this ontology is to
support COVID-19 cases and related respiratory
conditions using data from multiple brands of
computerized medical record systems. This work is
partially related to CODO. However, this ontology is
designed as a taxonomy consisting of classes such as
education for COVID-19, exposure to COVID-19,
definite and possible COVID-19, etc. This ontology
does not consist of any properties. This reduces the
semantic expressivity of the ontology.
DRUGS4COVID19
3
defines medications and
their relationships related to COVID-19. Some of the
key classes of the ontology are drug, effect, disease,
symptoms, disorder, chemical substance, etc. OVID-
19
4
is an ontology that consists of classes to enable
the description of COVID-19 datasets in RDF. Some
of the classes of this ontology are Dataset, Dataset of
the Johns Hopkins University, etc.
The World Health Organization’s (WHO)
COVIDCRFRAPID
5
ontology is a semantic data
model for the WHO's COVID-19 RAPID case record
form from 23 March 2020. This model provides
semantic references to the questions and answers of
the form.
5
https://bioportal.bioontology.org/ontologies/COVIDCRF
RAPID
CODO: An Ontology for Collection and Analysis of Covid-19 Data
77

The two ontologies that come closest to CODO
are Kg-COVID-19
6
(KG hub to produce a knowledge
graph for COVID-19 and SARS-COV-2.) and Linked
COVID-19 Data: Ontology
7
. However, both of these
ontologies have little semantic information in OWL
and are dependent on specific additional software to
utilize them.
CODO is an ontology that represents COVID-19
case data in a format based only on OWL and other
W3C standards which can be utilized by both other
ontologies and software systems. CODO provides
tracking of specific cases of the pandemic with details
such as how the patient is thought to have been
infected and potential additional contacts who may be
at risk due to their relationship to the infected
individual. CODO also provides tracking of clinical
tests, travel history, available resources, and actual
need (e.g., ICU bed, invasive ventilators), trend study
and growth projections.
2.2 FAIR Principles
The FAIR principles (Wilkinson 2016) are widely
seen as the best practice for scientific data. These
principles require that data be:
Findable. Data must have rich metadata and
unique and persistent identifiers.
Accessible. Metadata and data should be
understandable both to humans and machines.
Interoperable. Data and metadata should use
standards based languages that facilitate the use
of automated reasoning and federated queries.
Reusable. Data should leverage open industry
standard technology and domain vocabularies.
2.3 Knowledge Graphs
Knowledge graphs are widely recognized both by
industry and academia as the state of the market
technology for managing big data using FAIR
principles (Blumauer 2020).
Knowledge graphs are based on the following W3C
standards:
International Resource Identifiers (IRI)
Resource Description Framework
(RDF/RDFS)
Web Ontology Language (OWL)
Semantic Web Rule Language (SWRL)
SPARQL Protocol and RDF Query Language
(SPARQL)
6
https://github.com/Knowledge-Graph-Hub/kg-covid-19
An IRI looks very much like a URL. The primary
difference is that URLs typically point to resources
that are meant to be displayed in a browser. IRI’s are
more general than URLs and can describe resources
to a finer level of granularity than an HTML page. An
IRI can be any resource such as a class, a property, an
individual, etc. (DuCharme, 2011)
RDF is the foundation language for describing IRI
data as a graph rather than in relational or other types
of formats (W3C 2014).
RDFS is layered on top of RDF and provides basic
concepts such as classes, properties, and collections
(W3C 2014a).
OWL is layered on top of RDFS and provides the
semantics for knowledge graphs. OWL is an
implementation of Description Logic which is a
decidable subset of First Order Logic (W3C 2012).
OWL enables the definition of reasoners which are
automated theorem provers. OWL reasoners first
ensure that an ontology model is consistent. If the
model is not consistent the reasoner will highlight the
probable source of the inconsistency. If the model is
consistent reasoners can then deduce additional
information based on concepts described below such
as transitivity, inverses, value restrictions, etc. OWL
reasoners originated with the KL-One family of
knowledge representation languages and successors
to KL-One such as Loom. (MacGregor, 1991).
SWRL is a rule-based language that extends OWL
reasoners with additional constructs beyond what can
be described with OWL’s Description Logic
language (W3C 2004).
Finally, SPARQL allows federated queries across
heterogeneous sources of data. A SPARQL query
defines a graph pattern that is matched against the
available data sources and returns the data that
matches the pattern (DuCharme, 2011).
3 METHDOLOGY
This section provides a description of the CODO
ontology design and development methodology.
For designing an ontology, there are several
methodologies available in the literature. Some of the
state-of-the-art popular approaches are
METHONTOLOGY (Fernandez et al., 1997), TOVE
(Gruninger and Fox, 1995), DILIGENT (Vrandecic et
al., 2005), NeOn (Suárez-Figueroa et al, 2012),
UPON (De Nicola et al., 2005), YAMO (Dutta et al.,
2015), etc. The design approach of CODO has been
primarily influenced by YAMO, a step-by-step
7
https://zenodo.org/record/3765375#.XraWJmgzbIU
KEOD 2020 - 12th International Conference on Knowledge Engineering and Ontology Development
78
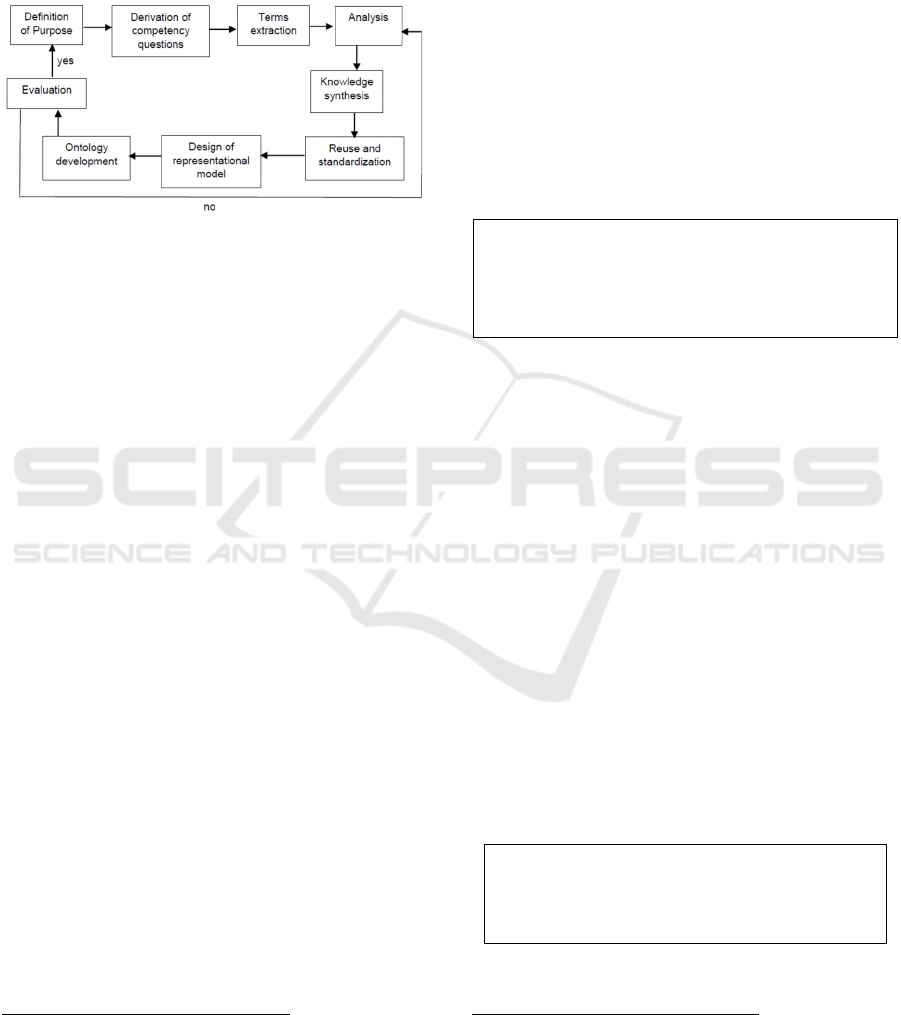
approach for building a formally defined large-scale
faceted ontology. The YAMO methodology also
provides a set of ontology design guiding principles
which is quite unique. The steps of the CODO
ontology design process are displayed in Figure 1 and
described below.
Figure 1: Steps of CODO ontology development process.
S1: Definition of purpose - this step describes the
purpose and application of the ontology. As discussed
above the purpose of the CODO ontology is to
facilitate publication of COVID-19 data as a
knowledge graph and to develop semantic services
and applications (e.g., decision support system,
advanced analytics) (Dutta, 2017). Also, to enable
various organizations (e.g., government agencies,
hospitals, researchers, data publishers, news
agencies, etc.) to annotate and describe COVID-19
information.
S2: Derivation of competency questions – Elaborate
the purpose defined in S1 into a set of competency
questions. Some of these competency questions are:
I. How many people recovered from COVId-
19 in place p until date t?
II. How many people died in country c?
III. Give me the travel history of patient p.
IV. Give me the COVID-19 patients p and their
relationship r, if any.
V. Give me the COVID-19 patients p who are
in family relationships f.
VI. Give me the primary reasons i for the
maximum number of COVID-19 patients p.
VII. Give me the most prevalent symptoms s of
Severe COVID-19 d.
VIII. Find all People p who are related to someone
r who has been diagnosed with Covid and
who has not yet been tested.
8
https://github.com/CSSEGISandData/COVID-19
9
https://www.isibang.ac.in/~athreya/incovid19/data.html
10
https://www.mohfw.gov.in/pdf/FinalGuidanceonMangae
mentofCovidcasesversion2.pdf
S3: Term extraction – in extracting the terms for the
ontology, we primarily referred to COVID-19
datasets on cases, patients, relations (e.g., family, co-
workers), geographic locations, and date-time
information. For this purpose, we referred to data
repositories, such as the COVID-19 Data Repository
by the Center for Systems Science and Engineering
(CSSE) at Johns Hopkins University
8
and the data
repository curated by Athreya et al.
9
We also used the
literature including government published documents
and guidelines. For example, guidance documents on
appropriate management of suspected/confirmed
cases of COVID-19
10
, WHO published literature
11
,
newspaper articles, etc. on CVID-19. Some of the
most significant extracted terms are:
patient, doctor, covid-19 dedicated facility, covid
care centre, dedicated covid health centre, covid-
19 clinical facility, mild and very mild covid-19,
severe covid-19, moderate covid-19, exposure to
civid-19, vital signs, test finding, symptom, SpO2,
cases, blood pressure, temperature
S4: Analysis - following the extraction of the terms,
this steps involves analysing the derived compound
and complex concepts and breaking them into their
elemental entities. The analysis is done based on the
definition and characteristic of each of the concepts
and then grouping them according to their similarity.
For example, analysing the terms covid care centre
(any facility, such as hotels/lodges/hostels/stadiums
for providing care to COVID-19 patients) and
dedicated health centre (hospitals that shall offer care
for all cases that have been clinically assigned as
moderate) based on their definition reveals that both
of them have a common point and can be grouped as
subclasses of the class for covid dedicated facility.
S5: Knowledge synthesis – this step involves
synthesizing and arranging the knowledge by
defining the relationships between the concepts. This
step lead to the discovery of concept hierarchies. For
example (the indention indicates the hierarchy)
Organization
COVID-19 dedicated facility
Covid care centre
Dedicated covid health centre
Dedicated covid hospital
11
https://www.who.int/news-room/q-a-detail/q-a-
coronaviruses
CODO: An Ontology for Collection and Analysis of Covid-19 Data
79
S6: Reuse and standardization – technology can only
go so far to enable integration and re-use. Ultimately,
what is required is to develop and re-use domain
vocabularies. We have followed this best practice in
the development of CODO. We have integrated
concepts from the following vocabularies into
CODO: Schema.org, Friend of a Friend (FOAF)
vocabulary
12
, SNOMED CT
13
and OBO.
14
Schema.org is used for modelling common
concepts such as gender and locations. FOAF is used
to model Agents, such as Person and Organization
classes and related properties. SNOMED CT and
OBO are used to model clinical findings and
symptoms.
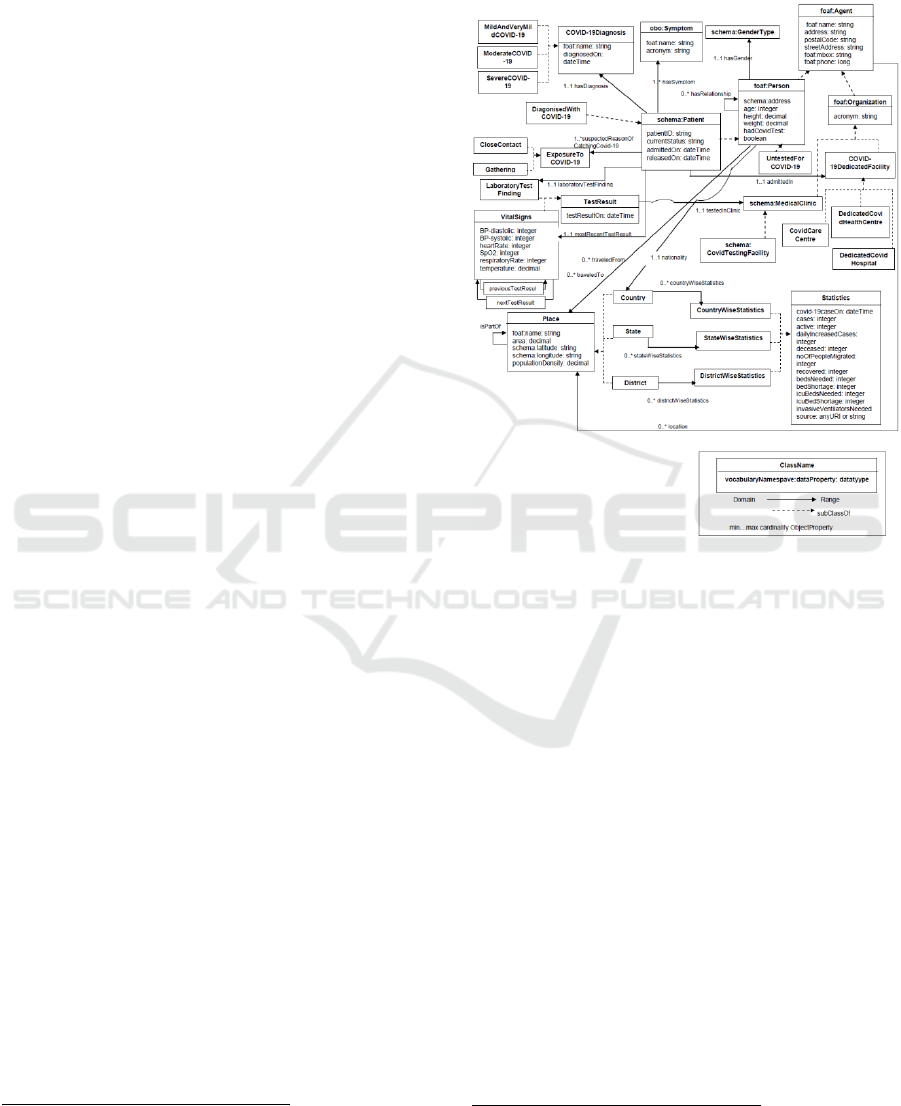
S7: Design of representational model – involves
structuring and modelling the domain knowledge
produced in the previous step. The idea is to model
the domain knowledge showing its various
components, such as classes, properties and their
relationships. This is important as in one side it
ensures the aggregation, substitution, improvement,
sharing and reapplication of the ontology (Dutta et al,
2015, Giunchiglia & Dutta, 2011), and on the other
side it provides a consolidated view of the ontology
and its coverage. Figure 2 shows a high-level view of
the CODO model.
S8: Ontology development – this step involves
developing the formal model using a formal logic
language. For developing CODO, we used OWL-DL,
a Description Logic ontology language. CODO was
designed using the Protégé ontology editor (Musen
2015) developed at Stanford University. In addition
to the core editor we utilized the Pellet reasoner,
SWRLTab, Cellfie, and Snap SPARQL plugins. The
details of the ontology are provided in Section 4.
S9: Evaluation – this step involves evaluating how
closely the ontology meets the design goals. It gauges
the technical competence of the ontology. There is no
easy and automatic way of evaluating an ontology.
The reasoners can verify the syntactic structure and
consistency of the ontology but cannot evaluate the
domain knowledge and knowledge structure. The
manual evaluation by domain experts is one of the
most prevalent methods (Lozano-Tello and Gomez-
Perez, 2004, Dutta et al., 2015).
To verify that the CODO ontology serves the
purpose it was designed for, we imported data on the
pandemic from the government of India using the
12
http://xmlns.com/foaf/spec/
13
http://www.snomed.org/
Cellfie Protégé plugin (described in section 5.1). We
also wrote SPARQL queries based on the competency
questions described in S2. An example SPARQL
query is illustrated in section 5.2.
Figure 2: Overview of the CODO model.
4 THE CODO ONTOLOGY
In this section we describe some of the important
classes, properties, and some sample individuals that
we developed to give users of the ontology examples
of the types of reasoning that can be automated with
the ontology.
The current version CODO1.2 is available here:
https://github.com/biswanathdutta/CODO. Also, the
HTML specification documents of the ontology is
available here: https://isibang.ac.in/ns/codo.
CODO1.2 consists of 50 classes, 62 object properties
and 45 data properties. The basic ontology has a
handful of sample individuals for illustrative
purposes. The first application of CODO on actual
data from the government of India has over 23,000
individuals representing cases of the pandemic in
India (the data dump is available here:
https://github.com/biswanathdutta/CODO).
14
http://www.obofoundry.org/
KEOD 2020 - 12th International Conference on Knowledge Engineering and Ontology Development
80

4.1 Properties and Reasoning
One of the main differences between OWL and other
object-oriented models is that properties in OWL are
first class entities that are not bundled with a specific
class. In traditional Object-Oriented Programming
(OOP) a property is defined as part of a class
definition. If the class is deleted so is the property. In
OWL properties are independent entities (W3C
2006).
Properties in OWL are equivalent to binary
relations in First Order Logic (FOL). They also have
a number of capabilities that relations in FOL have
and that can be automatically enforced by an OWL
reasoner.
Two examples of such capabilities leveraged by
CODO are symmetric and inverse properties. A
symmetric property is such that if the tuple <a, b> is
in the property then the tuple <b, a> must be as well.
An example of a symmetric property in CODO is
hasSpouse. If a Person p000001 hasSpouse p000004
then the reasoner automatically infers that p000004
hasSpouse p000001.
Inverses are defined such that if <a, b> is in a
property then <b, a> is in its inverse property. An
example of this in CODO are the hasChild and
isChildOf properties. These are inverse properties and
one merely has to assert that one of the properties
holds for two individuals and the reasoner will infer
that the appropriate inverse holds for the two
individuals as well.
Since OWL properties are FOL relations they are
sets (of binary tuples). Thus, just as classes can have
subclasses where the subclass is a subset of the
superclass so properties can have sub-properties
where all the tuples in the super-property are in the
sub-property but not necessarily vice versa.
One way this is leveraged in CODO is in the
hasRelationship property hierarchy (see figure 3).
The hasRelationship property captures some of
the ways that people can have interactions with each
other. It also has sub-properties that differentiate
hasCloseRelationship relations from others. A close
relationship is one where the two people are likely to
regularly live or work together such as parents and
children, co-workers, and roommates. This is
distinguished from other types of relationships such
as aunts and uncles where it is less likely that the two
individuals are in regular close contact.
Figure 3: The hasRelationship Property Hierarchy.
For example, in the test data for CODO we
asserted that p000001 hasDaughter p000007. The
reasoner automatically inferred that p000001
hasChild p000007 (because hasDaughter is a sub-
property of hasChild) and that p000001
hasCloseRelationship p000007 (because hasChild is
a sub-property of hasCloseRelationship). This
property hierarchy will be leveraged further as we
combine it with the capability to define necessary and
sufficient axioms for classes in the next section.
4.2 Defined Classes
OWL can be used to define axioms that are necessary
and sufficient for an individual to be a member of a
class. The OWL reasoners can use these axioms to
automatically restructure the class hierarchy as well
as to do significant additional reasoning about
individuals.
If one defines axioms for a class in the
SubClassOf field in Protégé these are necessary
axioms for the class. I.e., they must be true for any
individual that is a member of that class but it may not
be the case that every individual that fulfils that axiom
is a member of that class. When axioms are defined
in the EquivalentTo field in Protégé these axioms are
both necessary and sufficient conditions for that class.
I.e., any individual that satisfies those axioms is
automatically inferred to be an instance of that class.
Classes with necessary and sufficient axioms are
known as defined classes in OWL. In CODO we have
combined sub-properties with a defined class to
create a defined subclass of Person called
UrgentlyNeedsCovidTest. The necessary and
sufficient axioms for this class are:
CODO: An Ontology for Collection and Analysis of Covid-19 Data
81
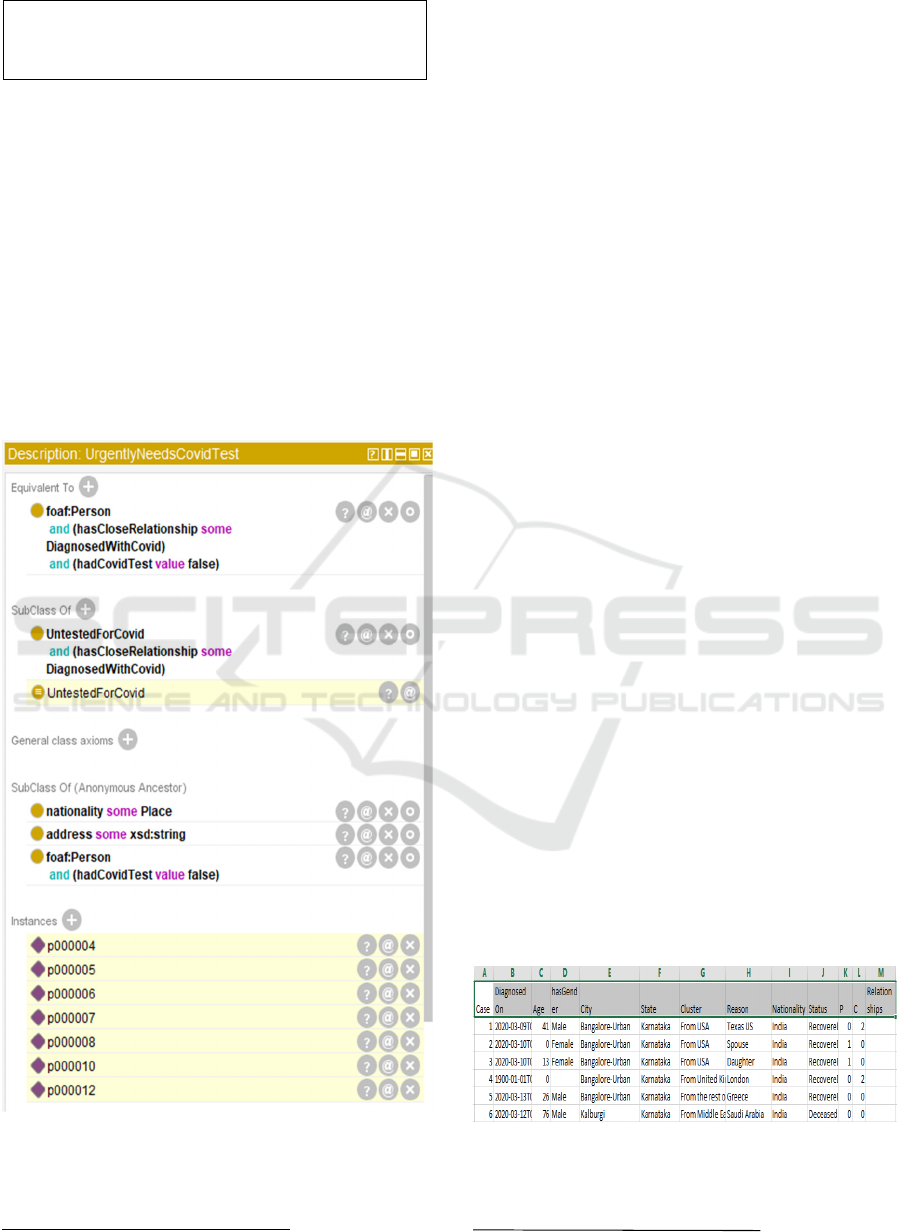
foaf:Person
and (hasCloseRelationship some
DiagnosedWithCovid) and (hadCovidTest value
false)
DiagnosedWithCovid is also a defined class with
necessary and sufficient conditions such that anyone
who has been diagnosed with the virus is a member
of that class. Thus, UrgentlyNeedsCovidTest defines
a class for anyone who has a close relationship that
has been diagnosed with Covid-19 and who has
themselves not yet had a Covid-19 test.
Figure 4 displays this defined class. The
individuals in the instances field are instances of the
Person class that the reasoner has inferred are also
instances of this defined class. Note: anything in
Protégé highlighted in yellow was not defined by
some input data but was inferred by the reasoner
based on the data and the axioms in the ontology.
Figure 4: UrgentlyNeedsCovidTest Defined Class.
15
https://www.mohfw.gov.in/
5 CODO EVALUATION
In this section we describe how we have evaluated the
CODO ontology. This evaluation was done by:
Populating the ontology with real world data
from the government of India.
Making the ontology available as a vocabulary
for others to use which has already occurred with
one system on the Bioportal site.
Developing SPARQL queries which implement
some of the use cases identified in our
development methodology.
Exporting the ontology to a commercial
triplestore product which provides capabilities
for much larger datasets and additional
visualization.
5.1 Data Population
In this section we describe how we have populated
the ontology with data from the Indian Ministry of
Health and Family Welfare website.
15
This data has
been collected into spreadsheets by Siva Athreya and
other researchers at the Indian Statistical Institute.
16
A snapshot of a datasheet is shown in Figure 5.
To integrate this data into the ontology we used
the Cellfie Protégé plugin (O'Connor 2010). Cellfie
allows the user to define transformation rules to
convert rows in a spreadsheet into instances of a class
in an ontology and property values for that instance
(see Table 1 for an example of a transformation rule).
We utilized Cellfie to import data about the
pandemic from the Indian province of Karnataka. Each
row in the spreadsheet corresponded to a case where a
specific patient was diagnosed with Covid. In the
CODO ontology each row was transformed into an
individual of the Patient class and values in each row
such as the age, sex, date of diagnosis, etc. were
transformed into the appropriate property values for
each patient.
Figure 5: A glimpse of the dataset.
16
https://www.isibang.ac.in/~athreya/incovid19/data.html
KEOD 2020 - 12th International Conference on Knowledge Engineering and Ontology Development
82
The resulting ontology had over 23,000 individual
patients with data from March to the beginning of
July 2020.
Table 1: Example Transformation rule.
Individual: @A*(mm:hashEncode
rdfs:label=("patient", @A*))
Types: Patient
Facts: 'diagnosed on' @B*(xsd:dateTime),
age @C*(xsd:decimal),
'has gender' @D*,
'city' @E*,
'state' @F*,
'travelled from' @G*,
nationality @I*,
status @J*,
'has caused any secondary infections'
@L*(xsd:boolean)
5.2 SPARQL Queries
The SPARQL query engine is roughly analogous to
OWL as SQL is to relational databases. However,
since the underlying structure of OWL are graphs
rather than tables, SPARQL constructs graph patterns
and then searches knowledge graphs for any
individuals that match the graph pattern. Like SQL,
SPARQL can do more than query, it can also delete,
insert, and transform data (DuCharme, 2011).
SPARQL has many features that provide
additional value beyond the capabilities described so
far. For one thing, SPARQL can integrate data from
multiple heterogeneous data sources. The beginning
of each SPARQL query starts with a list of
namespaces and the IRI where these namespaces can
be found. Hence, SPARQL can do queries across
broad data sets from multiple sources enabling a truly
federated virtual knowledge base. Since different data
sources may have different formats SPARQL can use
pattern matching to transform data from various
sources.
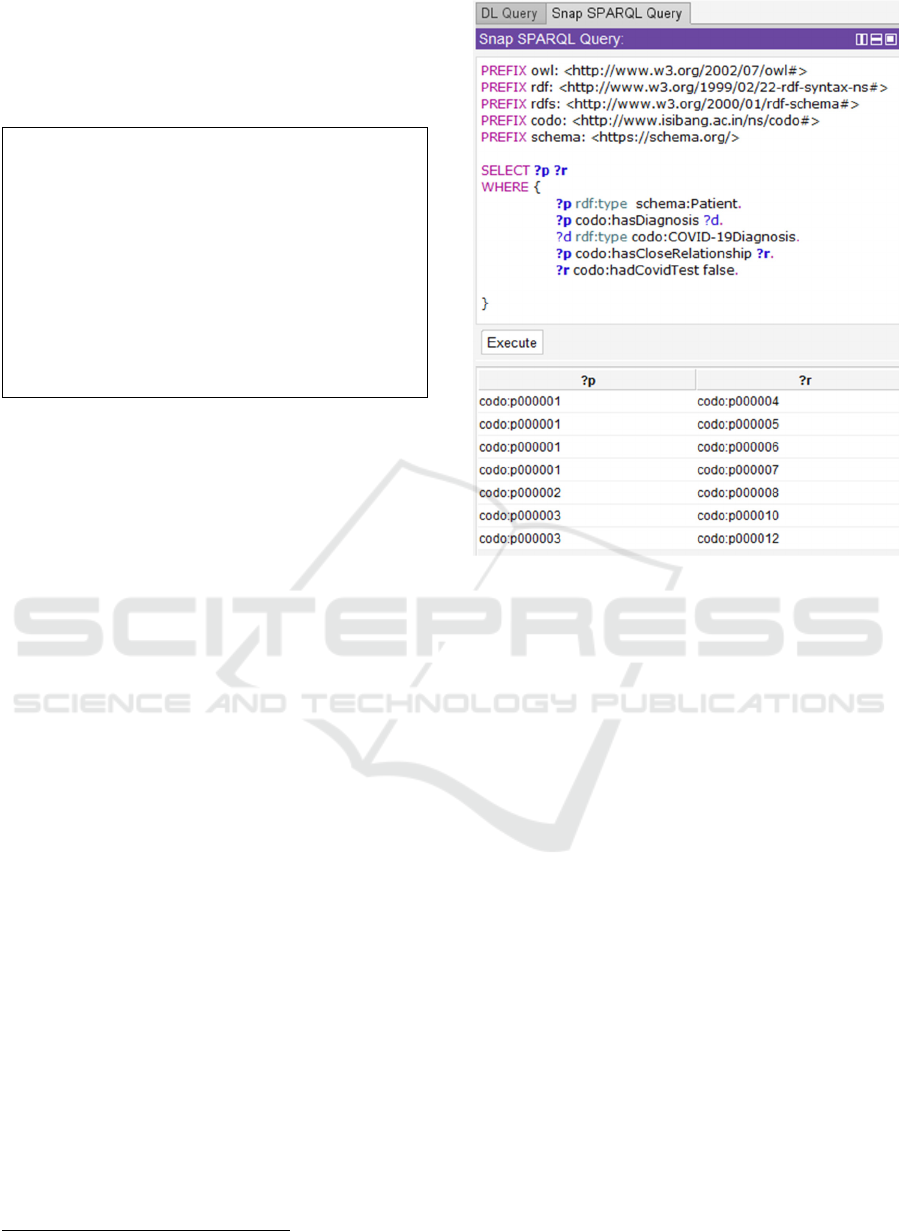
Figure 6 displays a SPARQL query using the Snap
SPARQL query plugin in Protégé. The above shows
the SPARQL syntax for the query “Find all People
who have a close relation to someone who has been
diagnosed with Covid and who has not yet been
tested.”
17
http://bioportal.bioontology.org/projects/Ping
Figure 6: CODO SPARQL Query.
The Prefixes first define the various namespaces
that the query will utilize and their IRIs. In this case
the query performs the same logic as the defined class
described in section 4.2. One advantage of using the
SPAQRL query is that in addition to seeing the
specific individuals who match the query (the ?r
column) we can also see the closely related individual
that has been diagnosed with Covid-19 (the ?p
column).
5.3 Utilization of CODO Vocabulary
One of the primary design goals for CODO was that
it could serve as a reusable vocabulary for other
projects. Although we have only recently published
the ontology on Github and Bioportal, we already
have one user from the Bioportal site: the Ping
COVID-19 risk detection system.
17
5.4 Triplestores and Visualization
Protégé is a modelling tool not a persistent storage
tool. Although it is possible to persist knowledge
graphs designed in Protégé with small to medium sets
of test data, to achieve the true power of knowledge
graph technology a triplestore product is required. A
CODO: An Ontology for Collection and Analysis of Covid-19 Data
83
triplestore is a database designed to store data as
graphs rather than as relational tables (Blumauer
2020).
Although the current test data in CODO can be
stored in files from Protégé we have already begun to
hit the limits of Protégé with the data we have
imported from the Indian government. We have
begun to utilize a triplestore environment in
anticipation of scaling up CODO to having data for
up to a million patients rather than the thousands
currently in the ontology. We have imported CODO
into the free version of the Allegrograph triplestore
product from Franz Inc. The free version is still
capable of supporting 5 million triples and also
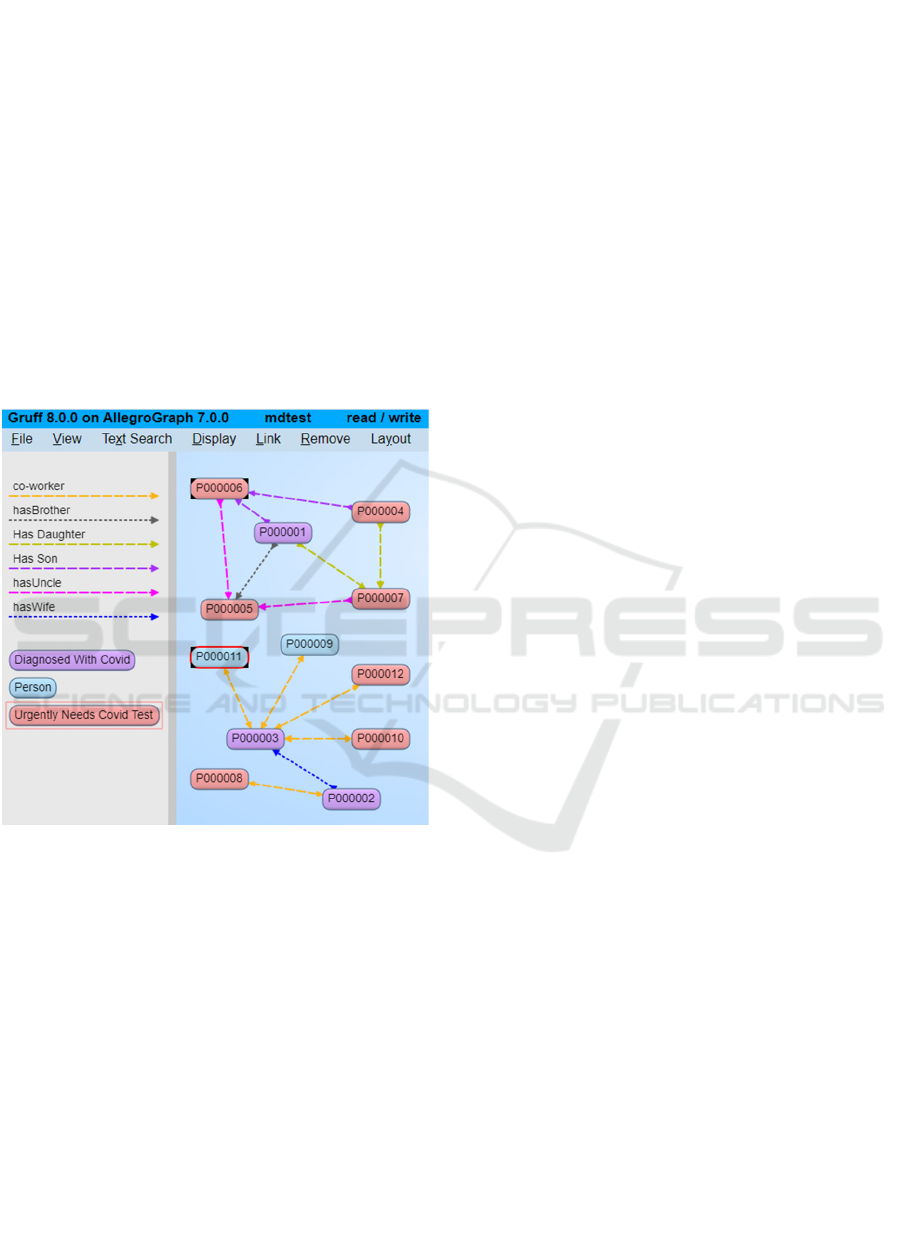
supports Allegro’s Gruff visualization tool. Figure 7
displays a small number of test data patients from the
current CODO ontology using Allegro’s Gruff tool.
Figure 7: CODO in Allegro’s Gruff Visualization Tool.
6 CONCLUSIONS
The CODO ontology is only the first step in providing
a knowledge graph model for COVID-19 based on
FAIR data principles. The current CODO ontology
has already found its use in a real world project called
Ping and in uploading thousands of cases from data
collected by the government of India. The main
limitation of the current work is it lacks a truly
rigorous evaluation of the developed ontology. In our
future work, we aim to evaluate the ontology by
health domain experts and also by applying the
Information Retrieval system evaluation technique.
In addition, we plan to enhance the current CODO
ontology by integrating many more COVID-19
datasets available on the Web, both from India and
world-wide. Finally, we plan to publish CODO using
a triplestore database published as a SPARQL
endpoint. This will provide capabilities to handle
much larger datasets. It will also enable SPARQL
queries that can integrate CODO with other
complimentary ontologies such as CIDO.
ACKNOWLEDGEMENTS
This work was conducted using the Protégé resource,
which is supported by grant GM10331601 from the
National Institute of General Medical Sciences of the
United States National Institutes of Health. Thanks to
Franz Inc. (http://www.allegrograph.com) and its
help with AllegroGraph and Gruff.
REFERENCES
Blumauer, Andreas, Nagy, Helmut, 2020. The Knowledge
Graph Cookbook. Edition Mono.
DuCharme, Bob, 2011. Learning SPARQL. O’Reilly.
Dutta, B., Chatterjee, U. and Madalli, D. P., 2015. YAMO:
Yet Another Methodology for Large-scale Faceted
Ontology Construction. Journal of Knowledge
Management. 19 (1): 6 – 24.
Dutta, B. (2017). Examining the interrelatedness between
ontologies and Linked Data. Emerald Library Hi Tech,
Vol. 35, no. 2, pp. 312-331.
De Nicola, A., Missikoff, M., and Navigli, R. (2005). A
proposal for a unified process for ontology building:
UPON. In International Conference on Database and
Expert Systems Applications, Springer, Berlin,
Heidelberg, 655-664.
Fernandez, M. and Gomez-Perez, A., Juristo, N. (1997).
Methontology: from ontological art towards ontological
engineering. In Proceedings of the AAAI97 Spring
Symposium Series on Ontological Engineering.
Gruninger, M. and Fox, M. (1995). Methodology for the
design and evaluation of ontologies. Workshop on
Basic Ontological Issues in Knowledge Sharing,
Montreal, Canada.
Giunchiglia, F. and Dutta, B. (2011). DERA: a Faceted
Knowledge Organization Framework. Technical
Report, # DISI-11-457, 3-18.
http://eprints.biblio.unitn.it/archive/00002104/
He, Y., Yu, H., Ong, E. et al. (2020). CIDO, a community-
based ontology for coronavirus disease knowledge and
data integration, sharing, and analysis. Sci Data 7, 181
(2020). https://doi.org/10.1038/s41597-020-0523-6
Lozano-Tello, A. and Gomez-Perez, A. (2004).
Ontometric: A method to choose the appropriate
ontology. Journal of Database Management, 15(2):1–
18.
KEOD 2020 - 12th International Conference on Knowledge Engineering and Ontology Development
84

MacGregor, Robert, 1991. Using a description classifier to
enhance knowledge representation. IEEE Expert. 6 (3):
41–46. doi:10.1109/64.87683
Musen, Mark, 2015. The Protégé Project: A Look Back and
a Look Forward. A.I. Matters 1(4).
https://dl.acm.org/doi/10.1145/2757001.2757003
M.J. O'Connor, C. Halaschek-Wiener, M. A. Musen.
Mapping Master: A Flexible Approach for Mapping
Spreadsheets to OWL. 9th International Semantic Web
Conference (ISWC), Shanghai, China, 2010.
Suárez-Figueroa, M. C., Gómez-Pérez, A. and Fernández-
López, M. (2012). The NeOn Methodology for
Ontology Engineering, in Ontology Engineering in a
Networked World, M. C. Suárez-Figueroa, A. Gómez-
Pérez, E. Motta, and A. Gangemi, Eds., ed Berlin,
Heidelberg: Springer Berlin Heidelberg, 9-34.
Vrandecic, D. and Pinto, S., Tempich, C., Sure, Y. (2005).
The DELIGENT knowledge processes. In Journal of
Knowledge Management, 9(5): 85-96.
W3C, 2004. SWRL: A Semantic Web Rule Language
Combining OWL and RuleML. W3C Member
Submission. https://www.w3.org/Submission/SWRL/
W3C, 2006. A Semantic Web Primer for Object-Oriented
Software Developers. https://www.w3.org/TR/sw-
oosd-primer/
W3C, 2012. Web Ontology Language Document Overview
(Second Edition). W3C Recommendation.
https://www.w3.org/TR/owl2-overview/
W3C, 2014. RDF 1.1: Concepts and Abstract Syntax. W3C
Recommendation. https://www.w3.org/TR/rdf11-
concepts/
W3C, 2014a. RDF Schema 1.1. W3C Recommendation.
https://www.w3.org/TR/rdf-schema/
Wilkinson, M., Dumontier, M., Aalbersberg, I. et al., 2016.
The FAIR Guiding Principles for scientific data
management and stewardship. Sci Data 3, 160018
(2016). https://doi.org/10.1038/sdata. 2016.18.
CODO: An Ontology for Collection and Analysis of Covid-19 Data
85
