
Table Interpretation and Extraction of Semantic Relationships to
Synthesize Digital Documents
Martha O. Perez-Arriaga
1
, Trilce Estrada
1
and Soraya Abad-Mota
2
1
Computer Science, University of New Mexico, 87131, Albuquerque, NM, U.S.A.
2
Electrical and Computer Engineering, University of New Mexico, 87131, Albuquerque, NM, U.S.A.
Keywords:
Table Understanding, Information Extraction, Information Integration, Semantic Analysis.
Abstract:
The large number of scientific publications produced today prevents researchers from analyzing them rapidly.
Automated analysis methods are needed to locate relevant facts in a large volume of information. Though
publishers establish standards for scientific documents, the variety of topics, layouts, and writing styles im-
pedes the prompt analysis of publications. A single standard across scientific fields is infeasible, but common
elements tables and text exist by which to analyze publications from any domain. Tables offer an additional
dimension describing direct or quantitative relationships among concepts. However, extracting tables infor-
mation, and unambiguously linking it to its corresponding text to form accurate semantic relationships are
non-trivial tasks. We present a comprehensive framework to conceptually represent a document by extracting
its semantic relationships and context. Given a document, our framework uses its text, and tables content
and structure to identify relevant concepts and relationships. Additionally, we use the Web and ontologies
to perform disambiguation, establish a context, annotate relationships, and preserve provenance. Finally, our
framework provides an augmented synthesis for each document in a domain-independent format. Our results
show that by using information from tables we are able to increase the number of highly ranked semantic
relationships by a whole order of magnitude.
1 INTRODUCTION
The rate at which scientific publications are produced
has been steadily increasing year after year. The es-
timated growth in scientific literature is 8 − 9% per
year in the past six decades (Bornmann and Mutz,
2015). This is equivalent to doubling the scientific lit-
erature every 9-10 years. Such massive production of
articles has enabled the rise of scientific text mining,
especially in the health sciences (Cohen and Hersh,
2005). The goal of this mining is to uncover non-
trivial facts that underlie the literature; however, these
facts emerge as correlations or patterns only after ana-
lyzing a large volume of documents. One of the most
famous works of literature-based mining led to dis-
cover that magnesium deficiency produces migraines
(Swanson, 1988). Some literature-based approaches
use word frequency and co-occurrence (Srinivasan,
2004), n-grams (Sekine, 2008), and semantic rela-
tionships between concepts using fixed patterns (Hris-
tovski et al., 2006). These methods, although effi-
cient, are lacking in three crucial aspects (1) they lead
to a large number of false conclusions, as they can-
not exploit context or disambiguate concepts; (2) they
rely on patterns and ignore structural and semantic re-
lationships, as their information extraction does not
consider relevant concepts with explicit relationships
expressed in tables; and (3) they represent their find-
ings for a specific domain, which cannot be exten-
sively and repeatedly exploited. Thus, the next gen-
eration of deep text mining techniques needs more
sophisticated methods for information extraction and
representation.
Extracting information from the vast scientific lit-
erature is challenging because even though confer-
ences and journals establish guidelines to publish
work, articles within a field are still reported with var-
ious formats (e.g., PDF, XML), layouts (e.g., one or
multi-column), and writing style. No unified vocab-
ulary exists. For instance, publications on healthcare
might use various names for the same concept (e.g.,
diabetes management and glycemic control). And the
practice of annotating articles with metadata and on-
tologies is far from widely adopted. Further, a unique
standard for publishing articles across scientific fields
is not feasible. Our work aims to fill this gap by pro-
viding a comprehensive mechanism to (1) conceptu-
ally representing a document by extracting and anno-
Perez-Arriaga, M., Estrada, T. and Abad-Mota, S.
Table Interpretation and Extraction of Semantic Relationships to Synthesize Digital Documents.
DOI: 10.5220/0006436902230232
In Proceedings of the 6th International Conference on Data Science, Technology and Applications (DATA 2017), pages 223-232
ISBN: 978-989-758-255-4
Copyright © 2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
223

tating its semantic relationships and context and (2)
preserving provenance of each and every relationship,
entity, and annotation included in the document’s syn-
thesis.
Our approach takes advantage of the structured
information presented in tables and the unstructured
text in scientific publications. Given a document,
our framework uses its tables’ content and structure
to identify relevant entities and an initial set of rela-
tionships. Additionally we use the document’s text to
identify metadata of the publication (e.g., Author, Ti-
tle, Keywords) and context. We use the publication’s
context, ontologies, and the Web to disambiguate the
conceptual framework of extracted entities, and we
refine the set of relationships by further searching for
these entities within the text. Our approach uses an
unsupervised method to rank the relevance of rela-
tionships in the context of the publication. Finally,
our approach organizes the metadata, entities and se-
mantic relationships in a domain-independent format
to facilitate batch analysis. The resulting document
can be used as a synthesis of a publication and can
be easily searched, analyzed, and compared as it uses
a controlled vocabulary, Web-based annotations, and
general ontologies to represent entities and relation-
ships. Our results show that the entities and semantic
relationships extracted provide enough information to
analyze a publication promptly.
The reminder of this paper is organized as follows:
Section 2 describes related work. Section 3 contains
our approach to understanding tables and extracting
semantic relationships from publications. Section 4
contains a working example using our approach. Sec-
tion 5 contains a set of experiments, results and dis-
cussion of our findings. Finally, Section 6 contains
our conclusion and future directions.
2 RELATED WORK
We review briefly work on table interpretation, an-
notation, disambiguation, semantic relationships and
summarization. Table interpretation is the process of
understanding a table’s content. There is work on in-
terpreting tables in digital documents with certain for-
mats (e.g., HTML, PDF) (Cafarella et al., 2008; Oro
and Ruffolo, 2008). Cafarella et al. present ‘webta-
bles’ to interpret tables from a large number of web
documents, enabling to query information from col-
lected tables (Cafarella et al., 2008).
Although tables and text contain relationships be-
tween concepts, external information sources to de-
scribe concepts in a publication are necessary. The
semantic web (Berners-Lee et al., 2001) contains en-
tities and relationships. DBpedia alone represents 4.7
billion relationships as triples (Bizer et al., 2009), in-
cluding the areas of people, books and scientific pub-
lications. Yet most triples from scientific publications
are missing in the semantic web, it can complement
scientific publications helping to explain concepts. To
understand tables, Mulwad et al. annotate entities
using the semantic web (Mulwad et al., 2010). Ex-
tracting tables from PDF documents poses more chal-
lenges because tables lack tags. Xonto (Oro and Ruf-
folo, 2008) is a system that extracts syntactic and se-
mantic information from PDF documents using the
DLP+ ontology representation language with descrip-
tors for objects and classes. Xonto uses lemmas and
part of speech tags to identify entities, but it lacks an
entity disambiguation process. Texus (Rastan et al.,
2015) is a method to identify, extract and understand
tables. Texus is evaluated with PDF documents con-
verted to XML. Their method performs functional and
structural analyses. However, it lacks disambiguation
and semantic analyses.
To disambiguate entities, some works (Abdal-
gader and Skabar, 2010) use a curated dictionary, such
as WordNet
1
. This suffices if publications only con-
tain concepts from this source. Others, like (Ferragina
and Scaiella, 2012) use richer sources like Wikipedia.
However, Wikipedia contains a large variety of ad-
ditional information, much of it increases noise. Our
work differs from these approaches in that we use DB-
pedia (Bizer et al., 2009), which is a curated ontology
derived from Wikipedia. In this case DBpedia offers
the best of both worlds: it is as vast as Wikipedia,
but, similarly to WordNet, it provides information in
a structured and condensed form following the con-
ventions of the Semantic Web.
A graphical tool, PaperViz (Di Sciascio et al.,
2017), allows to summarize references, manage meta-
data and search relevant literature in collections of
documents. Also, Baralis et al. (Baralis et al., 2012)
summarize documents with relevant sentences using
frequent itemsets. Still, researchers need to identify
concrete information with descriptions pertaining to a
defined context.
Regarding works on semantic relationships dis-
covery, Hristovski et al. find relationships from text
based on fixed patterns (Hristovski et al., 2006). The
open information extraction (IE) method has been
used successfully to find new relationships with no
training data (Yates et al., 2007). However, Fader et
al. state that IE can find incoherent and uninformative
extractions. To improve open IE, Reverb (Fader et al.,
2011) finds relationships and arguments using part of
speech tags, noun phrase sentences, and syntactic and
1
https://wordnet.princeton.edu/
DATA 2017 - 6th International Conference on Data Science, Technology and Applications
224
lexical constrains. Reverb uses a corpus built offline
with 500 million Web sentences to match arguments
in a sentence heuristically. In addition, Reverb identi-
fies a confidence for each relationship using a logistic
regression classifier with a training set of 1000 sen-
tences from the Web and Wikipedia. Reverb performs
better than the IE method Textrunner (Yates et al.,
2007). We use Reverb for our relationship extraction
from text because it finds new relationships and deter-
mines their importance with a confidence measure.
Generally, methods that extract semantic relation-
ships lack a way to represent their provenance. To
facilitate researchers’ work, it is important not only
to find important relationships from publications, but
also to systematically identify the specific sources
used for their extraction and annotation.
3 THE FRAMEWORK
We provide a comprehensive framework to interpret
the quantitative aspects of a document. Our approach
takes advantage of the rich source of information
found in tables, and their structure to identify concep-
tual entities and extract semantic relationships from
documents. The unstructured text provides a context,
to help in finding annotations of concepts, and meta-
data to characterize a publication. The entities and
relationships are used to annotate a scientific publica-
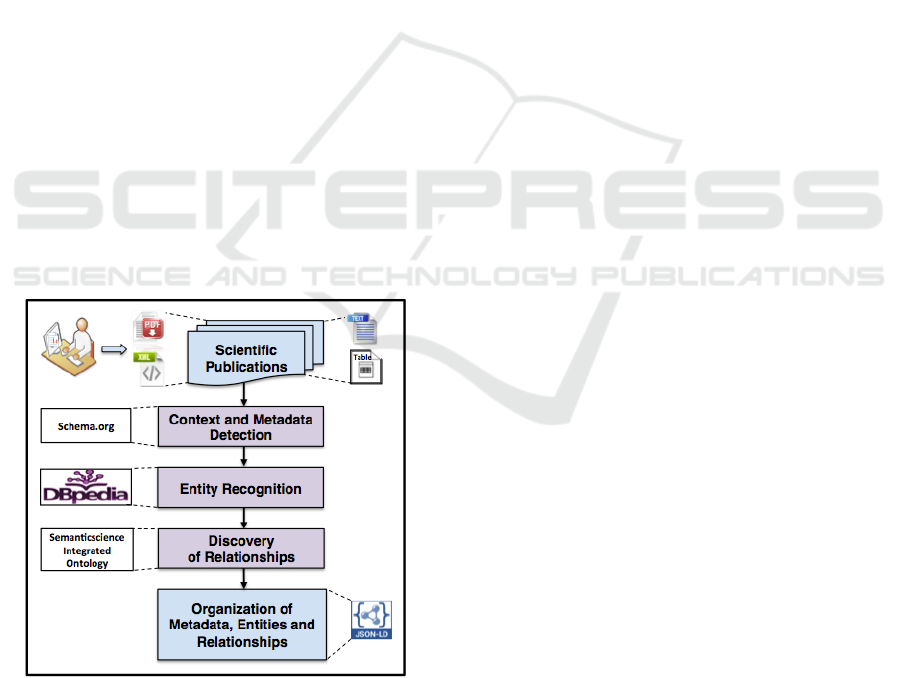
tion to facilitate its analysis (see Figure 1).
Figure 1: The framework for table interpretation and docu-
ment synthesis generation.
Our method receives as input digital publications
in PDF or XML formats. The publications undergo
processing to find its context and metadata to charac-
terize each publication. Then, we automatically rec-
ognize and extract tables’ content from each publi-
cation. The structured and summarized information
from tables is used to identify conceptual entities,
and the general ontology DBpedia is used to anno-
tate them. If an entity is undefined in the ontology,
we search the Web for a description of this entity.
Later, the entities are used to find structural and se-
mantic relationships from tables and text. We use the
SemanticScience Ontology (SIO) (Dumontier et al.,
2014) to express these relationships in a standard rep-
resentation (e.g., is a, is related to, is input of). Fi-
nally, this process produces a file containing meta-
data, entities and a set of semantic relationships found
in the publication. The output is presented in a special
JavaScript Object Notation (JSON) format to allow
interoperability even when publications belong to dif-
ferent domains. Since the extracted entities and rela-
tionships primarily belong to tables, this information
can be used to synthesize a publication’s quantitative
content. Also, because we preserve the context of a
publication and explicitly list semantic relationships,
our framework facilitates querying of particular infor-
mation, contrasting and comparing the claims in var-
ious documents, batch analysis, and as appropriate,
discovery of scientific facts.
3.1 Context and Metadata Detection
The first relevant pieces of information that one can
gather from a document are its metadata and con-
text. Metadata refers to the information that describes
the document itself, and includes data such as au-
thor, title, and keywords. Context refers to the par-
ticular field and topic addressed in the document.
Metadata and context can be easily extracted from
some publications that provide tags and keywords.
For these cases we obtain information directly us-
ing tags such as <author>, <article-title> and
<keywords>. However, many documents do not pro-
vide annotated data, and the extraction of metadata
and context becomes non-trivial. For documents lack-
ing a straightforward mechanism to extract their infor-
mation, we convert them into text format and search
for the most relevant concepts within the text. First,
to recover textual information, we convert a PDF doc-
ument to text format using PDFMiner (Shinyama,
2010). Then, to find the author and title of a publi-
cation, we perform pattern matching on the first page.
To recover keywords that define the context of a sci-
entific publication, we process the text (i.e., erasing
long, small and stop words) and apply term-frequency
inverse-document-frequency (TF-IDF) as explained
in (Ramos, 2003). For each word in the document,
we assess its relevance with a weight– measuring the
Table Interpretation and Extraction of Semantic Relationships to Synthesize Digital Documents
225

relative frequency of that word in the document com-
pared to its frequency in a large, but otherwise ran-
dom, collection of documents. In this case we use
Wikipedia
2
as the canonical collection of documents
because of its wide and diverse range of topics. To
search each word in Wikipedia, we use the API for the
search engine Bing
3
; by using a search engine rather
than a static collection we are sure always to retrieve
an up-to-date definition of each word. Bing’s results
provide the frequencies with which to calculate the
weight of each word. The resulting five concepts with
the best weights are defined as the keywords or con-
text of a publication.
Several standards exist to represent data to iden-
tify a publication (e.g., Dublin Core, Metadata Ob-
ject Description Schema). Though these standards
are useful to describe metadata for publications, we
require a broad and domain independent vocabulary.
Therefore we use the vocabulary schema.org
4
, which
was created to define and control general concepts by
important search engines (Ronallo, 2012) and con-
tains current concepts and categories commonly used
on the Internet. This vocabulary derived from the
Resource Description Framework (RDF) schema has
different hierarchical types, containing subclasses and
properties. This vocabulary is particularly useful for
our framework because it can represent publications’
metadata from different domains. Specifically we use
the properties ‘keywords’, ‘creator’ and ‘headline’ to
characterize a publication’s context, author and title
respectively. These properties are under the Schol-
arlyArticle type that belongs to the categories Thing,
CreativeWork, and Article.
3.2 Entity Recognition, Annotation, and
Disambiguation
We argue that to perform a comprehensive seman-
tic analysis of a document, it is important to take
advantage of the rich content and relationships ex-
pressed in tables and not only in text. Tables con-
tain structured cells organized in columns and rows.
Tables are useful to display summarized information
and are favored by scientists and researchers across
disciplines to present key results in publications (Kim
et al., 2012). Tables can be abstracted as an explicit
structural organization between their cells’ content by
column and row (see Table 1). Generally, header cells
define concepts or entities that describe the type of
information in the body of the table, (e.g., Country,
2
https://www.wikipedia.org/
3
http://www.bing.com/toolbox/bingsearchapi
4
https://schema.org
GDP, Population). Cells in the body of the table store
values representing a particular instance of their asso-
ciated entity (e.g. Canada). To take advantage of a ta-
ble’s structure and content, it is necessary to perform
table interpretation. Quercini and Reynaud define ta-
ble interpretation as 1) classifying the column meta-
data, that is, identifying the data category in a par-
ticular column (e.g., Country); 2) detecting concep-
tual entities within the table’s cells (e.g., Argentina,
Canada, Italy); and 3) finding structural relationships
between columns of the table (e.g., Country has at-
tribute GDP) (Quercini and Reynaud, 2013). How-
ever, depending on the format of the scientific pub-
lications, identifying and extracting this information
might present challenges. For scientific documents in
PDF, this interpretation is difficult due to the lack of
tags indicating even the existence of a table.
To identify and extract tables from PDF docu-
ments, we use TAO (Perez-Arriaga et al., 2016),
which identifies tables embedded in documents with
different layouts, and extracts and organizes their con-
tent by row. TAO includes a page number where the
table was found, a table number and metadata for each
cell (i.e., content, column number, coordinates, font,
size, data type, header or data label). TAO yields an
annotated document in JSON format.
To identify and extract table content from
well-formed XML documents, we use the tags
<table-wrap> and <table> indicating the presence
of tables. These tags are useful, but we cannot access
them directly. Therefore, we use Xpath (Berglund
et al., 2003) to detect the path of tags and locate a
table within a section of a document. Once we find
the tags, we organize its content by rows in JSON. To
determine if a cell is a header or data, we use the tags
<thead> and <tbody>. A regular expression detects
the datatype of a cell text. For simplicity, we detect
string and numeric data types. The table is enumer-
ated for organization purposes.
Entity Recognition. The organization of tables’ con-
tent by row supports our entity recognition process.
A header cell indicates that it groups other data cells.
Thus, it is more likely that it contains a concept. We
also focus our attention on data cells with type string
to perform entity recognition. Specifically, we use
Textblob (Loria, 2014), a semantic tool for Natural
Language Processing (NLP). Textblob receives text,
performs noun phrase analysis and returns the entity
or entities found. For example, for the text “Fig-
ure 2 shows the median curves for body mass in-
dex.”, Textblob recognizes the list of entities figure,
median curves, and body mass index.
Entity Annotation. After recognizing an entity,
we search the Web for a description to annotate
DATA 2017 - 6th International Conference on Data Science, Technology and Applications
226

it. For this step, we use DBpedia (Bizer et al.,
2009). DBpedia is a knowledge source that con-
tains billions of structured relationships as triples,
and is available in 125 languages. The English
version contains 4.22 million entities organized as
an ontology with properties (e.g., name: Diabetes,
type:disease). DBpedia’s naming convention uses a
capital letter for the first word, and underscore for
spaces between words. For consistency, we convert
an entity into this name convention. For instance,
we convert the entity “Diabetes Management” into
Diabetes_management. If an entity is found in
DBpedia, we use the DBpedia’s Universal Resource
Identifier (URI) as the entity’s annotation and the
property abstract as the entity’s description. A URI
in DBpedia contains the description of an entity, a
classification of its type (e.g., thing, person, country).
In addition, DBpedia allows us to find synonyms.
For instance, if we search for Glycemic_control,
DBpedia returns the entity Diabetes_management.
Entity Disambiguation. If an entity contains in
its abstract the words may ∨ can ∧ (mean ∨ re f er ∨
stand), it indicates a need to disambiguate an en-
tity. When an entity needs disambiguation or when
it is not found in DBpedia, we use a variation of the
Latent Semantic Indexing (LSI) analysis (Deerwester
et al., 1990) to find a Universal Resource Locator
(URL) with the closest meaning to the unresolved en-
tity. Price indicates that LSI works even on noisy
data to categorize documents using several hundred
dimensions (Price, 2003). For our framework we use
LSI + context. To set up a corpus for LSI, our frame-
work performs a Web search for documents related to
the unresolved entity and the context of its publication
(i.e., the publication’s keywords and entity to disam-
biguate). From this search, we: (1) select the top n
Web pages, with n = 100, and create a contextual-
ized document collection; (2) using the documents in
this collection, we build a matrix of term-document
frequencies v, where element v
i, j
represents the fre-
quency of the jth term in the ith document (or Web
page); (3) we eliminate stop words and normalize ∀v
vectors. We use this curated corpus along with LSI to
perform a search with respect to a vector q containing
sentences from the publication surrounding the unre-
solved entity to disambiguate. The URL whose corre-
sponding vector v
i
has the highest similarity to q is se-
lected as the entity annotation. The entity recognition
process enables us to identify concepts in headers,
entity instances, and structural relationships between
columns and rows from tables. As we explain in the
following section, entities are the building blocks of
semantic relationships and the key for their extraction.
3.3 Discovery of Relationships
Dahchour et al. define “Generic relationships”
as high-level templates for relating real-world enti-
ties. (Dahchour et al., 2005). More specifically, a
generic semantic relationship is a unidirectional bi-
nary relationship that represents static constraints and
rules, (i.e., classification, generalization, grouping,
aggregation). A binary relationship (R) contains the
basic semantics between a generic relationship and
two arguments. The relationship (a,R,b) indicates that
the arguments a and b are related by a relation R.
The arguments (a,b) can be entities, properties, or
values. A semantic relationship can be domain in-
dependent and represent information from different
areas. Throughout this work we use binary relation-
ships to express entity associations in a document. To
search for relationships, we divide this process into
two parts:
• Relationships in tables: We extract structural re-
lationships from header cells within a table. Also,
we use the entity’s annotations to describe con-
cepts in cells, as explained in Section 3.2.
• Relationships in tables and text: To find these re-
lationships, we use the entities found in a table
and relate them to its publication’s text.
The process for relationship identification is as
follows: First, we use the open information extrac-
tion tool Reverb (Fader et al., 2011), an unsuper-
vised method to extract relationships with a confi-
dence measure (see Section 2). We process a publi-
cation’s text with Reverb and select the relationships
with high confidence (≥ 0.70). Reverb is efficient, but
a relationship might be incomplete. For instance, it
finds, with 0.80 confidence, the relationship (obesity
and weight gain, are associated, with). Therefore,
we only use its output as a preliminary guide in our
relationship extraction procedure. Second, the pub-
lication’s text undergoes segmentation, which is the
process to separate the different sentences within the
text. Third, we determine the relevance of high con-
fidence relationships from Reverb by matching them
with the set of entities we deemed important. To en-
sure that the relationships are complete, we perform
pattern matching on the complete sentences surround-
ing the relationship text. For our previous example,
we get the relationship (obesity and weight gain, are
associated, with an increased risk of diabetes).
Finally, we use the Semanticscience Integration
Ontology (SIO) to formally represent relationships
in our framework. SIO defines relationships in the
Bioinformatics field between entities, such as objects,
processes and attributes. However, several definitions
can represent relationships from any other area of
Table Interpretation and Extraction of Semantic Relationships to Synthesize Digital Documents
227

study. We select the definitions of the most general re-
lationships from SIO as the basis to find relationships
in tables within publications regardless of their area.
To represent a relationship, we use pattern matching
to find the definition of a relationship. If one is found,
we store the identifier of the relationship’s definition
from SIO and the arguments composing a relation-
ship from a document. If a relationship is undefined
in SIO, our method generates its representation using
the verb found by Reverb. We store the ad-hoc rela-
tionships and use them for different publications.
3.4 Organization of Metadata, Entities
and Relationships
As the final phase, our approach organizes the meta-
data, entities, annotations and relationships into a
JSON file that synthesizes each publication in a stan-
dard and interoperable way.
The JSON - Linked Data (JSON-LD) format is
a representation of information with resources and
linked elements, and has been used to communicate
network messages (Lanthaler and G
¨
utl, 2012). This
format was created to facilitate the use of linked
data. A document in JSON-LD can define a type
of information, a set of relationships, not limited to
triples, and the location of other documents. An-
other advantage of this format is that it includes a
context of a document. We should not confuse a
JSON-LD document’s context with the context of a
publication (i.e., keywords). A JSON-LD context
refers to the location of a resource. For instance, for
a document that represents relationships from SIO,
the context is http://semanticscience.org/resource/.
This context indicates the location of the re-
lationships’ formal definitions. If a relation-
ship has a definition with identifier SIO 000001.
Then, the union of the context and the identi-
fier http://semanticscience.org/resource/SIO 000001
gives access to a specific resource (e.g., a definition
of a relationship).
We use JSON-LD to generate multiple records
per document. The records include the publication’s
metadata, entities with annotations, and the extracted
semantic relationships. These records in turn contain
other links, such as the ones used to annotate enti-
ties and to define relationships. By using JSON-LD
records, it is easy to either store them in files or as
records in NoSQL databases for querying.
4 WORKING EXAMPLE
To better describe our approach, we focus on a typical
example of a multi-column publication containing ta-
bles (See Figure 2). We use The continuing epidemics
of obesity and diabetes in the United States (Mokdad
et al., 2001).
Figure 2: The continuing epidemics of obesity and diabetes
in the United States - Publication used as example.
Context and Metadata Detection. To represent the
metadata of a publication, we search for the proper-
ties: creator, headline, and keywords. Once we extract
this information, we store it in our synthesis using the
schema.org representation as shown in Figure 3.
Figure 3: Representation of metadata for a publication.
The annotation "schema" indicates that the
concepts are defined from the schema.org vocabulary.
The property "@type" defines the category (i.e.,
Scholarly article); "creator" specifies the authors
of a publication; "headline" specifies the title; and
"keywords" defines its context. By using schema.org
we are able to build an explicit organization into
our data representation. This organization is easily
exploited for querying purposes.
Entity Recognition, Annotation, and Disambigua-
tion. The following example demonstrates the pro-
cess from table extraction to entity recognition and
annotation. We use partial data (see Table 1) from Ta-
ble 1: “Obesity and Diabetes Prevalence Among US
Adults, by Selected Characteristics, Behavioral Risk
Factor Surveillance System, 2000” in (Mokdad et al.,
2001).
The first row contains concepts in headers (e.g.,
DATA 2017 - 6th International Conference on Data Science, Technology and Applications
228

Table 1: Excerpt from “The continuing epidemics of obesity
and diabetes in the United States” (Mokdad et al., 2001).
Race Obesity, % (SE) Diabetes, % (SE)
Black 29.3(0.59) 11.1(0.39)
Hispanic 23.4(0.77) 8.9(0.59)
White 18.5(0.17) 6.6(0.11)
Other 12.0(0.68) 6.7(0.65)
Race, Obesity, Diabetes), and the other cells contain
particular values, which can also be an entity instance
or attribute. As mentioned earlier in Section 3.2, the
PDF publication undergoes a process for table recog-
nition using TAO. This process generates a JSON file
with the information of the table grouped by row. The
output for each cell includes column number, content,
coordinates of cell on a page, font, size, data type,
header or data labels. For didactic purposes we show
a fragment of TAO’s output in Figure 4.
Figure 4: TAO’s output for the first row in Table 1.
Using each cell’s content with data type string
(e.g., “Obesity % (SE)”), Textblob recognizes the ta-
ble’s entities as Race, Obesity and Diabetes. Then,
our method finds the entities’ annotations from DB-
pedia. For the entity Diabetes, the annotation is the
URI http://dbpedia.org/page/Diabetes mellitus and its
description follows:
“ Diabetes mellitus (DM), commonly referred
to as diabetes, is a group of metabolic diseases
in which there are high blood sugar levels over
a prolonged period...”
Discovery of Relationships. To describe the pro-
cess of discovering relationships, we use Table
1, which contains the entities Race, Obesity and
Diabetes. The header cell “Race” groups the data
cells “Black”, “Hispanic”, “White”, and “Other” on
the first column. From this column, we obtain four
instances for the entity race: Race ⇒ Black, Race
⇒ Hispanic, Race ⇒ White, Race ⇒ Other. These
four entity instances are used to find semantic rela-
tionships in the document’s text. One of such relation-
ships found in the text is (Obesity and weight gain,
are associated, with an increased risk of diabetes).
As explained before, we aim at using a standard rep-
resentation for all of our relationships. In this case the
relationship “are associated” is defined by SIO with
the label “is related to” and indexed by the SIO iden-
tifier SIO_000001. Additionally, this definition
5
con-
tains the description “A is related to B iff there is some
relationship between A and B”, and other properties.
To build the document’s synthesis, we save the rela-
tionship as a quintuple including identifier of the rela-
tionship, first argument, relationship label, second ar-
gument, and a relationship’s definition identifier. For
this example, the format is as follows:
id:1, argument A: “obesity and weight”, la-
bel: “is related to”, argument B: “with an in-
creased risk of diabetes”, sio id:SIO 000001.
Note that the identifier of a relationship within a
publication (e.g., id:1) is different from a relation-
ship’s definition identifier (e.g., sio id:SIO 000001).
Another example is the semantic relationship (the
prevalence of obesity and diabetes, has increased,
despite previous calls for action). In this case
SIO does not have a definition for the relationship
“has increased”. Still, we store the relationship for
further consultation and to enrich the entity ‘Obesity’.
Organization of Metadata, Entities and Relation-
ships. The first record in our synthesis contains con-
textual information, as the metadata shown in Fig-
ure 3. Additionally, it contains entities’ annotations,
descriptions, and identifiers. The record with infor-
mation of extracted relationships contains a unique
identifier and a context. Its context includes the loca-
tion of definitions of relationships from SIO and the
details of the extracted relationships from tables and
text (i.e., relationship identifier, arguments, relation-
ship definition’s identifier). For illustrative purposes,
we show a fragment of the records indicating annota-
tions for entities and relationships in Figure 5.
5 EVALUATION
We evaluated our approach quantitatively and quali-
tatively. To do so, we designed three sets of experi-
ments that evaluated our framework in terms of its (1)
accuracy to annotate and recognize entities, (2) abil-
ity to disambiguate entities, and (3) ability to identify
relevant semantic relationships between entities.
To assess our method, we use a dataset
that we call Pubmed. It comprises fifty
publications downloaded from the Web site
ftp://ftp.ncbi.nlm.nih.gov/pub/pmc/oa bulk/. This
collection contains 449 text pages with 133 tables.
The dataset includes various table formats and
document layouts (one and two-column).
5
http://semanticscience.org/resource/SIO 000001
Table Interpretation and Extraction of Semantic Relationships to Synthesize Digital Documents
229

Figure 5: Fragment of our table interpretation and publica-
tion’s synthesis.
5.1 Evaluation of Entity Extraction and
Annotation
Our first experiment evaluates our method’s ability to
recognize and annotate entities. First, we manually
extracted all the entities per table in our dataset as
the gold standard. The measures recall and precision
are used to obtain the average F1-measure for entity
recognition. Recall is the ratio between the correct
number of entities detected and the total number of
entities in a table. Precision is the ratio between the
correct number of entities detected and the total num-
ber of entities detected. We use the same measure-
ments for entities annotated and disambiguated.
To measure entity recognition, our gold standard
contains 2, 314 entities, which 1, 834 of them were
recognized. We obtained a recall of 79.2% and a pre-
cision of 94.3%, yielding a F1-measure of 86.1% for
recognition (see Table 2).
From the entities annotated, our method found
1, 262 (72.5%) of entities in DBpedia. From those,
785 (45.1%) obtained a direct annotation using DB-
pedia, and the rest (27.4%) needed disambiguation.
Then, our method used the LSI process described in
Section 3.2 to annotate a total of 955 entities. That is
54.9% of entities correctly recognized. From the total
1, 834 entities recognized, 1, 740 were correctly an-
Table 2: Experiment 1 to recognize and annotate entities.
Entity Recognition
Entities Recall Precision F1 measure
Recognized 0.79 0.94 0.86
Annotated 0.95 0.97 0.96
notated, yielding a recall of 94.8%, precision of 97%,
and F1-measure of 95.9% (see Table 2).
5.2 Evaluation of Entity
Disambiguation
For the second experiment, we evaluated the entity
disambiguation methods. In particular, we quantify
the effect of including information regarding the con-
text of the publication in the disambiguation process.
From the 1, 740 annotated entities, 955 needed disam-
biguation. From these, 838 correctly disambiguated
entities were discovered without context. The preci-
sion was 89% and recall 87%, yielding an F1-measure
of 88%. For the entities disambiguated using as con-
text the three more relevant keywords, there were 900.
The precision was 95% and the recall 94%, yielding
an F1-measure of 94.5%. Table 3 presents the results
of comparing disambiguation without and with con-
text.
Table 3: Experiment 2 to disambiguate entities.
Entity Disambiguation
Method Recall Prec. F1 NR urls
No context 0.87 0.89 0.88 12.3%
Context 0.94 0.95 0.94 5.8%
In addition, URLs were manually verified to de-
termine whether they were reliable or non-reliable
(See Table 3 column NR urls). The reliable URLs
include known organizations and domains. Non-
reliable URLs required further investigation. Al-
though this URL review does not ensure an exact de-
scription of an entity, it does ensure that a site or doc-
ument found is related to the entity, and consequently
to the publication. The non-reliable URLs found us-
ing no context were 117, that is 12.3% of the total dis-
ambiguated entities, while the number of non-reliable
URLs when including context were 55, that is 5.8%.
Therefore, the context reduced more than half of non-
reliable links.
Although the results with context are only
slightly better than non-context, the quality of the
URLs increased considerably using the context. For
example, for the entity Gain found in the work-
ing example from Section 4, the method with no
context found the URL http://gainworldwide.org/,
which is a site for global aid network. Us-
ing a context, our method found the URL
https://www.sciencedaily.com/releases/2010/02/100
222182137.htm, which is a site about weight gain
during pregnancy and increasing risk of gestational
diabetes. Although the first link belongs to an
organization, it does not relate to the entity Gain
DATA 2017 - 6th International Conference on Data Science, Technology and Applications
230

for this publication, while the second URL, explains
the risks of weight gain during pregnancy, which is
related to this specific publication.
Using context, we consistently found more reli-
able links, belonging to organizations, schools, gov-
ernment, clinics, dictionaries, and scientific and dig-
ital libraries, among others. In contrast, when con-
text was not included, we found commercial URLs
promoting services or products, unavailable sites, and
even some ill-intentioned links. To ensure reliability,
we could keep a second URL annotation for unavail-
able URLs.
5.3 Evaluation of Semantic
Relationships
The third experiment evaluated both quantitatively
and qualitatively the semantic relationships found by
our method. We report the total number of high rank-
ing (≥ 0.70) relationships derived either from tables
or text, and the average number of relationships per
article. In addition, a human judge evaluated qualita-
tively the relationships extracted from text and tables.
The judge detected complete versus incomplete rela-
tionships. A complete relationship indicates that the
components of a relationship show coherence, regard-
less of their accuracy. See results in Table 4.
Table 4: Experiment 3: finding semantic relationships.
Semantic Relationships
Method Rel. found Rel. complete
Text only 865 703
Tables 11,268 10,102
For this experiment, we found 11, 268 relationships
from tables and 865 from text. The average number
of relationships extracted per publication when using
table information is 225, while the average number of
relationships extracted using only text is 17 per pub-
lication.
A human judge analyzed the quality and com-
pleteness of relationships manually. From the total
of relationships extracted from text, 703 (81%) rela-
tionships were complete and the rest was labeled in-
complete. From the set of relationships derived from
tables, 10, 102 (89%) relationships were complete. To
further increase the number of relationships that we
can extract from a given article we could increase the
confidence threshold. However, there is the risk of
extracting common or irrelevant relationships.
Analyzing the results qualitatively, relationships
extracted from tables produced more complete infor-
mation. This confirms our initial intuition regarding
the relevance of structural information embedded in
tables. Even though our framework uses mostly con-
cepts from tables to extract relationships and generate
a synthesis, it can still be useful when a publication
lacks tables because it finds metadata and semantic
relationships from text. These relationships can be
found using a publication’s keywords. Regarding re-
lationships extracted from text only, our method im-
proved the completeness of relationships extracted by
Reverb.
6 CONCLUSION AND FUTURE
WORK
Because of their wide use and structured organiza-
tion, we propose using tables in digital publications
to recover summarized information and structural re-
lationships among conceptual entities in these docu-
ments. Additionally, we use text in publications to
find valuable information, such as context and de-
scription of concepts as semantic relationships.
We developed an integrated framework to extract
semantic relationships from electronic documents.
Our method takes advantage primarily of informa-
tion in tables, such as their content and structure, to
find relevant entities and relationships. Our frame-
work can seamlessly interpret documents in PDF and
XML format, which leverages multiple standards,
tools, Natural Language Processing, and unsuper-
vised learning to generate an end-to-end synthesized
analysis of a document. This synthesis contains rich
information organized in a standard, searchable, and
interoperable format that facilitates batch mining of
large document collections. In addition to interop-
erability and easiness of use, our output emphasizes
provenance, and it ensures that the user will be able
to trace back exactly the source of every extracted re-
lationship.
Our results demonstrate the importance of includ-
ing context to annotate and to identify semantic rela-
tionships of high quality. The results also support our
intuition regarding the usefulness of tables and show
that by using tables, it is possible to increase the num-
ber of highly ranked semantic relationships by one or-
der of magnitude.
In our future work, we plan to create a collection
of syntheses allowing users to consult them individu-
ally and globally; and to further evaluate data integra-
tion and interoperability. Our framework shall gener-
ate a network of the semantic relationships containing
context or entities of interest. Hence, it can extend ad-
ditional support to find important relationships among
documents from any domain.
Table Interpretation and Extraction of Semantic Relationships to Synthesize Digital Documents
231

REFERENCES
Abdalgader, K. and Skabar, A. (2010). Short-text similarity
measurement using word sense disambiguation and
synonym expansion. In Australasian Joint Conference
on Artificial Intelligence, pages 435–444. Springer.
Baralis, E., Cagliero, L., Jabeen, S., and Fiori, A. (2012).
Multi-document summarization exploiting frequent
itemsets. In Proc. of the 27th Annual ACM Sympo-
sium on Applied Computing, pages 782–786. ACM.
Berglund, A., Boag, S., Chamberlin, D., Fernandez, M. F.,
Kay, M., Robie, J., and Sim
´
eon, J. (2003). Xml
path language (xpath). World Wide Web Consortium
(W3C).
Berners-Lee, T., Hendler, J., Lassila, O., et al. (2001). The
semantic web. Scientific american, 284(5):28–37.
Bizer, C., Lehmann, J., Kobilarov, G., Auer, S., Becker, C.,
Cyganiak, R., and Hellmann, S. (2009). Dbpedia-a
crystallization point for the web of data. Web Seman-
tics: science, services and agents on the world wide
web, 7(3):154–165.
Bornmann, L. and Mutz, R. (2015). Growth rates of modern
science: A bibliometric analysis based on the number
of publications and cited references. Journal of the
Association for Information Science and Technology,
66(11):2215–2222.
Cafarella, M. J., Halevy, A. Y., Zhang, Y., Wang, D. Z.,
and Wu, E. (2008). Uncovering the relational web. In
WebDB. Citeseer.
Cohen, A. M. and Hersh, W. R. (2005). A survey of current
work in biomedical text mining. Briefings in bioinfor-
matics, 6(1):57–71.
Dahchour, M., Pirotte, A., and Zim
´
anyi, E. (2005). Generic
relationships in information modeling. In Journal on
Data Semantics IV, pages 1–34. Springer.
Deerwester, S., Dumais, S. T., Furnas, G. W., Landauer,
T. K., and Harshman, R. (1990). Indexing by latent
semantic analysis. Journal of the American society
for information science, 41(6):391.
Di Sciascio, C., Mayr, L., and Veas, E. (2017). Exploring
and summarizing document collections with multiple
coordinated views. In Proc. of the 2017 ACM Work-
shop on Exploratory Search and Interactive Data An-
alytics, pages 41–48. ACM.
Dumontier, M., Baker, C. J., Baran, J., Callahan, A., Che-
pelev, L. L., Cruz-Toledo, J., Nicholas, R., Rio, D.,
Duck, G., Furlong, L. I., et al. (2014). The seman-
ticscience integrated ontology (sio) for biomedical re-
search and knowledge discovery. J. Biomedical Se-
mantics, 5:14.
Fader, A., Soderland, S., and Etzioni, O. (2011). Identifying
relations for open information extraction. In Proc. of
the Conference on Empirical Methods in Natural Lan-
guage Processing, pages 1535–1545. Association for
Computational Linguistics.
Ferragina, P. and Scaiella, U. (2012). Fast and accurate an-
notation of short texts with wikipedia pages. IEEE
software, 29(1):70–75.
Hristovski, D., Friedman, C., Rindflesch, T. C., and Pe-
terlin, B. (2006). Exploiting semantic relations for
literature-based discovery. In AMIA.
Kim, S., Han, K., Kim, S. Y., and Liu, Y. (2012). Scientific
table type classification in digital library. In Proc. of
the 2012 ACM symposium on Document engineering,
pages 133–136. ACM.
Lanthaler, M. and G
¨
utl, C. (2012). On using json-ld to cre-
ate evolvable restful services. In Proc. of the Third In-
ternational Workshop on RESTful Design, pages 25–
32. ACM.
Loria, S. (2014). Textblob: simplified text processing. Sec-
ondary TextBlob: Simplified Text Processing.
Mokdad, A. H., Bowman, B. A., Ford, E. S., Vinicor, F.,
Marks, J. S., and Koplan, J. P. (2001). The continuing
epidemics of obesity and diabetes in the united states.
Jama, 286(10):1195–1200.
Mulwad, V., Finin, T., Syed, Z., and Joshi, A. (2010). Using
linked data to interpret tables. COLD, 665.
Oro, E. and Ruffolo, M. (2008). Xonto: An ontology-based
system for semantic information extraction from pdf
documents. In Tools with Artificial Intelligence, 2008.
ICTAI’08. 20th IEEE International Conference on,
volume 1, pages 118–125. IEEE.
Perez-Arriaga, M., Estrada, T., and Abad-Mota, S. (2016).
Tao: System for table detection and extraction form
pdf documents. In The 29th Florida Artificial Intel-
ligence Research Society Conference, pages 591–596.
AAAI.
Price, A. Z. R. J. (2003). Document categorization using
latent semantic indexing. In Proc. 2003 Symposium on
Document Image Understanding Technology, page 87.
UMD.
Quercini, G. and Reynaud, C. (2013). Entity discovery
and annotation in tables. In Proc. of the 16th Inter-
national Conference on Extending Database Technol-
ogy, pages 693–704. ACM.
Ramos, J. (2003). Using tf-idf to determine word relevance
in document queries. In Proc. of the First Instructional
Conference on Machine Learning.
Rastan, R., Paik, H.-Y., and Shepherd, J. (2015). Texus:
A task-based approach for table extraction and under-
standing. In Proc. of the 2015 ACM Symposium on
Document Engineering, pages 25–34. ACM.
Ronallo, J. (2012). Html5 microdata and schema. org.
Code4Lib Journal, 16.
Sekine, S. (2008). A linguistic knowledge discovery tool:
Very large ngram database search with arbitrary wild-
cards. In 22nd International Conference on on Com-
putational Linguistics: Demonstration Papers, pages
181–184. Association for Computational Linguistics.
Shinyama, Y. (2010). Pdfminer: Python pdf parser and an-
alyzer.
Srinivasan, P. (2004). Text mining: generating hypotheses
from medline. Journal of the American Society for
Information Science and Technology, 55(5):396–413.
Swanson, D. R. (1988). Migraine and magnesium: eleven
neglected connections. Perspectives in biology and
medicine, 31(4):526–557.
Yates, A., Cafarella, M., Banko, M., Etzioni, O., Broad-
head, M., and Soderland, S. (2007). Textrunner: open
information extraction on the web. In Proc. of The
Annual Conference of the North American Chapter of
the Association for Computational Linguistics, pages
25–26. Association for Computational Linguistics.
DATA 2017 - 6th International Conference on Data Science, Technology and Applications
232
