
A Comprehensive Study on Subgraph Crossover in Cartesian Genetic
Programming
Roman Kalkreuth
Department of Computer Science, TU Dortmund University, Otto-Hahn-Straße 14, Dortmund, Germany
Keywords:
Cartesian Genetic Programming, Crossover, Subgraph.
Abstract:
While tree-based Genetic Programming is often used with crossover, Cartesian Genetic Programming (CGP)
is mostly used only with mutation as the sole genetic operator. In contrast to comprehensive and fundamental
knowledge about crossover in tree-based GP, the state of knowledge in CGP appears to be still ambiguous
and ambivalent. Two decades after CGP was officially introduced, the role of recombination in CGP is still
considered to be an open and remaining question. Although some promising steps have been taken in the last
years, comprehensive studies are needed to evaluate the role of crossover in CGP on a large set of problems.
In this paper, we take a step forward on the crossover issue by comparing algorithms that utilize the subgraph
crossover technique which has been proposed for CGP to the traditional mutation-only CGP. Experiments on
well-known symbolic regression and Boolean function problems demonstrate that the use of algorithms that
utilize the subgraph crossover outperform the mutation-only CGP on well-known benchmark problems.
1 INTRODUCTION
Genetic programming (GP) can be understood as
a method that enables the automatic derivation of
programs for problem-solving. First work on GP
has been done by Forsyth (1981), Cramer (1985)
and Hicklin (1986). Later work by Koza (1990, 1992,
1994) significantly popularized the field of GP. GP
traditionally uses trees as program representation but
is not limited to this representation model. Over
two decades ago Miller, Thompson, Kalganova, and
Fogarty presented first publications on Cartesian Ge-
netic Programming (CGP) —an encoding model in-
spired by the two-dimensional array of functional
nodes connected by feed-forward wires of an FPGA
device (Miller et al., 1997; Kalganova, 1997; Miller,
1999). CGP offers a graph-based representation
which in addition to standard GP problem domains,
makes it easy to be applied to many graph-based ap-
plications such as electronic circuits, image process-
ing, and neural networks. In addition to tree-based
GP CGP has pivotal advantages:,
• CGP encodes a directed acyclic graph (DAG)
which allows the evolution of structures that can
be represented as DAGs. In this way, CGP also
facilitates evolving topologies.
• The maximal size of encoded solutions is
bounded, saving CGP to some extent from bloat
that is characteristic to GP
Standard CGP is mostly used with mutation as the
sole genetic operator and with a (1 + λ)-strategy. The
reason for this is that various standard genotypic
crossover techniques failed to improve the search per-
formance of standard CGP. In contrast to comprehen-
sive knowledge about crossover in tree-based GP, the
state of knowledge in CGP appears to be still ambigu-
ous and ambivalent. The state of knowledge about
crossover in CGP has been recently surveyed and the
role of crossover is still considered to be an open and
remaining question. Even if some progress has been
made in recent years, comprehensive and detailed
knowledge about crossover in CGP is still missing.
A promising step forward was made by the introduc-
tion of the subgraph crossover (Kalkreuth et al., 2017)
but this technique has not been comprehensively stud-
ied in the past. Therefore, this work follows up for-
mer work on the crosover question by investigating if
the search performance of CGP algorithms that utilize
the subgraph crossover can be more efficient as the
commonly used mutation-only CGP on a set of well-
known benchmark problems. This work also surveys
and analyzes relevant work which contributed to the
knowledge about crossover in standard CGP. Based
on our experiments and their results, we formulate
Kalkreuth, R.
A Comprehensive Study on Subgraph Crossover in Cartesian Genetic Programming.
DOI: 10.5220/0010110700590070
In Proceedings of the 12th International Joint Conference on Computational Intelligence (IJCCI 2020), pages 59-70
ISBN: 978-989-758-475-6
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
59

and analyze hypotheses that address important search
performance dogmas in the field of CGP. The primary
intention of this work is to shed more light on the role
of the subgraph crossover in CGP and to pave the way
for further analyses of the behavior of crossover based
CGP algorithms. We study the fitness space on a sub-
set of problems to obtain more understanding of our
findings.
Section 2 of this paper describes CGP and the sub-
graph crossover. Section 3 surveys previous work on
crossover in CGP, outlines the need for a comprehen-
sive study and hypotheses to be examined are formu-
lated. Section 4 is devoted to the experimental results
and the description of our experiments. We analyze
hypotheses that have been formulated in Section 3. In
Section 6 we discuss the results of our experiments.
Finally, Section 7 gives a conclusion and outlines fu-
ture work.
2 RELATED WORK
2.1 Cartesian Genetic Programming
In contrast to tree-based GP, CGP represents a ge-
netic program via genotype-phenotype mapping as an
indexed, acyclic, and directed graph. Originally the
structure of the graphs was a rectangular grid of n
r
rows and n
c
columns, but later work focused on a rep-
resentation with one row. The CGP decoding proce-
dure processes groups of genes and each group refers
to a function node of the graph. An exception are the
last genes of the genotype which represent the out-
puts. Each node is represented by two types of genes
which index the function number in the GP function
set and the node inputs. These nodes are called func-
tion nodes and execute functions on the input values.
The number of input genes depends on the maximum
arity n
a
of the function set. Given the number of out-
puts n
o
, the last n
o
genes in the genotype represent
the indices of the nodes, which lead to the outputs.
A backward search is used to decode the correspond-
ing phenotype. An example of the backward search
of the most popular one-row integer representation is
shown in Figure 1. The backward search starts from
the program output and processes all nodes which are
linked in the genotype. In this way, only active nodes
are processed during evaluation. The genotype in Fig-
ure 1 is grouped by the function nodes.
The first (underlined) gene of each group refers to
the function number in the corresponding function
set in the figure. The integer-based representation
of CGP phenotypes is mostly used with mutation
only. Early studies on the efficiency showed that sev-
Genotype
0 1 0 1 2 1 2 2 3
3
Phenotype
+
/
-
OP
IP1
IP2
Function
Lookup Table
Index Function
0
1
2
Addition
Subtraction
Division
Decode
Node
Number
2 3
4
OP
432
0
1
Figure 1: Example of the decoding procedure of a CGP
genotype to its corresponding phenotype. The nodes are
represented by two types of numbers which index the num-
ber in the function lookup table (underlined) and the inputs
(non-underlined) for the node. Inactive function nodes are
shown in gray color. The identifiers IP1 and IP2 stand for
the two input nodes with node index 0 and 1. The identifier
OP stands for the output node of the graph.
eral genetic crossover operators do not contribute to
the search performance of CGP. The number of in-
puts n
i
, outputs n
o
, and the length of the genotype is
fixed. Every candidate program is represented with
n
r
∗n
c
∗(n
a
+ 1) + n
o
integers. Even when the length
of the genotype is fixed for each candidate program,
the length of the corresponding phenotype in CGP is
variable, which can be considered as an advantage of
the CGP representation.
CGP is traditionally used with a (1+λ) selection
scheme of evolutionary algorithms. The new popu-
lation in each generation consists of the best individ-
ual of the previous population and the λ created off-
spring. The breeding procedure is mostly done by a
point mutation that swaps genes in the genotype of
an individual in the valid range by chance. An exam-
ple of a point mutation is given in Figure 2. The fig-
ure shows the flip of the value of a connection gene,
which causes a rewiring of the corresponding func-
tion node. Another point mutation is the flip of the
functional gene, which causes a change of functional
behavior of the corresponding function node.
The (1+λ)-CGP is often used with a selection strategy
called neutrality, the idea that genetic drift yields to
diverse individuals having equal fitness. The genetic
drift is implemented into the selection mechanism in a
way that individuals that have the same fitness as the
normally selected parent are determined, and one of
these same-fitness individuals is returned uniformly
at random.
ECTA 2020 - 12th International Conference on Evolutionary Computation Theory and Applications
60
0 0 0 41 1 0 2 2 3 3 3 3
Node
number
2 3 4 5 OP
IP1
IP2
+
-
*
/
OP
0
1
2
3
4
5
Parent
0 0 0 41 1 0 2 2 2 3 3 3
Node
number
2 3 4 5 OP
IP1
IP2
+
-
*
/
OP
0
1
2
3
4
5
Mutant
Mutation
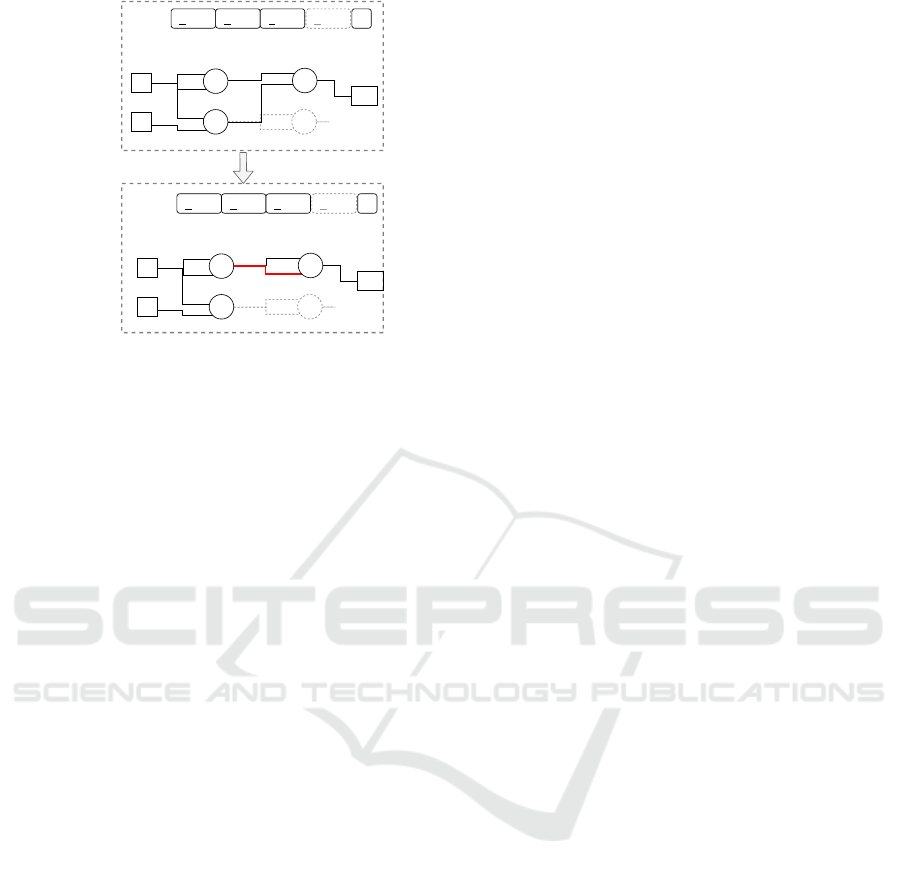
Figure 2: Example of the standard point mutation operator
in integer-encoded CGP. Genes of the genotype are selected
by chance, and their values are randomly flipped within the
legal range of possible values. The connection gene of node
4 is mutated from a value of 2 to a value of 3. This causes
a rewiring of the second input of node 4 from the output of
node 3 to the output node of node 2.
2.2 The Subgraph Crossover Technique
The subgraph crossover technique for CGP was in-
troduced by Kalkreuth et al. (2017) and is inspired
by the subtree crossover found in tree-based GP. To
recombine two directed acyclic graphs, the subgraph
recombination is performed by respecting the CGP
phenotype. Merely swapping parts of the genotype
would be a disastrous approach according to the re-
portings by Clegg et al. (2007). The phenotype of
each individual is represented by the active path of
the graph and is determined through the evaluation
process. Furthermore, the active path of a graph leads
to the semantic value of a certain individual in CGP.
As a consequence, the subgraph crossover exclusively
recombines the genetic material of the active paths.
The idea of the subgraph crossover is that it should re-
duce the disruption which is caused by the genotypic
single-point crossover in standard CGP and truly re-
combine subgraphs.
For the description of the subgraph crossover proce-
dure, let n
i
be the predefined number of inputs and
let n
f
be the predefined number of function nodes. In
CGP, the inputs are indexed from 0 to n
i
−1 and the
function nodes of each graph are indexed from n
i
to
n
i
+ n
f
−1. The nodes which lie between the input
and output nodes are denoted as function nodes. The
crossover is done with two parents which are denoted
as P
1
and P
2
. For the crossover procedure, the node
numbers of the active function nodes are necessary.
The node numbers of the active nodes of P
1
and P
2
are stored in two arrays M
1
and M
2
. The active nodes
are determined by the backward search in the evalua-
tion procedure.
To define one suitable crossover point, we define two
possible crossover points C
P1
and C
P2
of the two par-
ents. With information about the active nodes and
the length of the path, we can choose two possible
crossover points. The possible crossover points C
P1
and C
P2
are chosen by chance in the range of the ac-
tive function nodes which are stored in M
1
and M
2
.
The possible crossover points may not be input or out-
put nodes. A general crossover point C
P
is defined
by choosing the smaller crossover point from C
P1
and
C
P2
. The reason for this is that the subgraphs of the
parents, which will be placed in front of or behind
the crossover point of the offspring’s genome should
be balanced. The representation of CGP allows ac-
tive paths of an individual, which can start in the mid-
dle or back of the graph. The subgraph which will
be placed in front of the crossover point has to start
at more leading active nodes. If C
P
is defined as the
possible point C
P1
, the subgraph of P
1
in front of C
P
will be placed in front of C
P
in the offspring genome.
The subgraph behind C
P
of P
2
will be placed behind
C
P
in the offspring genome The crossover procedure
produces a new genome that represents the offspring
involving the phenotypes of both parents. In the case
that two children should be produced, the crossover
procedure is performed twice with two different gen-
eral crossover points. Since the representation of CGP
provides connections to any of the previous function
nodes of the graph, performing only the neighbour-
hood connect could result in a monotone data flow of
the resulting phenotype. An example of the crossover
procedure is illustrated in Figure 3
3 THE STATUS OF CROSSOVER
IN STANDARD CGP
According to Clegg et al. (2007), the first attempts of
recombination in standard CGP included the testing
of different genotypic crossover techniques. For in-
stance, the genetic material was recombined by swap-
ping parts of the genotypes of the parent individuals
or randomly exchanging selected nodes. Clegg et al.
(2007) reported that all four techniques failed to im-
prove the convergence of CGP and that merely swap-
ping the integers (in whatever manner) in the CGP
representation disrupts the search performance. Com-
pared to running CGP with mutation only, the addi-
tion of these crossover techniques hindered the per-
formance. The four methods were tested on the stan-
dard integer-based representation of CGP. In one of
the first empirical studies in CGP, Miller (1999) ana-
A Comprehensive Study on Subgraph Crossover in Cartesian Genetic Programming
61
0 2 1
*
*
+
2
4
4
x
1
Index Function
0 +
1 -
2 *
3 /
4
3 2 3
5
3
/
0
1
5
OP1
2 1 0
2 0 0
A
3
2
/
-
+
2
4
x
1
3
0
1
5
OP2
0 2 0
4
0 3 3
5
1 1 1
3 0 0
A
3
2
0 4 5
3 5 2
6
6
6
Parent P
1
Parent P
2
Parent P
1
Parent P
2
+
P
C2
:= 6
P
c
:= 3
Function Lookup Table
/
6
6
P
C1
:= 3
*
*
2
x
1
3
0
1
Offspring
+
4
+
5
OP2
/
6
0 2 0
4
0 2 1
5
2 1 0
2 0 0
A
3
2
3 5 2
6
6
Offspring
M
1
= {2,4}
M
2
= {2,3,5,6}
Subgraph S
1
Subgraph S
2
Random Connected Edge
Neighbourhood Connected Edge
+
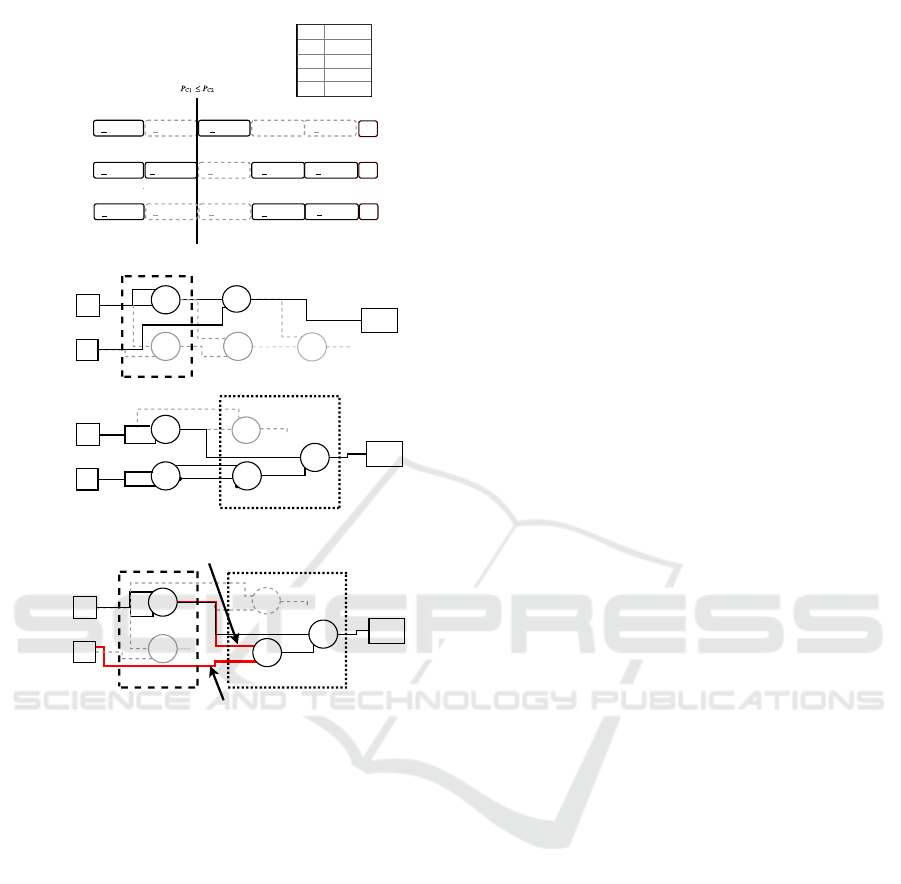
Figure 3: Example of the subgraph crossover technique.
The subgraph crossover basically works similar to the
single-point crossover except that the active nodes on both
sides of the crossover point are preserved. The crossover
point is chosen in a way that it is located between active
function nodes. At the top of the figure, the arrays with the
active nodes and crossover points are listed. Below this in-
formation, the genotypes and phenotypes of the parents and
the offspring are shown, and the parts of the crossover are
marked with dashed boxes.
lyzed the computational efficiency on Boolean func-
tion problems. More precisely, Miller analyzed and
studied the influence of population size on the effi-
ciency of CGP. The key finding was that extremely
low populations are most effective for the tested prob-
lems. The experiments of this study also revealed that
the use of a genotypic crossover reduces the compu-
tational effort only marginally. This was the motiva-
tion for the introduction of a real-valued representa-
tion and new crossover technique for CGP by Clegg
et al. (2007). The real-valued representation of CGP
represents the directed graph as a fixed-length list
of real-valued numbers in the interval [0,1]. The
genes are decoded to the integer-based representa-
tion by their normalization values (number of func-
tions or maximum input range). The recombina-
tion of two genotypes is performed by an arithmetic
crossover with a random weighting factor, which can
also be found in the field of real-valued Genetic Algo-
rithms. Clegg et al. (2007) showed that the new repre-
sentation in combination with crossover improves the
convergence behavior of CGP. However, for the later
generations, Clegg et al. (2007) found that the use
of crossover in real-valued CGP disrupts the conver-
gence on one of the two tested problems. Later work
by Turner (2012) presented results on three classes
of computational problems, digital circuit synthesis,
function optimisation and agent-based wall avoid-
ance. On these problems, it was found that the real-
valued representation together with the crossover op-
eration performed worse than standard CGP.
Kalkreuth et al. (2017) introduced and investigated a
subgraph crossover in CGP. They evaluated the utility
of the new crossover operator for a range of crossover
rates on a suite of benchmark problems in circuit de-
sign, symbolic regression and image filter design. Of-
ten high crossover rates were beneficial and in all
cases a nonzero crossover rate performed better than
without crossover. However, they did not compare
their results with standard CGP so it still remains un-
clear whether crossover can significantly outperform
mutational CGP.
Husa and Kalkreuth (2018) proposed the block
crossover and performed parameter sweeps with the
(1 + λ)-strategy versus a genetic algorithm using
block crossover on a suite of Boolean functions and
symbolic regression problems. The results of the
comparative study demonstrated that the traditional
(1+λ)-CGP algorithm cannot be stated as the univer-
sally predominant algorithm for CGP. The outcome of
the study gave significant evidence that the (1 + λ)-
CGP cannot be considered as the most efficient CGP
algorithm in the Boolean function domain, although
it seems to be often a good choice. The experiments
on the 1-resilient Boolean function demonstrated that
the (1 + λ)-CGP may indeed be significantly inferior
to the other CGP algorithms. In this way, the outcome
of the study gave the first evidence, that it is possi-
ble for crossover operators to outperform the standard
(1+λ) strategy. da Silva and Bernardino (2018) intro-
duced a new form of crossover for CGP when a single
genotype representation is used and the desired model
has multiple outputs. The proposed method modifies
the standard evolutionary strategy commonly adopted
in CGP by combining the subgraphs of the best out-
puts of the parent and its offspring in order to generate
a new fittest individual. Recently, Kalkreuth (2019)
ECTA 2020 - 12th International Conference on Evolutionary Computation Theory and Applications
62

presented a study on two phenotypic mutation tech-
niques for CGP which are called Insertion and Dele-
tion. The subgraph crossover was used within a
(µ + λ)-strategy for a search performance comparison
between CGP and Evolving Graphs with Graph Pro-
gramming (EGGP) (Atkinson et al., 2018), another
state-of-the-art technique for evolving graphs. The
search performance of the (µ + λ)-strategy with sub-
graph crossover and Insertion and Deletion mutation
has been found very effective, when compared to the
search performance of EGGP.
3.1 The Need for a Comprehensive
Study
Husa and Kalkreuth (2018) pointed out that it is diffi-
cult to obtain well-performing parameter settings of
crossover algorithms and complicates fair compar-
isons in CGP. The reason for that is that crossover
based CGP algorithms require the configuration of
more parameters as the (1 + λ)-CGP. Husa and
Kalkreuth (2018) also stated that merely relying on
the results of the meta-evolution can lead to ineffec-
tive parameter settings which result in unfair com-
parisons. Since the meta-evolution is done by an
evolutionary algorithm, it is possible that the meta-
evolution evolves towards a local optimum, which can
result in ineffective parameter settings. Therefore, a
solid and precise parameter tuning requires additional
steps which will be described in the following section.
Another reason for a new comparative study is that
a comparison with a bigger set of benchmark prob-
lems is needed to shed more light on the question of
crossover in CGP. Husa and Kalkreuth (2018) tested
only 8 problems in two major GP benchmark fields.
In a very recent survey about the status and the fu-
ture of CGP, Miller (2020) addressed open questions
in CGP. Regarding the open question of crossover in
CGP, Miller stated that crossover is still very underde-
veloped in CGP and that the subgraph crossover needs
to be comprehensively studied in the future.
3.2 Formulation of Hypotheses
With this work we want to shed more light into some
important dogmas, claims and research questions in
the field of CGP, which we address with the formula-
tion and analysis of the following hypotheses:
Hypothesis 1 (Crossover). Crossover does not contribute
to the search performance of integer-based standard CGP.
Hypothesis 2 (Redundancy). Extremely large genotypes
perform most effectively in CGP.
Hypothesis 3 (Population size). Small populations perform
most effectively in CGP.
4 EXPERIMENTS
4.1 Experimental Setup
We performed experiments in the problem domain
of symbolic regression and Boolean function. To
evaluate the search performance of the tested algo-
rithms, we measured the number of fitness evalu-
ations until the CGP algorithm terminated success-
fully (fitness-evaluations-to-success) and the best fit-
ness value, which was found after a predefined num-
ber of generations (best-fitness-of-run). In addition
to the mean values of the measurements, we calcu-
lated the standard deviation (SD) and the standard er-
ror of the mean (SEM). To classify the significance
of our results, we used the Mann-Whitney-U-Test.
The mean values are denoted a
†
if the p-value is less
than the significance level 0.05 and a
‡
if the p-value
is less than the significance level 0.01 compared to
the (1 + 4)-CGP. Note that the mean values are only
denoted with the significance level marker if the re-
sult of a certain algorithm is better than the result of
the (1 + 4)-CGP. We performed 100 independent runs
with different random seeds.
Parameter Tuning. To compare various CGP al-
gorithms fairly, we performed meta-evolution exper-
iments. Moreover, to find effective parameter set-
tings for the respective algorithms we used a meta-
evolutionary algorithm, which evolved sets of effec-
tive settings. These sets were validated for the par-
ticular problem, and the best collection of parameters
was selected afterward. To determine efficient param-
eter settings for fair comparisons, we utilized an ap-
proach to parameter tuning for CGP, which has been
used by Kaufmann and Kalkreuth (2017). The param-
eter tuning is done in three steps: In the first place,
a set of well-performing parameter settings is deter-
mined by a meta evolutionary algorithm. The meta-
evolution is repeated several times, and the best set of
settings is used for further validation and fine-tuning.
The determined set of parameters is validated manu-
ally, and the best configuration is chosen for further
tuning. Finally, the best performing configuration is
manually fine-tuned. The algorithms which were used
in our study are listed in Table 1.
Table 1: List of the CGP algorithms.
Identifier Description
(1 + 4)-CGP Traditional (1 + 4)-CGP algorithm
(1 + λ)-CGP Traditional (1 + λ)-CGP algorithm
(µ + λ)-CGP (µ + λ)-algorithm with subgraph crossover
Canonical-CGP Canonical genetic algorithm with
tournament selection and subgraph crossover
A Comprehensive Study on Subgraph Crossover in Cartesian Genetic Programming
63
4.2 Benchmarks
Symbolic Regression. We chose nine symbolic re-
gression problems from the work of McDermott et al.
(2012) for better GP benchmarks. The functions of
the problems are shown in Table 2. A training data set
U[a,b,c] refers to c uniform random samples drawn
from a to b inclusive and E[a,b,c] refers to a grid
of points evenly spaced with an interval of c, from
a to b inclusive. The Koza function set consisted of
eight mathematical functions (+, −, ∗, /, sin, cos,
ln(|n|), e
n
) and the Keijzer function set of five math-
ematical functions (+, ∗,
1
n
, −n ,
√
n ). The fitness
of the individuals was represented by a cost func-
tion value. The cost function was defined by the
sum of the absolute difference between the real func-
tion values and the values of an evaluated individual.
Let T =
x
p
P
p=1
be a training dataset of P random
points and f
ind
(x
p
) the value of an evaluated indi-
vidual and f
ref
(x
p
) the true function value. Let C :=
∑
P
p=1
|f
ind
(x
p
) − f
ref
(x
p
)| be the cost function. When
the difference of all absolute values becomes less than
0.01, the algorithm is classified as converged. We
evaluated the more simple symbolic regression prob-
lems Koza 1, 2 & 3 with the fitness-evaluation-to-
termination method. We defined a maximum num-
ber of 8 ·10
7
fitness evaluations for these three ex-
periments. The reason for choosing these three prob-
lems is the fact that we can find an ideal solution more
likely on average than the other more complex bench-
mark problems, which require a huge amount of fit-
ness evaluations to find an ideal solution. The remain-
ing more complex problems were evaluated with the
best-fitness-of-run method. We measured the best fit-
ness after a budget of 10000 fitness evaluations.
Table 2: Symbolic regression problems of the first experi-
ment.
Problem Objective Function Vars Training Set Function Set
Koza-1 x
4
+ x
3
+ x
2
+ x 1 U[-1,1,20] Koza
Koza-2 x
5
−2x
3
+ x 1 U[-1,1,20] Koza
Koza-3 x
6
−2x
4
+ x
2
1 U[-1,1,20] Koza
Nguyen-4 x
6
+ x
5
+ x
4
+ x
3
+ x
2
+ x 1 U[-1,1,20] Koza
Nguyen-5 sin(x
2
)cos(x)−1 1 U[-1,1,20] Koza
Nguyen-6 sin(x) + sin(x + x
2
) 1 U[-1,1,20] Koza
Nguyen-7 ln(x + 1) + ln(x
2
+ 1) 1 U[0,2,20] Koza
Keijzer-6
∑
x
i
1/i 1 E[1,50,1] Keijzer
Pagie-1 1/(1 + x
−4
) + 1/(1 + y
−4
) 2 E[-5,5,0.4] Koza
Boolean Functions. In the Boolean domain, we
chose seven Even-Parity problems with n = 3 to 9
Boolean inputs. The goal was to find a program that
produces the value of the Boolean even parity depend-
ing on the n independent inputs. The fitness was rep-
resented by the number of fitness cases for which the
candidate solution failed to generate the correct value
of the Even-Parity function. We also investigated
Table 3: Boolean function problems for the search perfor-
mance evaluation.
Problem Number of Inputs Number of Outputs
Parity-3 3 1
Parity-4 4 1
Parity-5 5 1
Parity-6 6 1
Parity-7 7 1
Parity-8 8 1
Parity-9 9 1
Adder 1-Bit 3 2
Adder 2-Bit 5 3
Subtractor 2-Bit 4 4
Multiplier 2-Bit 4 4
Mulitplier 3-Bit 6 6
Table 4: Parameter space explored by meta evolution for the
fundamental CGP algorithms.
(1 + 4)-CGP
number of nodes [10,4000]
mutation rate[%] [1,20]
(1 + λ)-CGP
λ [2,150]
number of nodes [10,4000]
mutation rate[%] [1,20]
Canonical-CGP
number of nodes [10,4000]
mutation rate[%] [1,20]
crossover rate[%] [10,100]
population size [10,1000]
tournament size [2,20]
(µ + λ)-CGP
µ [2,150]
λ [2,1000]
number of nodes [10,4000]
mutation rate[%] [1,20]
crossover rate[%] 20, 50, 70, 90
multiple output problems as the 2-Bit-Adder, 2-Bit-
Subtractor, 2-Bit- and 3-Bit-Multiplier. These sorts
of problems differ markedly from the parity problems,
and the multiple output multiplier has been proposed
as a suitable alternative. As a result, we receive a di-
verse set of problems in this domain.
To evaluate the fitness of the individuals on the multi-
ple output problems, we defined the fitness value of an
individual as the number of different bits to the corre-
sponding truth table. When this number became zero,
the algorithm terminated successfully. We evaluated
the majority of the Boolean function problems with
the fitness-evaluations-to-success method. However,
since the Even-Parity-8 & 9 and the 3-Bit-Multiplier
are very complex and computing-intensive problems,
we evaluated these problems with the best-fitness-of-
run method with a budget of 10000 fitness evalua-
tions. The set of benchmark problems with the cor-
responding number of inputs and outputs is shown in
Table 3.
ECTA 2020 - 12th International Conference on Evolutionary Computation Theory and Applications
64
4.3 Meta Evolution
We tuned significant parameters for all utilized CGP
algorithms on the set of problems which are shown in
Table 4. The ranges for the number of nodes are ori-
ented with the parameter settings found in Kaufmann
and Kalkreuth (2017). We used the meta-evolution
extension package of the Java Evolutionary Compu-
tation Research System (ECJ)
1
. For the meta-level,
we used a basic genetic algorithm (GA). The setting
of the meta-level GA is shown in Table 5.
Table 5: Configuration of the meta-level GA.
Property Setting
Maximum generations 200
Population size 50
Mutation rate 1/n
Mutation tape gaussian mutation
Tournament selection size 4
Crossover rate 0.7
Crossover type intermediate recombination
Evaluation method best-fitness-of-run
Number of trials 4
Tables 6 and 7 show the results of the meta-evolution.
The results in the Boolean domain reveal an effective
parametrization with small population size and an ex-
tremely high number of function nodes in the geno-
type. In the symbolic regression domain, the number
of function nodes is on average smaller compared to
the Boolean function domain. For the image operator
design problems, extremely high levels of redundancy
seem to be a good choice for the mutation-only CGP
algorithms. For the Canonical-CGP a smaller number
of function nodes seems to be an appropriate choice.
4.4 Search Performance Evaluation
Tables 8 and 9 show the results of the algorithm com-
parison in the Boolean domain. As visible, the re-
sults show no overall dominant and outstanding CGP
algorithm in this problem domain. However, on the
high order Parity-even problems, the (1+ 4)-CGP and
the (1+λ)-CGP seem to be a good choice. The re-
Table 6: Results of the meta evolution for the Boolean function problems.
Problem Algorithm Number Mutation Crossover µ λ Population Tournament
of nodes rate[%] rate[%] size size
Parity-3
(1 + 4)-CGP 2000 1 – – – – –
(1 + λ)-CGP 2000 1 – – 2 – –
Canonical-CGP 2000 1 70 – – 10 2
(µ + λ)-CGP 3000 2 70 2 2 – –
Parity-4
(1 + 4)-CGP 1500 1 – – – – –
(1 + λ)-CGP 1500 1 – – 2 – –
Canonical-CGP 1500 1 70 – – 5 2
(µ + λ)-CGP 3000 1 90 2 2 – –
Parity-5
(1 + 4)-CGP 2000 1 – – – – –
(1 + λ)-CGP 2000 1 – – 2 – –
Canonical-CGP 1000 1 70 – – 5 2
(µ + λ)-CGP 2000 1 70 2 2 – –
Parity-6
(1 + 4)-CGP 2000 1 – – – – –
(1 + λ)-CGP 2000 1 – – 2 – –
Canonical-CGP 2000 1 70 – – 5 2
(µ + λ)-CGP 2000 1 90 4 2 – –
Parity-7
(1 + 4)-CGP 2500 1 – – – – –
(1 + λ)-CGP 2500 1 – – 2 – –
Canonical-CGP 2500 1 70 – – 5 2
(µ + λ)-CGP 2500 1 90 4 2 – –
Parity-8
(1 + 4)-CGP 2000 1 – – – – –
(1 + λ)-CGP 2000 1 – – 2 – –
Canonical-CGP 2000 1 70 – – 5 2
(µ + λ)-CGP 2000 1 70 2 2 – –
Parity-9
(1 + 4)-CGP 2000 1 – – – – –
(1 + λ)-CGP 2000 1 – – 2 – –
Canonical-CGP 2000 1 70 – – 5 2
(µ + λ)-CGP 2000 1 70 2 2 – –
Adder-1Bit
(1 + 4)-CGP 150 8 – – – – –
(1 + λ)-CGP 150 5 – – 2 – –
Canonical-CGP 200 6 50 – – 20 4
(µ + λ)-CGP 150 3 50 2 2 – –
Adder-2Bit
(1 + 4)-CGP 100 3 – – – – –
(1 + λ)-CGP 150 1 – – 2 – –
Canonical-CGP 150 2 90 – – 20 4
(µ + λ)-CGP 150 1 50 2 2 – –
Mult.-2Bit
(1 + 4)-CGP 1500 1 – – – – –
(1 + λ)-CGP 1500 1 – – 2 – –
Canonical-CGP 1000 1 70 – – 10 2
(µ + λ)-CGP 2000 1 70 2 8 – –
Mult.-3Bit
(1 + 4)-CGP 2000 2 – – – – –
(1 + λ)-CGP 1000 1 – – 2 – –
Canonical-CGP 1000 1 30 – – 5 2
(µ + λ)-CGP 2000 1 30 2 2 – –
Subtr.-2Bit
(1 + 4)-CGP 2000 1 – – – – –
(1 + λ)-CGP 2000 1 – – 8 – –
Canonical-CGP 1500 1 90 – – 5 2
(µ + λ)-CGP 1500 1 70 2 2 – –
1
https://cs.gmu.edu/ eclab/projects/ecj/
A Comprehensive Study on Subgraph Crossover in Cartesian Genetic Programming
65
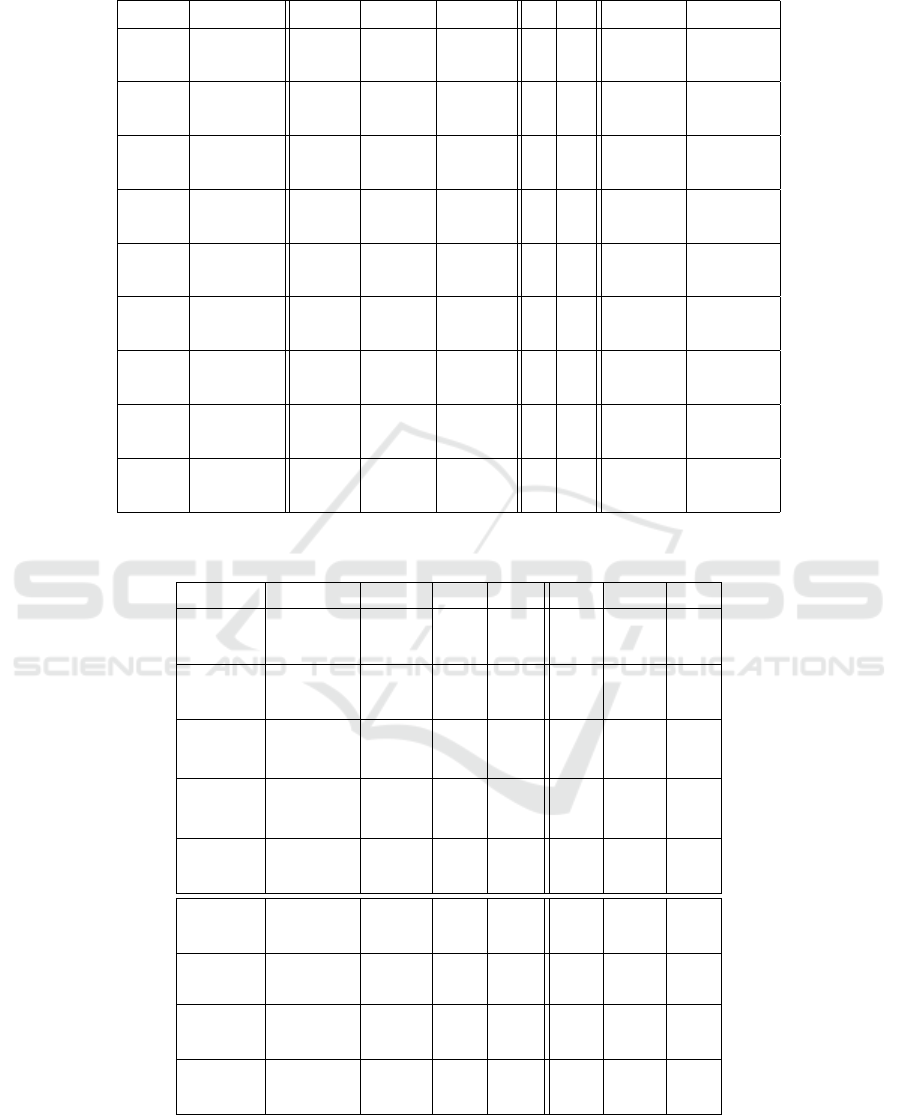
Table 7: Results of the meta evolution for the symbolic regression problems.
Problem Algorithm Number Mutation Crossover µ λ Population Tournament
of nodes rate[%] rate[%] size size
Koza-1
(1 + 4)-CGP 10 20 – – – – –
(1 + λ)-CGP 10 20 – – 8 – –
Canonical-CGP 10 20 70 50 4
(µ + λ)-CGP 10 20 70 4 16 – –
Koza-2
(1 + 4)-CGP 10 20 – – – – –
(1 + λ)-CGP 10 20 – – 8 – –
Canonical-CGP 10 20 70 50 4
(µ + λ)-CGP 10 20 90 4 16 – –
Koza-3
(1 + 4)-CGP 10 20 – – – – –
(1 + λ)-CGP 10 20 – – 8 – –
Canonical-CGP 10 20 70 50 4
(µ + λ)-CGP 10 20 70 1 8 – –
Nguyen-4
(1 + 4)-CGP 120 10 – – – – –
(1 + λ)-CGP 100 10 – – 16 – –
Canonical-CGP 220 9 90 – – 50 5
(µ + λ)-CGP 200 1 70 10 200 – –
Nguyen-5
(1 + 4)-CGP 60 7 – – – – –
(1 + λ)-CGP 60 7 – – 16 – –
Canonical-CGP 100 6 25 – – 10 2
(µ + λ)-CGP 300 5 70 10 250 – –
Nguyen-6
(1 + 4)-CGP 100 10 – – – – –
(1 + λ)-CGP 100 10 – – 16 – –
Canonical-CGP 20 20 90 – – 50 7
(µ + λ)-CGP 300 1 70 10 250 – –
Nguyen-7
(1 + 4)-CGP 200 2 – – – – –
(1 + λ)-CGP 1000 2 – – 16 – –
Canonical-CGP 500 3 70 – – 250 7
(µ + λ)-CGP 200 10 90 10 200 – –
Keijzer-6
(1 + 4)-CGP 2000 3 – – – – –
(1 + λ)-CGP 2000 5 – – 16 – –
Canonical-CGP 100 5 70 – – 250 10
(µ + λ)-CGP 700 5 70 10 250 – –
Pagie-1
(1 + 4)-CGP 1500 7 – – – – –
(1 + λ)-CGP 1500 7 – – 16 – –
Canonical-CGP 500 5 90 – – 200 7
(µ + λ)-CGP 500 8 75 25 125 – –
Table 8: Results of the algorithm comparison for the Boolean function problem evaluated by the number of fitness evaluations
(FE) to termination.
Problem Algorithm Mean FE SD SEM 1Q Median 3Q
Parity-Even-3
(1 +4)-CGP 3177 3417 ±343 1246 2136 3760
(1 +λ)-CGP 2495 2919 ±293 846 1534 2872
Canonical-CGP 3107 3070 ±307 1201 2104 3907
(µ +λ)-CGP 1565
‡
1517 ±152 602 1168 1892
Parity-Even-4
(1 +4)-CGP 15420 14152 ±1422 6292 10358 17726
(1 +λ)-CGP 16523 19168 ±1926 6095 11276 18557
Canonical-CGP 54967 47042 ±4727 24813 40612 71851
(µ +λ)-CGP 11135
‡
8447 ±845 5117 8527 14085
Parity-Even-5
(1 +4)-CGP 45542 33947 ±3411 21524 36834 61222
(1 +λ)-CGP 34375
‡
28146 ±2828 20685 27104 38941
Canonical-CGP 28413
‡
25538 ±2566 23388 19640 34876
(µ +λ)-CGP 43476 2055 ±1022 23814 36188 57182
Parity-Even-6
(1 +4)-CGP 199989 142915 ±14291 107418 163234 242573
(1 +λ)-CGP 118768
‡
73682 ±7368 65766 91577 156639
Canonical-CGP 242986 161762 ±16257 134518 200196 309346
(µ +λ)-CGP 110158
‡
75163 ±7516 63908 90676 135148
Parity-Even-7
(1 +4)-CGP 478055 301113 ±30111 268210 393362 605372
(1 +λ)-CGP 441857 328539 ±32853 226272 352254 545197
Canonical-CGP 631568 548180 ±54818 293613 453204 750792
(µ +λ)-CGP 358420
‡
246131 ±24613 189278 303988 451667
Adder-1Bit
(1 +4)-CGP 1895 1856 ±186 634 1252 2494
(1 +λ)-CGP 1415 1532 ±154 508 1057 1640
Canonical-CGP 2155 2018 ±202 882 1521 2907
(µ +λ)-CGP 1393
†
1311 ±132 496 954 1784
Adder-2Bit
(1 +4)-CGP 85667 84355 ±8478 29506 58650 110794
(1 +λ)-CGP 73417 58589 ±5888 33367 53654 94009
Canonical-CGP 225652 200384 ±20038 78247 158130 271717
(µ +λ)-CGP 68375 43229 ±5361 32998 64006 98052
Multiplier-2Bit
(1 +4)-CGP 11583 10469 ±1046 5020 8524 14498
(1 +λ)-CGP 17664 21664 ±2177 5400 9233 19262
Canonical-CGP 30489 25700 ±2582 13384 22696 22916
(µ +λ)-CGP 11055
‡
13281 ±1334 3635 6693 13220
Subtractor-2Bit
(1 +4)-CGP 11029 13975 ±13975 4642 6986 11878
(1 +λ)-CGP 8377
‡
9958 ±1000 2989 6111 9579
Canonical-CGP 35829 41822 ±4203 10346 20056 40698
(µ +λ)-CGP 15161 22388 ±2250 5291 9671 16705
ECTA 2020 - 12th International Conference on Evolutionary Computation Theory and Applications
66
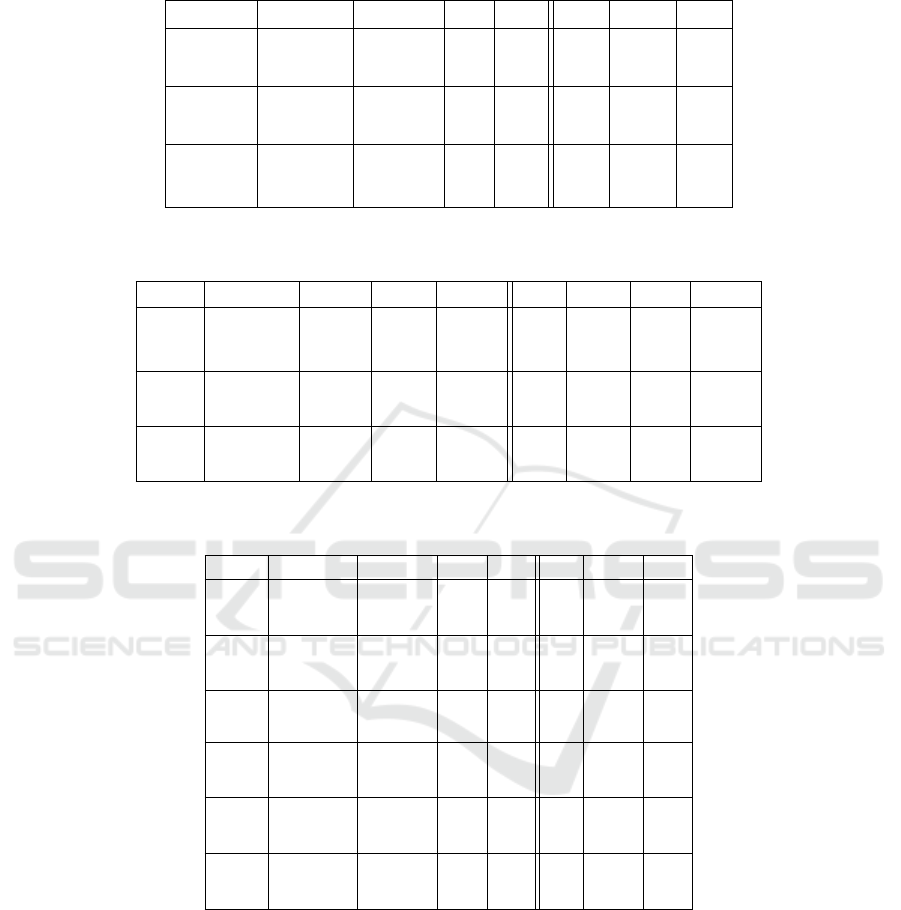
Table 9: Results of the algorithm comparison for the complex Booleans problems evaluated with the best-fitness-of-run
method.
Problem Algorithm Mean SD SEM 1Q Median 3Q
Best Fitness
Parity-8
(1 + 4)-CGP 104, 24 8,50 ±0,86 99 105 110
(1 + λ)-CGP 92,9
‡
11,56 ±1,15 85 94,5 102,25
Canonical-CGP 113,28 5, 01 ±0, 50 110,75 114 117
(µ + λ)-CGP 107,03 7, 49 ±0, 74 103 108 112
Parity-9
(1 + 4)-CGP 230, 38 8,74 ±0,87 224,75 230 236
(1 + λ)-CGP 225,69
‡
11,45 ±1,14 220 228 234
Canonical-CGP 228,37 9, 02 ±0, 90 223,75 229 235, 25
(µ + λ)-CGP 226,63 13,71 ±1,37 220, 5 230 236
Multiplier-3Bit
(1 + 4)-CGP 46,39 7,57 ±0,75 41 47 51
(1 + λ)-CGP 42,31
‡
8,67 ±0,86 36 43 48
Canonical-CGP 52, 9 8,40 ±0,84 47 53 59
(µ + λ)-CGP 43,98
†
8,86 ±0,88 38 44 49
Table 10: Results for the algorithm comparison for the problems Koza 1, 2 & 3 evaluated by the number of fitness evaluations
(FE) to termination.
Problem Algorithm Mean FE SD SEM 1Q Median 3Q Unfinished
runs
Koza-1
(1 +4)-CGP 8675635 16681422 ±1668142 441477 1814344 7045961 2
(1 +λ)-CGP 7370880
‡
17384354 ±1738435 204400 1050936 4294170 3
Canonical-CGP 663822
‡
838546 ±83854 135162 337950 710275 0
(µ +λ)-CGP 7780751
‡
15830735 ±1583073 197284 1830312 6318740 3
Koza-2
(1 +4)-CGP 8264426 19894512 ±1989451 150140 888884 4378756 6
(1 +λ)-CGP 8191549 20275790 ±2027579 94290 559028 4710848 1
Canonical-CGP 444118
‡
95000 ±286700 627550 29650 78800 0
(µ +λ)-CGP 5729778 11021660 ±1102166, 238156 1320880 5878696 1
Koza-3
(1 +4)-CGP 600153 1214527 ±121452 39076 177418 443038 0
(1 +λ)-CGP 753551 2535215 ±253521 29528 120368 431318 0
Canonical-CGP 32870
‡
57156 ±10435 2488 6700 32713 0
(µ +λ)-CGP 926857 3473467 ±347347 28548 121040 362180 0
Table 11: Results of the algorithm comparison algorithm for the symbolic regression problems evaluated with the best-fitness-
of-run method.
Problem Algorithm Mean SD SEM 1Q Median 3Q
Best Fitness
Nguyen-4
(1 +4)-CGP 0,68 0,55 ±0,05 0,34 0, 58 0, 77
(1 +λ)-CGP 0, 61 0, 46 ±0,04 0, 35 0, 54 0, 74
Canonical-CGP 0,50
†
0,28 ±0,04 0,31 0, 47 0,60
(µ +λ)-CGP 0, 60
†
0,40 ±0,04 0,36 0, 54 0,76
Nguyen-5
(1 +4)-CGP 0,45 0,42 ±0,04 0,06 0, 32 0, 81
(1 +λ)-CGP 0, 39 0, 33 ±0,03 0, 08 0, 27 0, 63
Canonical-CGP 0,29
‡
0,27 ±0,03 0,05 0, 20 0,40
(µ +λ)-CGP 0, 28
‡
0,25 ±0,02 0,06 0, 19 0,45
Nguyen-6
(1 +4)-CGP 0,54 0,66 ±0,06 0,16 0, 29 0, 61
(1 +λ)-CGP 0, 50 0, 67 ±0,06 0, 15 0, 22 0, 50
Canonical-CGP 0,31
‡
0,31 ±0,03 0,15 0, 24 0,40
(µ +λ)-CGP 0, 61 0, 67 ±0,06 0, 16 0, 35 0, 67
Nguyen-7
(1 +4)-CGP 0,79 0,48 ±0,05 0,45 0, 67 1, 06
(1 +λ)-CGP 0, 71 0, 45 ±0,04 0, 44 0, 67 0, 76
Canonical-CGP 0,60
‡
0,35 ±0,03 0,36 0, 60 0,68
(µ +λ)-CGP 0, 62
‡
0,40 ±0,04 0,42 0, 63 0,68
Keijzer-6
(1 +4)-CGP 3,78 2,61 ±0,26 2,16 3, 24 4, 59
(1 +λ)-CGP 3, 38 2, 52 ±0,25 2, 41 3, 03 3,158
Canonical-CGP 2,81
†
1,13 ±0,11 1,78 2, 90 3,75
(µ +λ)-CGP 2, 88
†
1,09 ±0, 1 2,25 3, 14 3, 15
Pagie-1
(1 +4)-CGP 128, 18 48,19 ±4, 81 87,81 119,09 161,08
(1 +λ)-CGP 120, 75 44,95 ±4, 49 86,14 120,91 155,06
Canonical-CGP 98, 52
‡
50,57 ±5,08 59,04 85,31 130,04
(µ +λ)-CGP 99, 74
‡
41,246 ±4,12 65,32 95,79 131,76
sults of our experiments in the symbolic regression
domain are shown in Table 10 and 11. It is visi-
ble that the Canonical-CGP algorithm performs better
than the mutation-only CGP algorithms on all tested
problems. Furthermore, as visible (1 + 4)-, (1 + λ)-
and (µ + λ)-CGP reported runs in which no solution
was found.
4.5 Redundancy and Fitness Space
Analysis
We investigated the three symbolic regression prob-
lems Koza 1, 2 & 3 in more detail due to the com-
paratively low number of 10 function nodes which
has been determined with the parameter tuning. With
the experiments which are described in this subsec-
tion, we intended to investigate the role of continuous
and discrete fitness spaces for the evolutionary search
CGP. We first measured the search performance of
A Comprehensive Study on Subgraph Crossover in Cartesian Genetic Programming
67
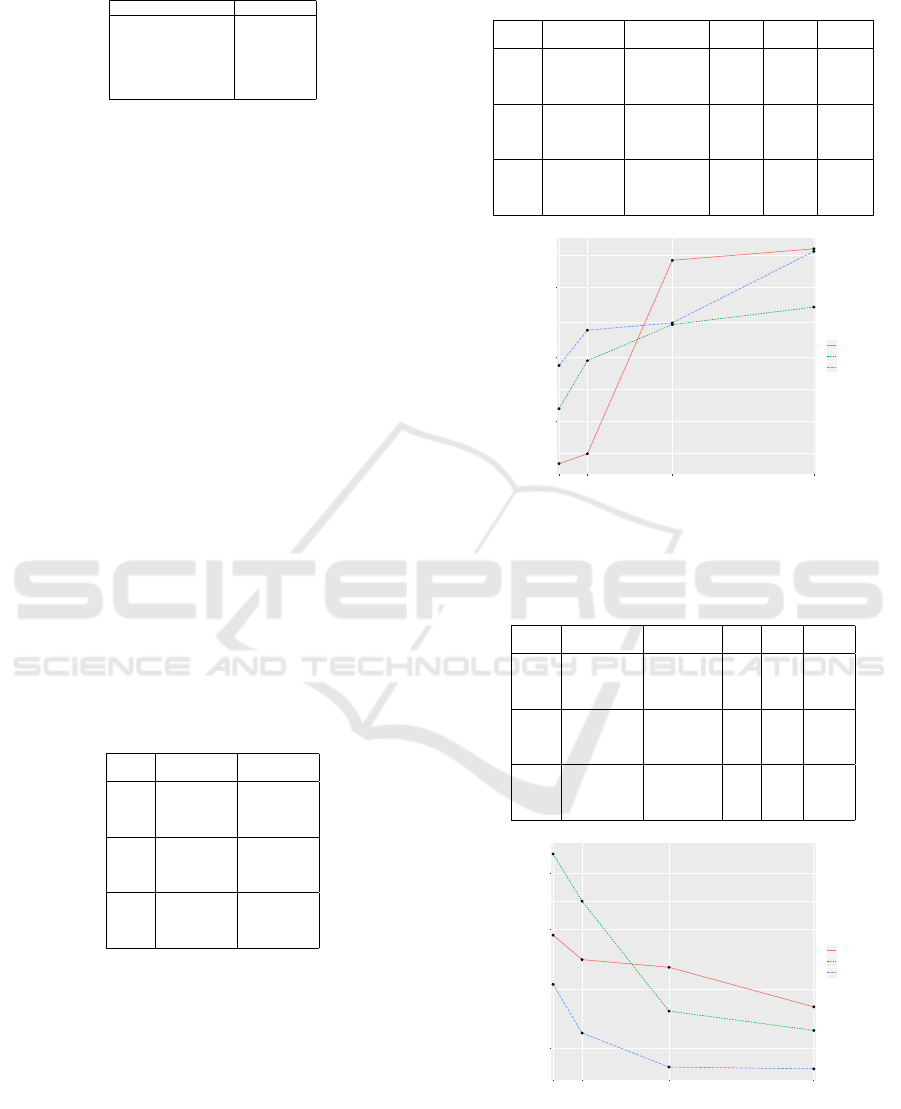
Table 12: Configuration of the (1 + 4)-CGP.
Property (1 + 4)-CGP
Maximum node count 10/20/50/100
Mutation rate [%] 20/10/8/6
Number of inputs 2
Number of outputs 1
Population size 5
Function set +, −, ∗, /
the three symbolic regression problems with genotype
lengths of 10, 20, 50, and 100 function nodes. The al-
gorithm configuration for the experiments is shown in
Table 12. We performed 100 runs for each experiment
and measured the search performance by the number
of generations until the ideal solution was found. We
utilized the (1 + 4)-CGP algorithm. When this fitness
function is used, the fitness values can vary in a range
R
>=0
. Since the range of this fitness function is not
fixed, we evaluated the size of the fitness space for
genotype lengths with 10, 20, 50 and 100 function
nodes by sampling 10
6
random genotypes. We eval-
uated the fitness of each genotype and stored its fre-
quency in a hash map. Afterward, we used the size of
the hash map to conclude the magnitude of the space
of fitness values. We also investigated all bench-
mark functions with discrete fitness. In these types
of experiments, the fitness function was similar to the
continuously fitness function, with the exception that
the fitness values were discretized within a range of
whole numbers from 0 to 20. In this way, the exper-
iments covered the investigation of the search perfor-
mance for different lengths of the genotype with con-
tinuous and discrete fitness. The respective mutation
rates have been determined empirically.
Table 13: Number of fitness values for various genotype
lenght for the problems Koza 1, 2 & 3.
Problem Number of Number of
function nodes fitness values
Koza-1
10 15655
20 53695
50 172244
100 306242
Koza-2
10 15646
20 54133
50 173122
100 307665
Koza-3
10 15859
20 54823
50 173523
100 307412
Table 14 shows and Figure 4 illustrates the stylized
search performance behavior for various genotype
lengths when continuous fitness is used. It is visible
that when the increase of the length of the genotype,
the search performance decreases. Table 13 shows the
results of the analysis of the fitness space for the three
Koza problems. It can be seen that the size of the fit-
ness space increases when the length of the genotype
is increased. Table 15 shows and Figure 15 illustrates
the search performance behavior for various genotype
lengths when discrete fitness is used. In this case, it
Table 14: Results for the symbolic regression problems
Koza 1, 2 &¸ 3 when continuous fitness is used.
Problem Number of Mean SD SEM Median
function nodes Fitness Evaluations
Koza-1
10 456547 749589 ±74958 47852
20 536685 814927 ±81493 56704
50 1373228 662682 ±66268 1586900
100 1407718 709660 ±70966 1934054
Koza-2
10 466726 667196 ±66719 123156
20 695164 768904 ±76890 281468
50 808779 800203 ±80020 525106
100 942653 877873 ±87787 709382
Koza-3
10 638451 ±75615 52473 259490
20 774234 812167 ±81216 476620
50 867915 826392 ±82639 540420
100 1154141 902271 ±90227 1852700
1e+05
3e+05
1e+06
10 20 50 100
Number of function nodes
Median number
of fitness evaluations
Koza−1
Koza−2
Koza−3
Figure 4: Stylized search performance behavior for various
genotype lengths when continuous fitness is used.
Table 15: Results for the symbolic regression problems
Koza 1, 2 & 3 when discrete fitness is used.
Problem Number of Mean SD SEM Median
function nodes fitness evaluations
Koza-1
10 3694 3300 ±330 2840
20 3346 3870 ±387 2268
50 3300 4165 ±416 2114
100 2798 3812 ±381 1470
Koza-2
10 8589 8768 ±876 5984
20 5478 5518 ±552 3880
50 3076 5154 ±515 1414
100 1998 2469 ±246 1184
Koza-3
10 2500 2852 ±285 1808
20 2011 2238 ±223 1156
50 1212 1220 ±122 846
100 1145 1107 ±110 832
1000
3000
5000
10 20 50 100
Number of function nodes
Median number
of fitness evaluations
Koza−1
Koza−2
Koza−3
Figure 5: Stylized search performance behavior for various
genotype lengths when discrete fitness is used.
is visible that increasing the length of the genotype
leads to an increase of the search performance.
ECTA 2020 - 12th International Conference on Evolutionary Computation Theory and Applications
68

5 ANALYSIS OF HYPOTHESES
Hypothesis 1 (Crossover). Crossover does not
contribute to the search performance of integer-based
standard CGP.
The results of our experiments clearly show that two
algorithms that utilize subgraph crossover outper-
formed the (1 + 4)-CGP and (1 + λ)-CGP on various
problems in two different problem domains. Based
on these findings we can disprove that crossover can-
not contribute to the search performance of standard
integer-based CGP.
Hypothesis 2 (Redundancy). Extremely large geno-
types perform most effective in CGP.
Our redundancy and fitness space analysis demon-
strated, that the rise in the genotype length increased
the size of the space of fitness values rapidly on three
tested symbolic regression problems. Our experi-
ments also showed that the increase of the genotype
length led to a deterioration of the search performance
on all three tested problems. Therefore, we can state
that the dogma that extremely large genotypes per-
form most effectively in CGP cannot be generalized.
Hypothesis 3 (Population size). Small populations
perform most effective in CGP.
Our results in the symbolic regression domain show
that the claim that small population sizes generally
perform most effectively in CGP cannot be general-
ized. In the Boolean domain, the results and findings
of former studies seem to be coherent. However, it
has been demonstrated that this claim does not hold
for the symbolic regression and image operator de-
sign domain. Our results give clear evidence that the
use of medium and high population sizes can lead to a
significantly better search performance than the tradi-
tional (1+4)-CGP in the symbolic regression domain.
6 DISCUSSION
Our experiments demonstrate that the subgraph
crossover can contribute to the search performance
by using a canonical GA or (µ + λ)-strategy. Further-
more, the results of our experiments indicate that the
predominance of the (1 + 4)-CGP and (1 + λ)-CGP
algorithms cannot be generalized in the Boolean
domain. However, for the high order parity problems,
the performance of the (1+λ)-CGP seems to be solid.
Overall, the results for the (1+λ)-CGP in the boolean
domain are coherent with previous studies (Miller and
Smith, 2006; Kaufmann and Kalkreuth, 2017). Our
experiments in the symbolic regression indicate that
the use of the subgraph crossover is beneficial and can
contribute significantly to the search performance in
these problem domains. Especially the performance
of the Canonical-CGP algorithm was superior to the
(1 + 4)-CGP on all tested problems in this problem
domain. Furthermore, our comparison of the whole
evolutionary process for the more simple benchmark
problems Koza 1, 2 & 3 revealed a big gap of the
search performance between the (1 + 4)-CGP and the
Canonical-CGP on these problems. Furthermore, the
Canonical-CGP finished all runs successfully within
the given budget of fitness evaluations. The results
of our redundancy and fitness space analysis indicate
that there might be a correlation between the size
of the space of fitness values and the search perfor-
mance of a respective CGP algorithm. Since our
results also show that the parameter settings vary for
different problem domains it opens up the question,
which conditions or types of problems require bigger
or smaller population sizes and in which way the
subgraph crossover contributes to the evolutionary
search. The same question arises for the length of the
genotype. A preliminary assumption could be that
the fitness landscape of certain problems requires
more exploration abilities to overcome local optima
or to explore a bigger and denser continuous fitness
space. However, since the experiments of Miller and
Smith (2006) mostly focused on Boolean function
problems which are evaluated with discrete fitness
more research is needed to investigate the behavior
of CGP algorithms in continuous fitness spaces.
Our results also demonstrate that the ideal
parametrization of CGP depends on the prob-
lem. In this way our results confirm the results of
a former study by Turner and Miller (2015) which
investigated different genotype lengths up to 100,000
nodes. In all cases there was an ideal number of
nodes which varied depending on the problem. The
ideal genotype lengths for the tested problems varied
between 50 and 3000 function nodes.
7 CONCLUSION AND FUTURE
WORK
A comprehensive study has been presented which
demonstrates that the use of subgraph crossover
is beneficial on various problems in two different
problem domains. The results of the experiments
also clearly show that popular performance dogmas
of CGP are not coherent in another problem do-
main. However, our experiments demonstrate that
former findings of the effective use of low population
A Comprehensive Study on Subgraph Crossover in Cartesian Genetic Programming
69

sizes and extremely large genotypes are valid in the
Boolean domain. In the symbolic regression domain,
our experiments revealed that this dogma cannot be
generalized and showed that comparatively smaller
genotypes and bigger populations can also perform
effectively in CGP. This study paves the way for fur-
ther studies on the behavior of subgraph crossover
based CGP algorithms. Therefore, our future work
will focus on exploration analysis in fitness and phe-
notype space of CGP. We will also focus on theoret-
ical work for crossover-based algorithms which will
be similar to the runtime analysis of Kalkreuth and
Droschinsky (2019) for mutational-only CGP.
REFERENCES
Atkinson, T., Plump, D., and Stepney, S. (2018). Evolving
graphs by graph programming. In Castelli, M., Sekan-
ina, L., Zhang, M., Cagnoni, S., and Garcia-Sanchez,
P., editors, EuroGP 2018: Proceedings of the 21st Eu-
ropean Conference on Genetic Programming, volume
10781 of LNCS, pages 35–51, Parma, Italy. Springer
Verlag.
Clegg, J., Walker, J. A., and Miller, J. F. (2007). A new
crossover technique for cartesian genetic program-
ming. In GECCO ’07: Proceedings of the 9th annual
conference on Genetic and evolutionary computation,
volume 2, pages 1580–1587, London. ACM Press.
Cramer, N. L. (1985). A representation for the adaptive
generation of simple sequential programs. In Grefen-
stette, J. J., editor, Proceedings of an International
Conference on Genetic Algorithms and the Applica-
tions, pages 183–187, Carnegie-Mellon University,
Pittsburgh, PA, USA.
da Silva, J. E. H. and Bernardino, H. (2018). Cartesian ge-
netic programming with crossover for designing com-
binational logic circuits. In 7th Brazilian Confer-
ence on Intelligent Systems, BRACIS 2018, S
˜
ao Paulo,
Brazil, October 22-25, 2018, pages 145–150. IEEE
Computer Society.
Forsyth, R. (1981). BEAGLE a Darwinian approach to pat-
tern recognition. Kybernetes, 10(3):159–166.
Hicklin, J. (1986). Application of the genetic algorithm to
automatic program generation. Master’s thesis, Uni-
versity of Idaho.
Husa, J. and Kalkreuth, R. (2018). A comparative study on
crossover in cartesian genetic programming. In Eu-
roGP 2018: Proceedings of the 21st European Con-
ference on Genetic Programming, volume 10781 of
LNCS, pages 203–219, Parma, Italy. Springer Verlag.
Kalganova, T. (1997). Evolutionary approach to design
multiple-valued combinational circuits. In Proceed-
ings. of the 4th International conference on Applica-
tions of Computer Systems (ACS’97), pages 333–339,
Szczecin, Poland.
Kalkreuth, R. (2019). Two new mutation techniques for
cartesian genetic programming. In Proceedings of
the 11th International Joint Conference on Compu-
tational Intelligence, IJCCI 2019, Vienna, Austria,
September 17-19, 2019, pages 82–92. ScitePress.
Kalkreuth, R. and Droschinsky, A. (2019). On the time
complexity of simple cartesian genetic programming.
In Proceedings of the 11th International Joint Con-
ference on Computational Intelligence, IJCCI 2019,
Vienna, Austria, September 17-19, 2019, pages 172–
179. ScitePress.
Kalkreuth, R., Rudolph, G., and Droschinsky, A. (2017).
A new subgraph crossover for cartesian genetic pro-
gramming. In EuroGP 2017: Proceedings of the 20th
European Conference on Genetic Programming, vol-
ume 10196 of LNCS, pages 294–310, Amsterdam.
Springer Verlag.
Kaufmann, P. and Kalkreuth, R. (2017). Parametrizing
cartesian genetic programming: An empirical study.
In KI 2017: Advances in Artificial Intelligence - 40th
Annual German Conference on AI, Dortmund, Ger-
many, September 25-29, 2017, Proceedings, volume
10505 of Lecture Notes in Computer Science, pages
316–322. Springer.
Koza, J. (1990). Genetic Programming: A paradigm for ge-
netically breeding populations of computer programs
to solve problems. Technical Report STAN-CS-90-
1314, Dept. of Computer Science, Stanford Univer-
sity.
Koza, J. R. (1992). Genetic Programming: On the Pro-
gramming of Computers by Means of Natural Selec-
tion. MIT Press, Cambridge, MA, USA.
Koza, J. R. (1994). Genetic Programming II: Automatic
Discovery of Reusable Programs. MIT Press, Cam-
bridge Massachusetts.
McDermott, J., White, D. R., Luke, S., Manzoni, L.,
Castelli, M., Vanneschi, L., Jaskowski, W., Kraw-
iec, K., Harper, R., De Jong, K., and O’Reilly, U.-
M. (2012). Genetic programming needs better bench-
marks. In GECCO ’12: Proceedings of the fourteenth
international conference on Genetic and evolutionary
computation conference, pages 791–798, Philadel-
phia, Pennsylvania, USA. ACM.
Miller, J. F. (1999). An empirical study of the efficiency of
learning boolean functions using a cartesian genetic
programming approach. In Banzhaf, W., Daida, J.,
Eiben, A. E., Garzon, M. H., Honavar, V., Jakiela, M.,
and Smith, R. E., editors, Proceedings of the Genetic
and Evolutionary Computation Conference, volume 2,
pages 1135–1142, Orlando, Florida, USA. Morgan
Kaufmann.
Miller, J. F. (2020). Cartesian genetic programming: its sta-
tus and future. Genetic Programming and Evolvable
Machines, 21(1):129–168.
Miller, J. F. and Smith, S. L. (2006). Redundancy and com-
putational efficiency in cartesian genetic program-
ming. IEEE Transactions on Evolutionary Computa-
tion, 10(2):167–174.
Miller, J. F., Thomson, P., and Fogarty, T. (1997). Design-
ing electronic circuits using evolutionary algorithms.
arithmetic circuits: A case study. In Genetic Algo-
rithms and Evolution Strategies in Engineering and
Computer Science, pages 105–131. Wiley.
Turner, A. J. (2012). Improving crossover techniques in a
genetic program. Master’s thesis, Department of Elec-
tronics, University of York.
Turner, A. J. and Miller, J. F. (2015). Neutral genetic drift:
an investigation using cartesian genetic programming.
Genet. Program. Evolvable Mach., 16(4):531–558.
ECTA 2020 - 12th International Conference on Evolutionary Computation Theory and Applications
70
