
GEMO: Grammatical Evolution Memory Optimization System
Meghana Kshirsagar
1a
, Rushikesh Jachak
2b
, Purva Chaudhari
2c
and Conor Ryan
1d
1
Biocomputing Developmental Systems, University of Limerick, Irland
2
Department of Computer Science, Government College of Engineering, Aurangabad, India
Keywords: Evolutionary Computation, Memory Optimization, Grammatical Evolution, Multi-Objective Optimization,
Autoregressive Time Series Forecasting.
Abstract: In Grammatical Evolution (GE) individuals occupy more space than required, that is, the Actual Length of
the individuals is longer than their Effective Length. This has major implications for scaling GE to complex
problems that demand larger populations and complex individuals. We show how these two lengths vary for
different sizes of population, demonstrating that Effective Length is relatively independent of population size,
but that the Actual Length is proportional to it. We introduce Grammatical Evolution Memory Optimization
(GEMO), a two-stage evolutionary system that uses a multi-objective approach to identify the optimal, or at
least, near-optimal, genome length for the problem being examined. It uses a single run with a multi-objective
fitness function defined to minimize the error for the problem being tackled along with maximizing the ratio
of Effective to Actual Genome Length leading to better utilization of memory and hence, computational
speedup. Then, in Stage 2, standard GE runs are performed restricting the genome length to the length
obtained in Stage 1. We demonstrate this technique on different problem domains and show that in all cases,
GEMO produces individuals with the same fitness as standard GE but significantly improves memory usage
and reduces computation time.
1 INTRODUCTION
Evolutionary Algorithms have gained a lot of
popularity to automatically generate programs,
especially Koza’s Genetic Programming (GP) (Ryan,
1998). Although powerful, GP has some restrictions,
specifically the use of single types. This issue has
been tackled by several other grammar-based
flavours of GP, the most commonly used of which is
Grammatical Evolution (GE) (Ryan, 1998), which
employs linear binary strings to generate programs in
any arbitrary language using a Backus-Naur Form
(BNF) Grammar.
The computational complexity of an Evolutionary
Algorithm depends to a large extent on the
complexity of the fitness function. Genetic Algorithm
(GA) tries to obtain optimal values for the objective
function by either maximizing or minimizing a
solution to the problem. However, there is also the
a
https://orcid.org/0000-0002-8182-2465
b
https://orcid.org/0000-0001-6036-0030
c
https://orcid.org/0000-0002-4613-937X
d
https://orcid.org/0000-0002-7002-5815
issue of space complexity, which is typically caused
by large individuals or populations, or both. This can
lead to poor utilization of memory which in turn
increases computational time. We address this by
using a Multi-Objective Approach based system,
GEMO which minimizes error and maximizes
memory utilization.
Many researchers have explored the idea of multi-
objective optimization using fitness, size and
diversity as objectives. In a multi-objective
optimization approach, the idea of non-dominated
solutions along with Pareto optimal individuals is
considered to achieve all the specified objectives.
Pareto fronts have been used to obtain a set of
individuals that can optimize multi-objective Genetic
Algorithms (Deb, 2001). Efforts have also been made
to improve the multi-objective optimization
algorithms by redefining search and selection criteria.
(Eddy, 2001) has used a distinct point metric and
184
Kshirsagar, M., Jachak, R., Chaudhari, P. and Ryan, C.
GEMO: Grammatical Evolution Memory Optimization System.
DOI: 10.5220/0010106501840191
In Proceedings of the 12th International Joint Conference on Computational Intelligence (IJCCI 2020), pages 184-191
ISBN: 978-989-758-475-6
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

cluster metrics as defined by (Wu, 2000) to optimize
multi-objective problems. The distinct point indicates
the number of unique individuals in the solution space
whereas cluster metric measures the number of
unique individuals in the cluster, which is calculated
by dividing the number of individuals with the
number of distinct individuals. Bleuler et al (Bleuler,
2008) considered fitness of solution and program size
as two objectives and applied a bi-objective
optimization using a Pareto-based method in which
individuals with a smaller code size were preferred
over similar performing individuals.
GE presents an extra challenge due to disconnect
between Actual and Effective Lengths; individuals
with short Effective Lengths don’t necessarily result
in short Actual Lengths. Indeed, Section 4
demonstrates that the Actual Lengths grow at a higher
rate than Effective Lengths which leads to poor
memory usage.
2 GRAMMATICAL EVOLUTION
GE is a combination of a GA and GP. Programs or
phenotypes are evolved through the process of
mapping using a variable length genotype (also
referred to as chromosome) and a formal grammar
which is written in BNF (Backus, 1963) (Neill, 2003).
A chromosome consists of binary strings (or genes)
of certain length (number of binary digits) where each
gene is a variable or parameter under consideration.
The entire length of an individual is known as the
Actual Length while the number of codons used to
generate phenotype is referred to as the Effective
Length (Nicolau, 2012). If the Effective Length is less
than the Actual Length then the remaining unused
codons, the tail of the genome, are not used in
deriving expressions.
However, if an individual does not have enough
codons to map to a valid phenotype structure, then the
mapping process terminates, and the individual is
considered to be invalid. Alternatively, to mitigate
this issue, GE can use the concept of wrapping, which
re-uses the same genome sequence, in an attempt to
completely map an individual. The number of times
an individual is wrapped is referred to as the
Wrapping Factor (WF).
To demonstrate this concept, consider the
following grammar. As with all BNF grammars, it
can be defined as the tuple <S, N, T, P>, where T is
the set of terminals, i.e., items that can appear in
syntactically valid programs, N is a set of non-
terminals, i.e. intermediate constructs that don’t
appear in syntactically valid programs and P is a set
of production rules that maps the non-terminals into
terminals. S is the start symbol, from which all
individuals grow from; in this case, it is the non-
terminal <algo>.
<algo> ::= <var><op><algo> | <var>
<op> ::= + | - | * | /
<var> ::= 0
|
1
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
9
Grammar 1: Simple Arithmetic Calculator.
The above grammar consists of 3 non-terminals
(<algo>, <op> and <var>) which are used to map
fourteen terminal symbols (0, 1, ... 9, +, -, *, /).
The Effective and Actual Length required in four
different scenarios of mapping in GE are shown in
Table 1. Memory utilization can be defined as the
ratio of Effective Length to Actual Length. In all the
scenarios, the maximum length of the genome needs
to be defined carefully, as too large an Actual Length
results in a waste of memory (Scenario 1) while too
short of an Actual Length results in genomes that are
unable to map program structures completely
(Scenario 2). Although wrapping can be applied
(Scenario 3) to address this issue, the process of
mapping becomes trapped in infinite loops failing to
evolve an individual completely (Scenario 4).
Table 1: Actual Length and Effective Length under
different scenarios of mapping using Grammar 1.
Scenario
Input
Genome
String
generated
Actual
Lengt
h
Effectiv
e
Length
Sufficient
Genome
Length
(4, 13, 8,
4, 14, 23,
20, 5)
3 + 4 8 5
Insufficient
Genome
Length
(6, 13, 9,
4, 27, 15,
20, 12)
Invalid
Phenotype
8 n/a
Insufficient
Genome
Length
with
Wrapping
(6, 13, 9,
4, 27, 15,
20, 12)
3 - 7 * 2 - 9 8 11
Infinite
loop
problem in
Wrapping
(4, 14, 5,
2, 19, 23)
Invalid
Phenotype
6 n/a
Inappropriate definition of genome lengths can
lead to poor memory usage and computational
overhead; for example, some of the experiments in
Section 6 demonstrate that for some problems less
than 15% of the Actual Length is used for mapping
GEMO: Grammatical Evolution Memory Optimization System
185
in standard GE. To make matters even worse, this
usage degrades over time, so the longer a run is, the
worse the situation gets.
3 PROBLEM DOMAINS
We examine three different problem domains: Time
Series Analysis (Ryan, 2020), Symbolic Regression
and a Boolean Logic Problem.
3.1 Autoregression
Autoregressive Time Series Forecasting is a type of
regression model, linear for this case, which is used
to predict a variable based on a linear combination of
input values. The general form of the equation is
described as:
Y = A
0
+ A
1
* X
1
+ A
2
* X
2
(1)
This method is employed on the time series data
where a number of input variables are taken as
observations from previous time steps called lagged
variables. To predict the value for the next time step
(t+1), the problem can be formulated by considering
the values of previous time steps as:
X
(
t+1
)
= B
0
+ B
1
* X
(t
-1
)
+ .... + B
n
* X
(t
-n
)
(2)
As the regression model forecasts data from the
same input variable, it is referred to as
AutoRegressive Time Series Forecasting.
y = < intercept > + < expr >
< expr > ::= (< expr > + < expr >) |
(< expr > - < expr >) |
(< constant > * < var >)
< constant > ::= 0.< num > | -0.< num >
< intercept > ::= < num >.< num > |
-< num >.< num >
< num > ::= 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
< num >< num1 >
< num1 > ::= 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9
< var > ::= X.shift(< lag > + 1)
< lag > ::= < num >
Grammar 2: Grammar for AutoRegression.
Grammar 2 generates values for each of
<intercept>, <constant> and <lag>. The
<intercept> generates a value which is not a function
of time while <constant> derives smoothing
coefficients for lag variables ranging between -1 to 1.
The <lag> variable is the input value to the forecaster
as a function of time.
We examine four
AutoRegressive datasets, which we term AR1 (Daily
Dublin Waste), AR2 (Hourly Riders), AR3 (Daily
Temperature) and AR4 (Monthly Dublin Waste).
Each of these uses Root Mean Square Error as its
error metric.
1
(3)
3.2 Symbolic Regression
We also examine two classic GP Symbolic
Regression problems, namely the Vladislavleva4
(Ryan, 1998), generally considered to be at the higher
end of the difficulty range, and the infamous Quartic
Polynomial problem, which we include to
demonstrate the impact of GEMO even on easy
problems. We have used grammar as mentioned in
(Ryan, 1998) for experimentation. The error metric
for these problems is defined in Table 2.
Table 2: Error Metric and Domain for Symbolic Regression
Problems.
Problem Error metric Domain
Vladislavleva
4
Mean Square Error
1
0.05 to
6.05
Quartic
Polynomial
Mean Average Error
1
-1 to 1
3.3 Boolean Logic
The third domain we examine is Boolean Logic
which takes true or false as an input. For this domain,
we choose the 11-Multiplexer problem as employed
by Koza (Ryan, 1998) and use their grammar for
experimentation. Error metric used in this case is
Hamming Error as defined in equation 4.
!
(4)
ECTA 2020 - 12th International Conference on Evolutionary Computation Theory and Applications
186
4 MEMORY AND RUNTIME
ANALYSIS
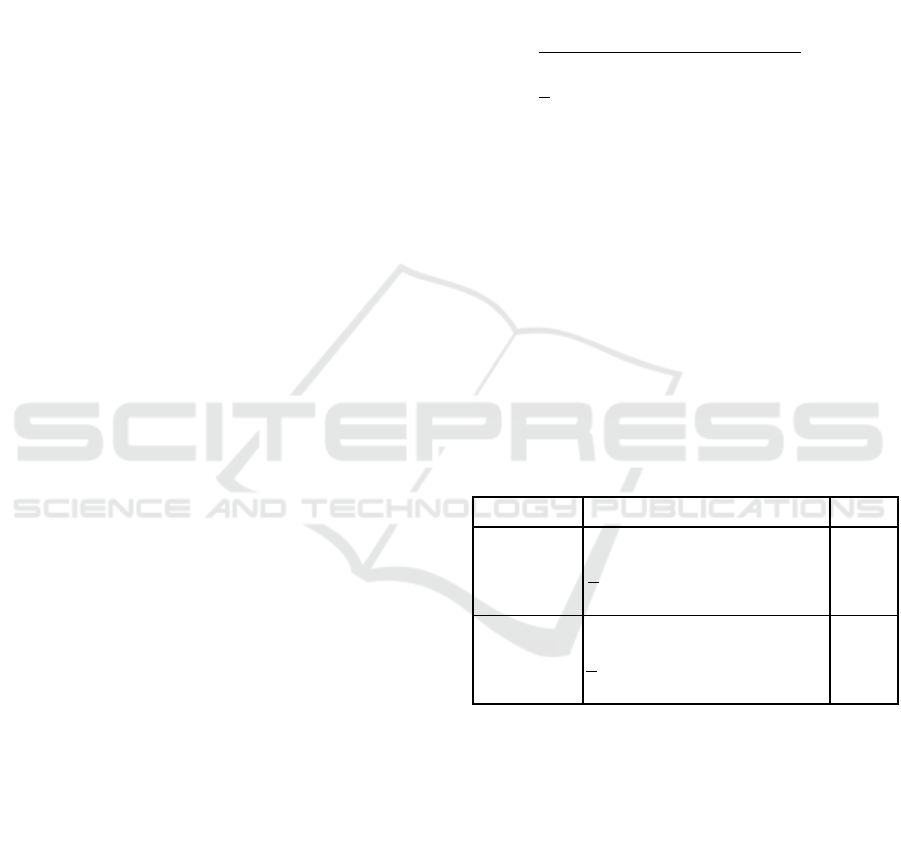
Figure 1 shows the effect of varying population size
on the Effective and Actual Genome Lengths on AR1.
We noticed that in all experiments, regardless of
domain, there appears to be something of a steady
state value for Effective Length, which is independent
of the population size, while the Actual Length
increases with population size leading to higher space
complexity. The computational complexity (CC) for
EAs can be established in terms of evolutionary
parameters as:
CC = (WF *GL * UI * G) (5)
Where WF is Wrapping Factor, GL is Genome
length, UI is the number of unique individuals, and G
is the number of generations. This indicates that the
computational complexity is directly proportional to
the length of genome as shown in equation 5.
Considering WF, UI and G to be constant for an
experiment, CC of GE program is upper bounded by
GL shown in equation 6.
CC = O(GL) (6)
Notice that in the plot below, both Actual and
Effective Lengths drop after the first generation. This
is due to the difficulty which GE often encounters
with invalid individuals in the first generation, as
noted by (Ryan, 2003).
Figure 1: Effect of Varying Population Size on Effective
and Actual Genome Length on Problem AR1.
Memory utilization is 13%, 29% and 32%
respectively; it either remains constant or degrades at
later generations, as the algorithm evolves, which
leads to increase in computation time. The maximum
memory utilization for standard GE is less than 33%,
which means more than two-thirds of the memory is
wasted, which makes it difficult to scale GE platform
to complex problems.
Therefore, it is essential to define an appropriate
length of genome to reduce memory wastage and
computational complexity of an algorithm without
affecting its objective fitness.
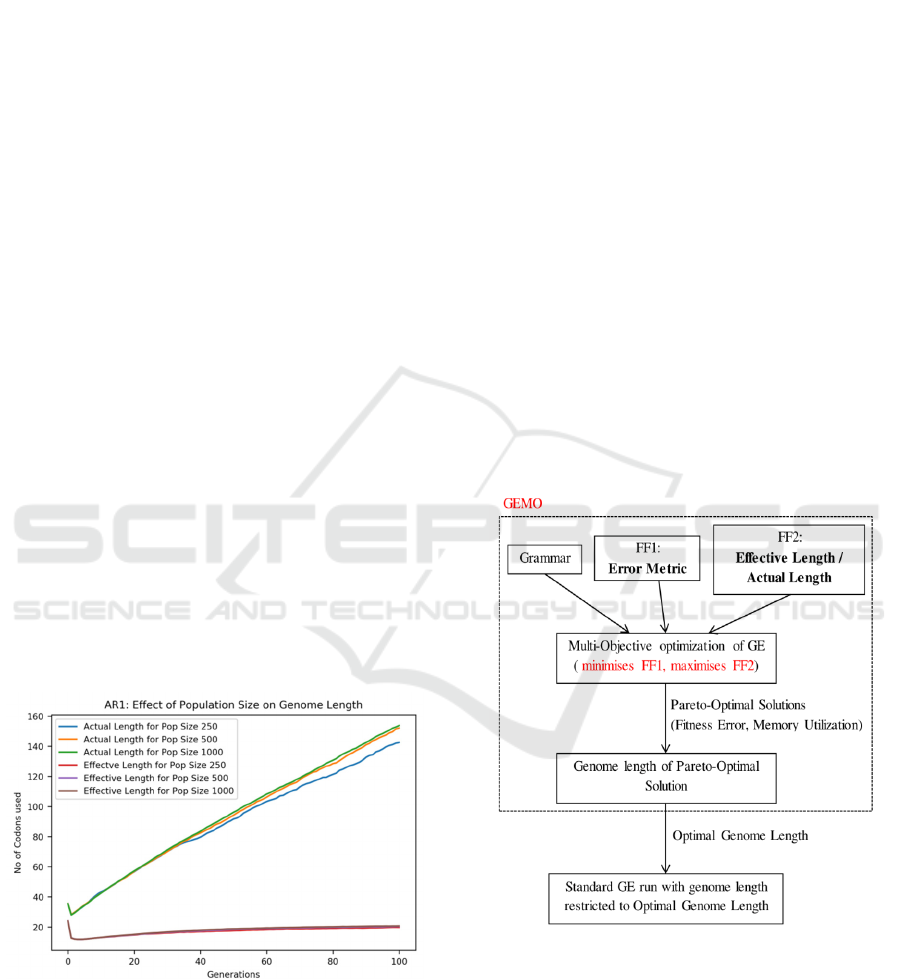
5 GEMO
GEMO applies a multi-objective approach as shown
in Figure 2 to optimize error metric and memory
utilization.
1. Fitness Function 1: Minimize the error
metric given for a specific problem as
discussed in section 3.
2. Fitness Function 2: Maximize the memory
utilization which is the ratio of effective to
Actual Length as described in equation 7.
maximize (Effective Length / Actual
Length)
(7)
Figure 2: Methodology diagram for GEMO system (FF1:
Fitness Function 1, FF2: Fitness Function 2).
GEMO uses two dimensional pareto vectors,
where each objective refers to the pareto variable in
one of the dimensions. Since this is a multi-objective
optimization problem, it tries to optimize all of the
objectives which could be contradictory and hence, a
trade-off needs to be considered while selecting
individuals to satisfy each objective. The Pareto-
GEMO: Grammatical Evolution Memory Optimization System
187
optimal individuals are calculated using the crowding
distance value of a solution, which provides an
estimate of the density of individuals surrounding that
particular solution. The set of solutions are sorted
according to each objective function, and crowding
distance is calculated as the average distance of its
two neighbouring solutions. To promote diversity in
the set of solutions, individuals with the highest and
lowest objective fitness are always selected.
Since, GEMO tries to minimize error metric and
maximize memory utilization, pareto fronts are sorted
in descending order in terms of given error metric and
for the same error, it is sorted in ascending orders for
the memory utilization. We then select individuals
with the low error and maximum memory utilization
as optimal solutions. The genome lengths obtained
from these optimal solutions are passed as a
parameter to Standard GE to restrict the length of
genome.
Note that this doesn’t guarantee that we will find
the absolutely minimally sized genome that can
generate useful solutions but, as Section 6
demonstrates, the memory savings are still enormous.
There has also been some work (Ryan, 2003) that
shows that GE sometimes relies on having unused
tails to aid evolution, meaning that if we restrict it too
much, we might hamper its progress.
The size of the
genotype suggested this way is subsequently
validated in Section 7.2 through statistical tests by
taking the amount of wrapping into consideration.
6 EXPERIMENTAL RESULTS
The evolutionary parameters used in all experiments
in this paper are
{ Population
Size
:
250, 500, 1000; Maximum Number
of Generations
: 100;
Crossover Type
:
Single Point
;
Crossover Probability
: 0.95;
Mutation Type
:
Int Flip
Codon
;
Mutation Probability: 0.01; Selection Type
:
Tournament
;
Initialization Method
:
Position Independent
Growth
;
Initial Depth
: 7;
Maximum Depth: 10; Number
of Runs
: 100. }.
Position independent growth (Fagan, 2016) is
used to initialize the population with a maximum tree
depth of 10, with an initial maximum depth of 7.
Tournament selection (Fang, 2010), followed by
single point crossover with probability of 0.95 and
integer flip codon mutation with probability of 0.01
are incorporated in the framework. The above values
are selected, since optimal results were achieved
within Standard Runs of GE using this set of
configuration and hence served as a benchmark for
our framework. All the experiments are carried out
using PonyGE2 (Fenton, 2017). Recall that GEMO is
a two stage process. The same parameters are used in
both stages; the first stage is run once, to determine
the length of individual to use. The second stage is
run 100 times to obtain statistically valid results.
Section 7.1, contains some experiments
demonstrating that the use of a single run in the first
stage is reasonable.
6.1 Selection of Pareto-Optimal
Solutions
Figure 4.a illustrates pareto fronts solutions for the
error metric and memory utilization (ratio of
Effective to Actual Length) obtained for each
generation Individuals having least regression error
(X axis, to the left) and highest memory utilization
(Y-axis, to the top).
T
he individual with the lowest
regression error (4410) and the highest memory
utilization of 1.0 is selected as a pareto-optimal
solution indicated through dark brown colour.
Figures 4.b, plot the Effective Length of pareto
individuals obtained in 4.a. The Effective length of
the pareto-optimal solutions is selected as optimal
genome length for that particular problem. If
solutions with multiple genome lengths are obtained,
individuals with least effective length are preferred.
We report these as (fitness, effective_length) tuples.
The optimal Effective Length of 21 indicated
through dark brown colour is selected from optimal
pareto fronts obtained in Figure 4.a.
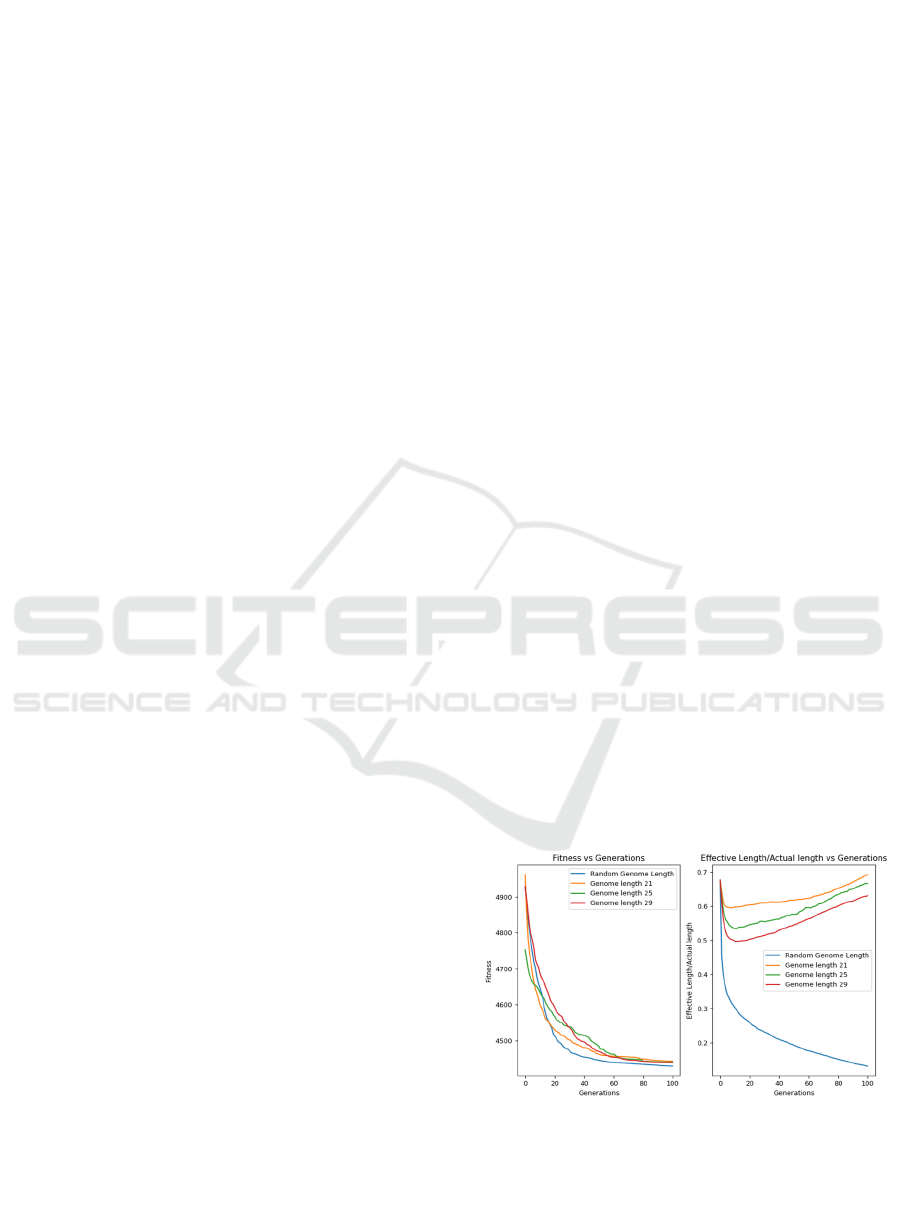
6.2 Fitness Performance – AR1
We look in detail at the performance of GEMO on
AR1.
Figure 3: Left: Comparison of Fitness Vs Generations for
unrestricted genome length and optimal solutions on AR1.
Right: Comparison of the ratio of Effective to Actual
Length and Fitness vs Generations for unrestricted genome
length and optimal solutions.
ECTA 2020 - 12th International Conference on Evolutionary Computation Theory and Applications
188
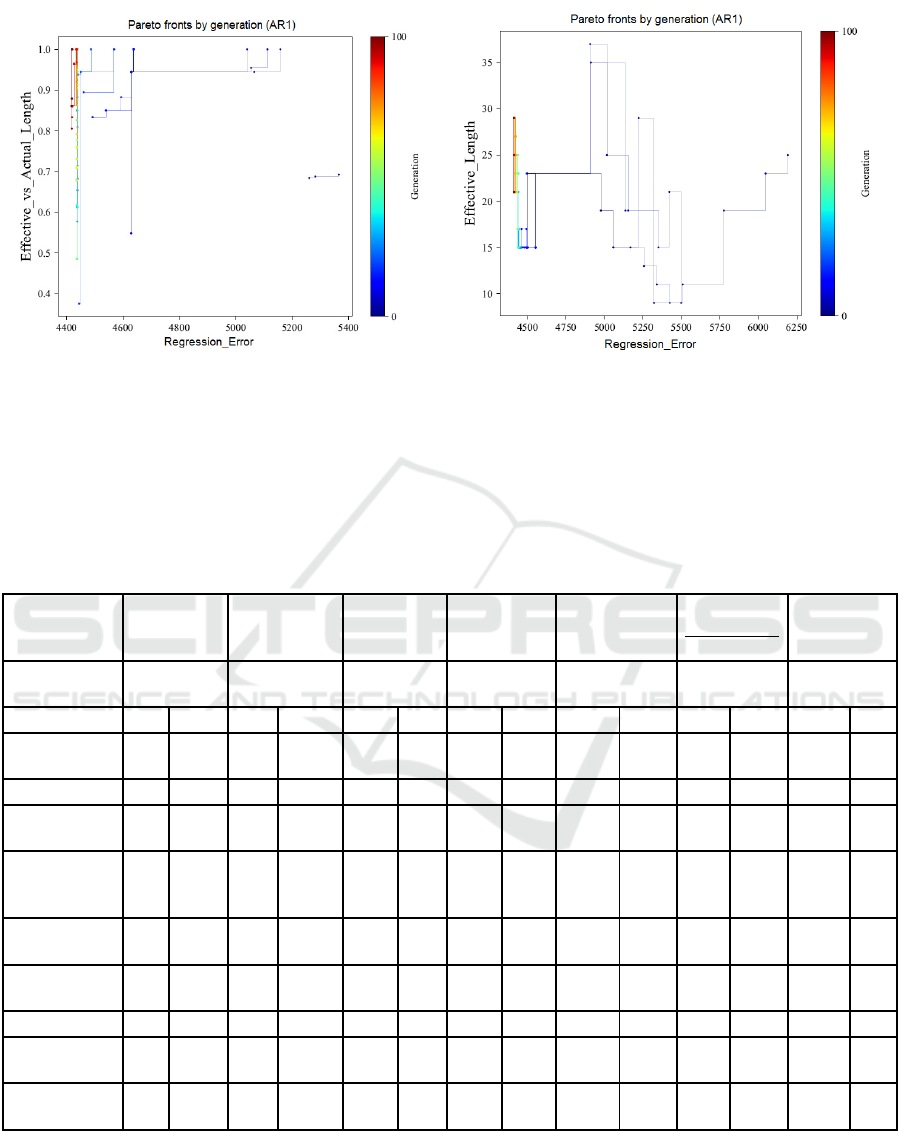
(a) AR1 (b) AR1
Figure 4: Results for GEMO for AR1. Graph on the left indicate the ratio of Effective Length to Actual Length vs Fitness.
The number of generations is indicated by a color map; notices that later generation are on the left of the graph; this is because
of the Pareto plot which has fitter individuals closer to the origin. These individuals are then replotted using the graph on the
right hand side to identify the shortest Effective Length. We report these as (fitness, effective_length) tuples, and obtain
pareto-optimal individuals (4410, 21) for AR1.
Table 3: Results for Single and Multi-Objective Optimal solutions for AR1, AR2, AR3, AR4, Quartic Polynomial,
Vladislavleva4, 11-Multiplexer averaged over 100runs (UL: Unrestricted Length, OL: Optimal Length obtained from GEMO
stage 1). There is no statistically significant difference in any fitness results.
Dataset
AR1
(Kilograms)
AR2 (Number
of Riders)
AR3
(Degree
Celsius)
AR4
(Kilograms)
Quartic
Polynomial
(X
4
+X
3
+X
2
+X)
Vladislavleva4
∑
11-
Multiplexer
Number of
Instances
1,000 18,290 3,652 136 10,000 10,000 2,048
Method UL OL UL OL UL OL UL OL UL OL UL OL UL OL
Effective
Length
69 14.49 23.61 10.91 14.45 11.68 14.71 10.3 8.49 2.68 74 6.68 394.05 33.76
Actual Length 512
21
212
16.22 105.34 19.98 111.43 21.32 340.09 35.36 252 12.83 1214.85 76.38
Decrease in
Actual Length
-
24.38X
-
13.07X
-
5.27X
-
5.2X
-
9.6X
-
19.64X
-
15.3X
Memory
Utilization
(in percentage)
13.47
69
11.14
67.33
13.72
58.5
13.2
48.3
2.43
7.57
29.45
52.06
32.47
42.83
Utilization
Improvement
-
5.12X
-
6.04X
-
4.26X
-
3.66X
-
3.37X
-
1.76X
-
1.31X
Computation
Time (seconds)
92
79.32
139
103
96
80
104
79
9.65
7.69
40
22
54
13
Speedup -
1.15X
-
1.34X
-
1.2X
-
1.31X
-
1.25X
-
1.81X
-
4.15X
Mean Best
Fitness
4428 4447 28.11 28.11 0.51 0.52 18,061 18,432 0.02 0.12 0.028 0.027 617 645
Standard
Deviation
11 4 13.67 11.39 0.54 0.49 3,024 2,673 0.05 0.057 0.0031 0.0007 49 35
Crucially, the computation time to obtain the
solutions for AR1 with minimal regression error is
less with genome length restricted to 21 when
compared to unrestricted length genome and exhibits
69% memory utilization as compared to 13.8% for
unrestricted as shown in Figure 3. Table 3 shows that
the Optimal Length Genome individuals achieve
similar accuracy when compared to individuals with
GEMO: Grammatical Evolution Memory Optimization System
189

Unrestricted Genome Length in terms of Best, Mean
and Worst Case Fitness values, but has a lower
standard deviation resulting in the optimal value
being achieved more consistently. In each of the other
three cases the overall fitness was either the same
(AR2 or AR3) or better (AR4), while the experiments
employing GEMO always use significantly less
memory (between 3.6 and 6 times less) and run faster
as shown in Table 3.
6.3 Fitness Performance Results –
Symbolic Regression and Boolean
Logic
In addition to the Vladislavleva4 we also considered
the simple Quartic Polynomial problem. The results
are shown in Table 3; in each of the cases, GEMO
performed essentially the same in terms of fitness (no
statistically significant difference) but did so with
substantially less memory and in less time; moreover,
the memory that was used is utilized better, as noted
in the Memory Utilization row.
These results show that the Quartic Polynomial
problem used 9.69 times less memory and showed a
speed up of 1.25, while in Vladislavleva4, the decrease
in memory required was 19.64 times less, with a speed
up of 1.76. Similarly, with the 11-Multiplexer
experiments, the memory utilization improvement was
15.3 and the speed up 4.15.
Notice that the optimal
length identified by the GEMO for each of these
problems is, in fact, substantially longer than the
required Effective Length, since it doesn’t have 100%
memory utilization while Standard GE run is being
performed. This is due to the stochastic nature of GE.
However, this indicates that simply imposing a
reasonable constraint on GE (where reasonable in this
case means long enough to accommodate the genome
needed) has a large impact on memory usage. It also
suggests that further optimization in GEMO could
yield even better results.
7 ANALYSIS
We perform two different analyses on the GEMO. As
noted above, the first stage of GEMO uses just a
single run; Section 7.1, below, describes experiments
to test the variance across multiple runs to ascertain
how reasonable an approach this is. The following
section determines if the lengths determined by Stage
1 are too short, by performing an analysis of the
impact of removing wrapping.
7.1 Variability of Stage 1
Due to the heuristic nature of GE, we performed
multiple runs of GEMO, and compared results of
Mean Efficient Length with that of a single run of
GEMO and no statistically significant difference was
observed on the used datasets.
7.2 Sufficiency of Lengths Obtained
To validate whether the obtained set of genome
length is sufficient or not, we ran another set of
experiments in which individuals were allowed to
evolve by taking the Wrapping factor into
consideration. This will establish if we genuinely
have discovered usable genome lengths, or if GE is
simply exploiting the wrapping operator to make it
appear as though we have. We conduct statistical
significance tests which allow us to support or reject
a claim being made at different significance levels. As
all significance tests begin with a null hypothesis H
0,
the null hypothesis of samples from two populations
with and without wrapping is formulated as shown
below:
Null Hypothesis H
0
: There is no statistical
difference between Mean Best Fitness of Datasets
with wrapping and without wrapping.
Table 4: Number of times the Null Hypothesis is rejected
at significance level 0.01, 0.05, 0.1 out of 100 trials.
Problems
Significance Level
0.01 0.05 0.10
AR1
1 4 11
Vladislavleva4
7 14 26
Quartic Polynomial
7 18 27
11-Multiplexer
6 15 22
The number of times null hypothesis rejected out
of 100 trials for a sample size of 30 is shown in Table
4 for a significance level (alpha) of 0.01, 0.05 and
0.10. It can be seen that on average, the fitness values
evaluated without considering wrapping factor are
equal to the fitness values evaluated considering
wrapping factor at alpha equal to 0.01, 0.05 and 0.10
respectively.
Hence, we fail to reject the null hypothesis for all
of the problems listed in Table 4. Therefore, it can be
stated GEMO does not rely on wrapping and that the
length it produces is optimal and does not force GE to
resort to wrapping.
ECTA 2020 - 12th International Conference on Evolutionary Computation Theory and Applications
190

8 CONCLUSIONS
Optimal use of resources and low computation time
is an important aspect for any framework. We have
introduced Grammatical Evolution Memory
Optimization, GEMO, a multi-objective based
framework for memory optimization. It defines
memory utilization as one of the objectives to explore
individuals with genome length that maximizes
memory utilization along with its low error metric.
The result obtained from GEMO is used to constrain
the maximum length of the genome in an otherwise
standard GE run. GEMO was tested on three
benchmark domains, including Time Series
Forecasting, Symbolic Regression and Boolean
Logic. Experimental results validated that GEMO had
statistically similar fitness results, but it exhibited
significant increase in memory utilization, as well as
a decrease in computational overhead. The system
also ruled out the possibility that wrapping is the
reason for GEMO succeeding with shorter genomes
by conducting experiments both with and without
wrapping and no statistically significant difference
was observed.
We can safely conclude that this strategy will be
useful for large, multidimensional datasets where we
can be sure of optimizing memory and speedup.
However further experimentation needs to be carried
out in future to check their viability for small datasets.
The results further showed that the maximum Actual
Lengths suggested by GEMO were, in general, longer
than they needed to be. Future work will explore this
to see if it is feasible to expand the framework to
produce shorter maximum genome, maximum
utilization of memory and to establish the cost/benefit
trade off of spending more time on this part of the
search.
ACKNOWLEDGEMENTS
This work is supported in part by the Science
Foundation of Ireland grant #16/IA/4605.
REFERENCES
Ryan C., Collins J., Neill M.O. (1998) Grammatical
evolution: Evolving programs for an arbitrary language.
In: Banzhaf W., Poli R., Schoenauer M., Fogarty T.C.
(eds) Genetic Programming. EuroGP 1998. Lecture
Notes in Computer Science, vol 1391. Springer, Berlin,
Heidelberg
Deb K. Multi-objective optimization using evolutionary
algorithms. Chichester, UK: John Wiley and Sons,
2001.
Eddy, J., & Lewis, K. (2001, September). Effective
generation of Pareto sets using genetic programming. In
Proceedings of ASME design engineering technical
conference (Vol. 132).
Wu, J., and Azarm, S. (January 1, 2000). "Metrics for
Quality Assessment of a Multiobjective Design
Optimization Solution Set ." ASME. J. Mech. Des.
March 2001; 123(1): 18–25.
Bleuler S., Bader J., Zitzler E. (2008) Reducing Bloat in GP
with Multiple Objectives. In: Knowles J., Corne D.,
Deb K., Chair D.R. (eds) Multiobjective Problem
Solving from Nature. Natural Computing Series.
Springer, Berlin, Heidelberg
J. W. Backus, F. L. Bauer, J. Green, C. Katz, J. McCarthy,
P. Naur, A. J. Perlis, H. Rutishauser, K. Samelson, B.
Vauquois, J. H. Wegstein, A. van Wijngaarden, M.
Woodger, Revised report on the algorithmic language
ALGOL 60, The Computer Journal, Volume 5, Issue 4,
1963, Pages 349-367.
M. O'Neill, C. Ryan, “Grammatical Evolution:
Evolutionary Automatic Programming in an Arbitrary
Language”, Kluwer Academic Publishers, 2003.
M. Nicolau, M. O'Neill and A. Brabazon, "Termination in
Grammatical Evolution: grammar design, wrapping,
and tails," 2012 IEEE Congress on Evolutionary
Computation, Brisbane, QLD, 2012, pp. 1-8.
Ryan, C.; Kshirsagar, M.; Chaudhari, P. and Jachak,
Rushikesh (2020). GETS: Grammatical Evolution
based Optimization of Smoothing Parameters in
Univariate Time Series Forecasting. In Proceedings of
the 12th International Conference on Agents and
Artificial Intelligence - Volume 2: ICAART, ISBN 978-
989-758-395-7, ISSN 2184-433X, pages 595-602.
Ryan C., Keijzer M., Nicolau M. (2003) On the Avoidance
of Fruitless Wraps in Grammatical Evolution. In:
Cantú-Paz E. et al. (eds) Genetic and Evolutionary
Computation — GECCO 2003. GECCO 2003. Lecture
Notes in Computer Science, vol 2724. Springer, Berlin,
Heidelberg
David Fagan, Michael Fenton, Michael O’Neill, “Exploring
Position Independent Initialisation in Grammatical
Evolution”, IEEE Congress on Evolutionary
Computation, At Vancouver, Canada, 2016.
Yongsheng Fang and Jun Li, “A Review of Tournament
Selection in Genetic Programming”, Advances in
Computation and Intelligence, 5th International
Symposium, ISICA 2010 Wuhan, China, October 22-
24, 2010,LNCS Springer Proceedings.
Michael Fenton, James McDermot, David Fagan, Stefan
Forstenlechner, Erik Hemberg, Michael O’Neill,
“PonyGE2: Grammatical Evolution in Python”,
GECCO ’17 , Berlin, Germany, July 15-19, 2017.
GEMO: Grammatical Evolution Memory Optimization System
191
